CircZBTB46 Protects Acute Myeloid Leukemia Cells from Ferroptotic Cell Death by Upregulating SCD
Abstract
Simple Summary
Abstract
1. Background
2. Materials and Methods
2.1. CircRNA Microarray Analysis
2.2. Human Samples
2.3. Cell Lines and Cell Culture
2.4. RNA Extraction, and Quantitative Reverse Transcription–PCR (qRT–PCR)
2.5. RNase R and Actinomycin D Treatment Assays
2.6. Small Interfering RNAs (siRNAs), Short Hairpin RNAs (shRNAs), Lentiviral Vector Construction, and Cell Transfection
2.7. Cell Counting Kit-8 (CCK-8) Assay
2.8. Cell Cycle Analysis
2.9. Cell Death Assay
2.10. Lipid Reactive Oxygen Species (ROS) and Malondialdehyde (MDA) Detection
2.11. RNA-Binding Protein Immunoprecipitation (RIP)
2.12. RNA Pulldown Assay
2.13. Western Blot (WB) Analysis and Antibodies
2.14. Animal Experiments
2.15. Bioinformatics Analysis
2.16. Statistical Analysis
3. Results
3.1. CircZBTB46 Is Upregulated in AML and Associated with the Stages of AML
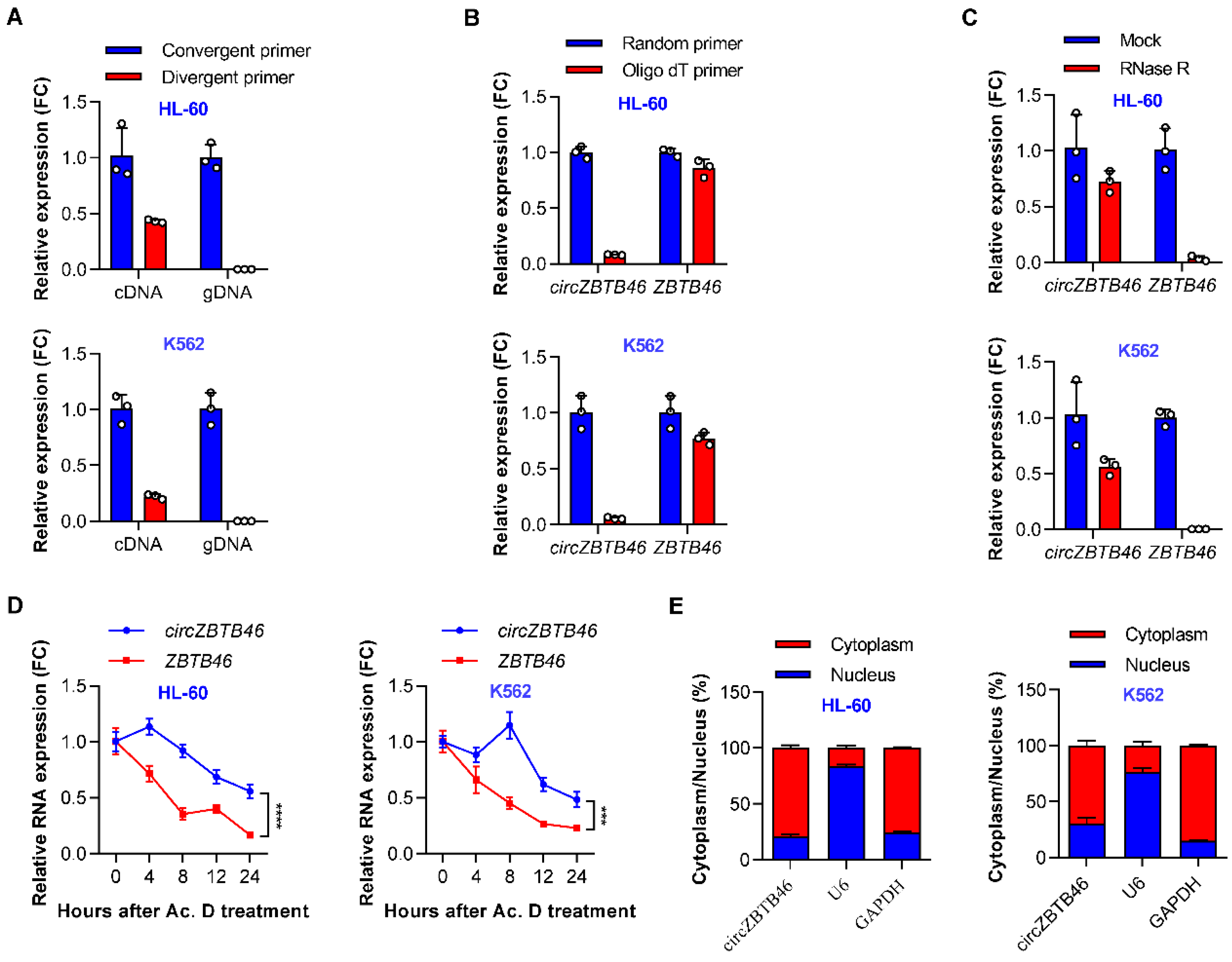
3.2. Identification and Validation of circZBTB46 in AML
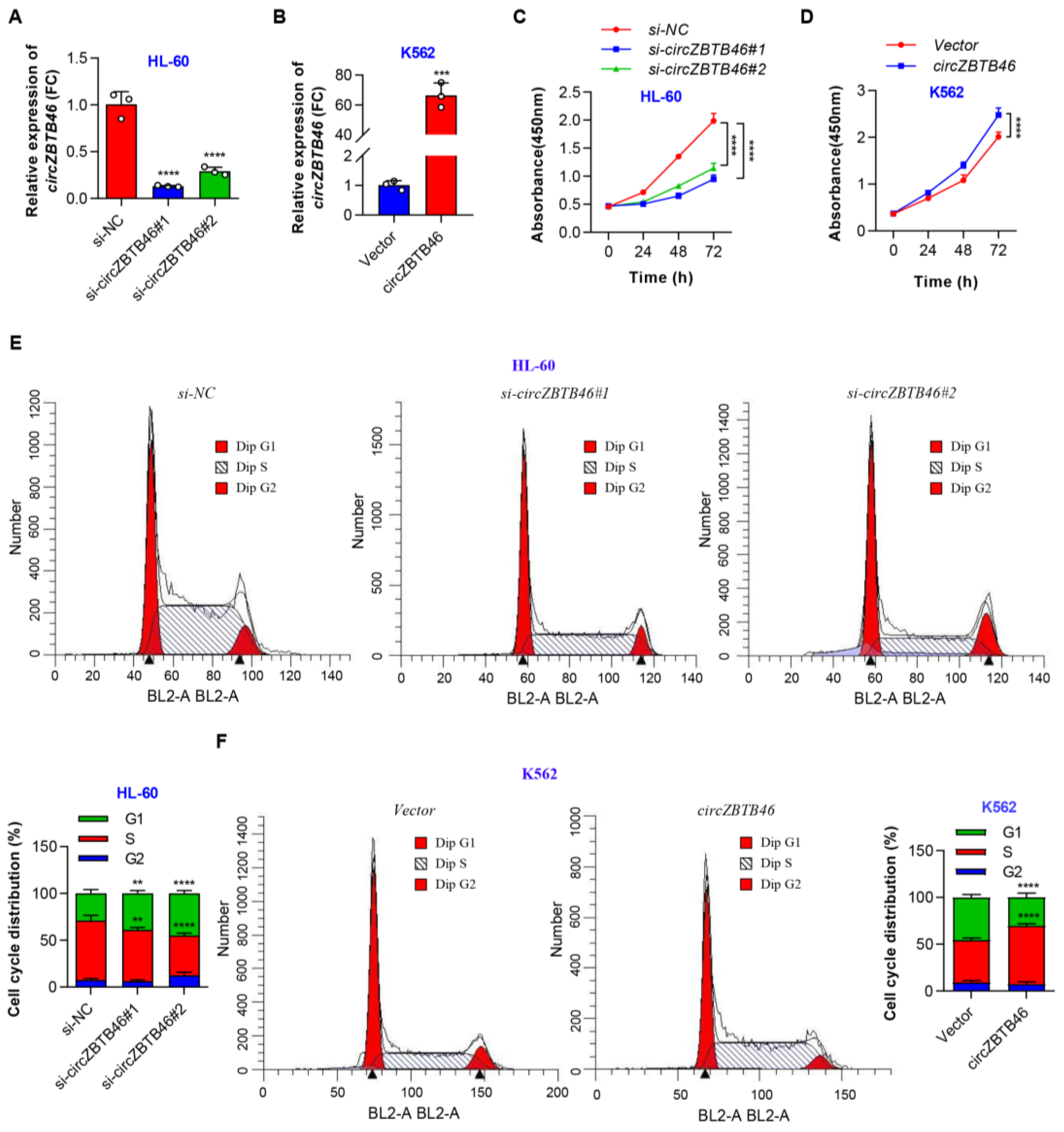
3.3. CircZBTB46 Promotes Cell Proliferation and Cell Cycle Progression of AML
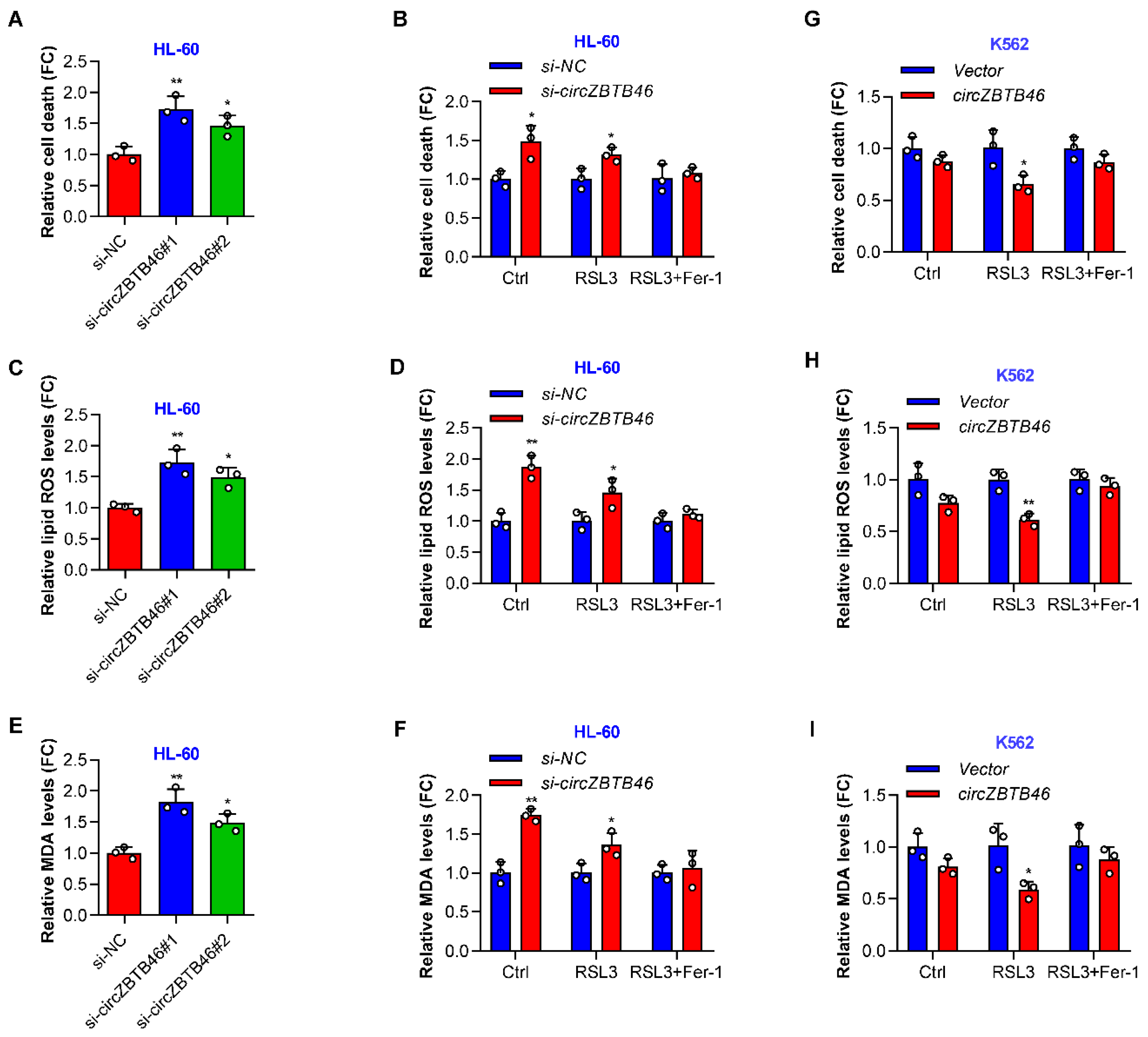
3.4. CircZBTB46 Protects AML Cells against Ferroptosis
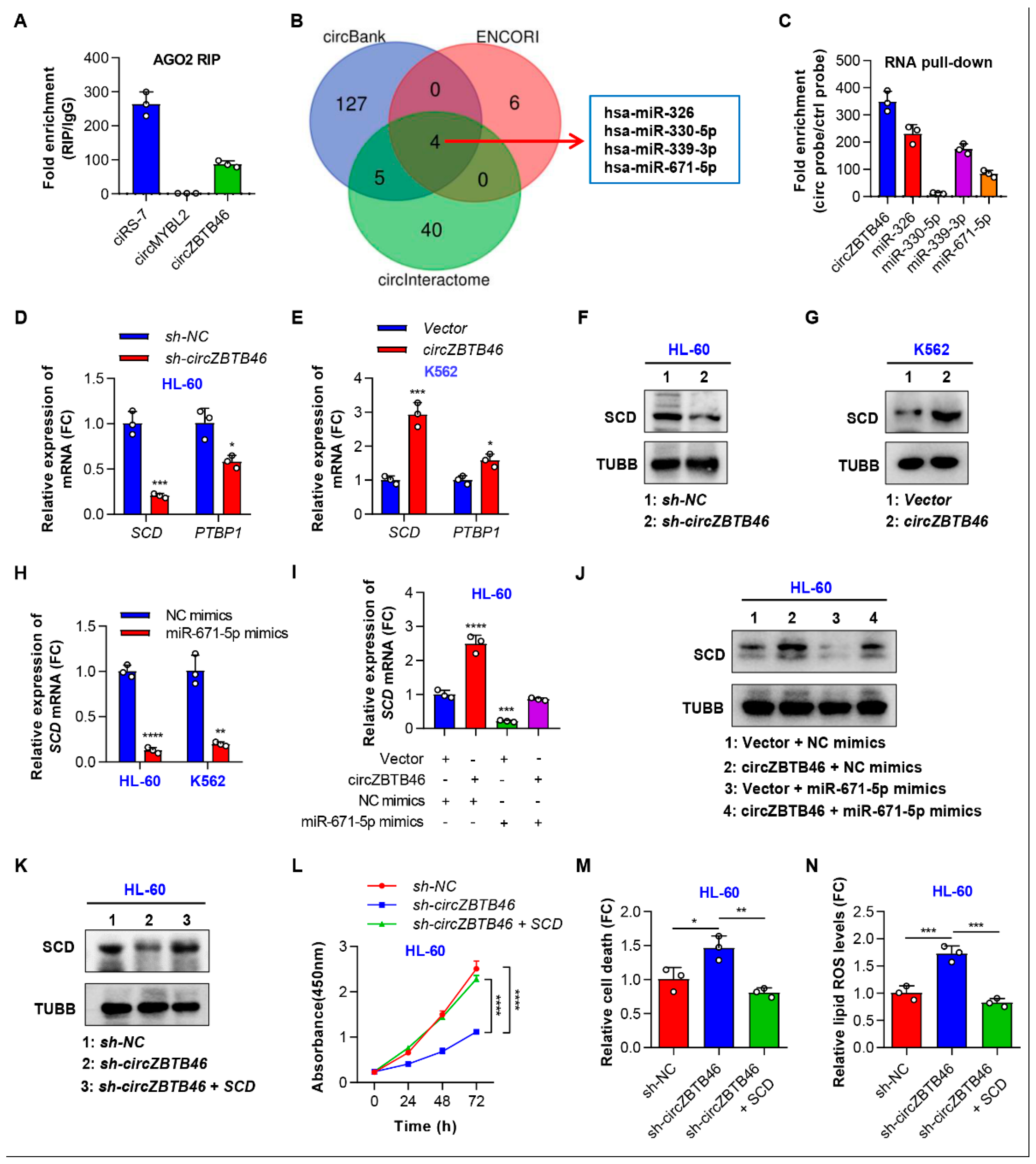
3.5. CircZBTB46 Exerts Its Oncogenic Effect by Upregulating SCD
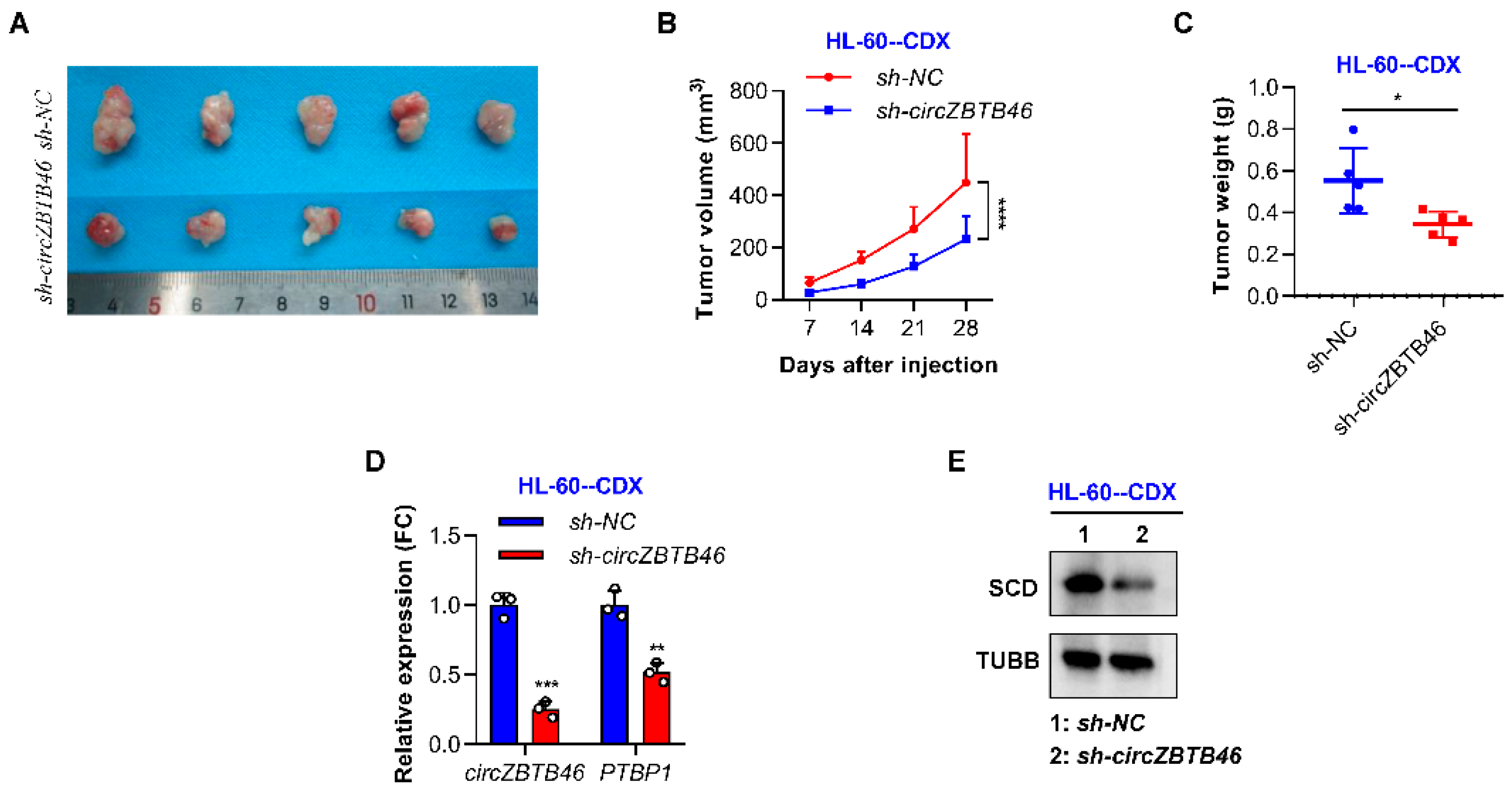
3.6. CircZBTB46 Knockdown Impairs the Tumorigenesis of AML Cells In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Löwenberg, B. Towards precision medicine for AML. Nat. Rev. Clin. Oncol. 2021, 18, 577–590. [Google Scholar] [CrossRef]
- Newell, L.F.; Cook, R.J. Advances in acute myeloid leukemia. BMJ (Clin. Res. Ed.) 2021, 375, n2026. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Estey, E.H. Acute myeloid leukemia: 2019 update on risk-stratification and management. Am. J. Hematol. 2018, 93, 1267–1291. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef]
- Zhou, X.; Zhan, L.; Huang, K.; Wang, X. The functions and clinical significance of circRNAs in hematological malignancies. J. Hematol. Oncol. 2020, 13, 138. [Google Scholar] [CrossRef]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Singh, V.; Uddin, M.H.; Zonder, J.A.; Azmi, A.S.; Balasubramanian, S.K. Circular RNAs in acute myeloid leukemia. Mol. Cancer 2021, 20, 149. [Google Scholar] [CrossRef] [PubMed]
- Shang, Z.; Ming, X.; Wu, J.; Xiao, Y. Downregulation of circ_0012152 inhibits proliferation and induces apoptosis in acute myeloid leukemia cells through the miR-625-5p/SOX12 axis. Hematol. Oncol. 2021, 39, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, X.; Cai, M.; Luo, A.; He, Y.; Liu, S.; Zhang, X.; Yang, X.; Xu, L.; Jiang, H. CircRNF220, not its linear cognate gene RNF220, regulates cell growth and is associated with relapse in pediatric acute myeloid leukemia. Mol. Cancer 2021, 20, 139. [Google Scholar] [CrossRef]
- Doll, S.; Proneth, B.; Tyurina, Y.Y.; Panzilius, E.; Kobayashi, S.; Ingold, I.; Irmler, M.; Beckers, J.; Aichler, M.; Walch, A.; et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017, 13, 91–98. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, R.Z.; Saez, B.; Sharda, A.; van Gastel, N.; Yu, V.W.C.; Baryawno, N.; Scadden, E.W.; Acharya, S.; Chattophadhyay, S.; Huang, C.; et al. Aldehyde dehydrogenase 3a2 protects AML cells from oxidative death and the synthetic lethality of ferroptosis inducers. Blood 2020, 136, 1303–1316. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Chai, W.; Xie, M.; Yang, M.; Yu, Y.; Cao, L.; Yang, L. HMGB1 regulates erastin-induced ferroptosis via RAS-JNK/p38 signaling in HL-60/NRAS(Q61L) cells. Am. J. Cancer Res. 2019, 9, 730–739. [Google Scholar]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Sun, Y.M.; Wang, W.T.; Zeng, Z.C.; Chen, T.Q.; Han, C.; Pan, Q.; Huang, W.; Fang, K.; Sun, L.Y.; Zhou, Y.F.; et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 2019, 134, 1533–1546. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R. Ferroptosis turns 10: Emerging mechanisms, physiological functions, and therapeutic applications. Cell 2022, 185, 2401–2421. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Kang, R.; Klionsky, D.J.; Tang, D. Ferroptosis: Machinery and regulation. Autophagy 2021, 17, 2054–2081. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296. [Google Scholar] [CrossRef]
- Weber, S.; Parmon, A.; Kurrle, N.; Schnütgen, F.; Serve, H. The Clinical Significance of Iron Overload and Iron Metabolism in Myelodysplastic Syndrome and Acute Myeloid Leukemia. Front. Immunol. 2020, 11, 627662. [Google Scholar] [CrossRef]
- Birsen, R.; Larrue, C.; Decroocq, J.; Johnson, N.; Guiraud, N.; Gotanegre, M.; Cantero-Aguilar, L.; Grignano, E.; Huynh, T.; Fontenay, M.; et al. APR-246 induces early cell death by ferroptosis in acute myeloid leukemia. Haematologica 2022, 107, 403–416. [Google Scholar] [CrossRef]
- Du, J.; Wang, T.; Li, Y.; Zhou, Y.; Wang, X.; Yu, X.; Ren, X.; An, Y.; Wu, Y.; Sun, W.; et al. DHA inhibits proliferation and induces ferroptosis of leukemia cells through autophagy dependent degradation of ferritin. Free Radic. Biol. Med. 2019, 131, 356–369. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liu, Y.; Miao, Z.; Cheng, S.; Zhu, Y.; Wu, Y.; Fan, X.; Yang, J.; Li, X.; Guo, L. Neratinib inhibits proliferation and promotes apoptosis of acute myeloid leukemia cells by activating autophagy-dependent ferroptosis. Drug Dev. Res. 2022, 83, 1641–1653. [Google Scholar] [CrossRef]
- Lai, X.; Sun, Y.; Zhang, X.; Wang, D.; Wang, J.; Wang, H.; Zhao, Y.; Liu, X.; Xu, X.; Song, H.; et al. Honokiol Induces Ferroptosis by Upregulating HMOX1 in Acute Myeloid Leukemia Cells. Front. Pharmacol. 2022, 13, 897791. [Google Scholar] [CrossRef]
- Dong, L.H.; Huang, J.J.; Zu, P.; Liu, J.; Gao, X.; Du, J.W.; Li, Y.F. CircKDM4C upregulates P53 by sponging hsa-let-7b-5p to induce ferroptosis in acute myeloid leukemia. Environ. Toxicol. 2021, 36, 1288–1302. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Long, F.; Lin, Z.; Li, L.; Ma, M.; Lu, Z.; Jing, L.; Li, X.; Lin, C. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol. Cancer 2021, 20, 26. [Google Scholar] [CrossRef]
- Lin, Z.; Long, F.; Zhao, M.; Zhang, X.; Yang, M. The role of circular RNAs in hematological malignancies. Genomics 2020, 112, 4000–4008. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jin, P.; Zhang, Y.; Wang, K. CircSPI1 acts as an oncogene in acute myeloid leukemia through antagonizing SPI1 and interacting with microRNAs. Cell Death Dis. 2021, 12, 297. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, J.; Huang, S.; Li, F.; Huang, J.; Li, X.; Ling, Q.; Ye, W.; Wang, Y.; Yu, W.; et al. Development and validation of a novel circular RNA as an independent prognostic factor in acute myeloid leukemia. BMC Med. 2021, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Cheng, F. Circular RNA circCRKL inhibits the proliferation of acute myeloid leukemia cells via the miR-196a-5p/miR-196b-5p/p27 axis. Bioengineered 2021, 12, 7704–7713. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Liu, J.; Jin, Y.; Wang, W. Circular RNA circ_0004277 Inhibits Acute Myeloid Leukemia Progression Through MicroRNA-134-5p / Single stranded DNA binding protein 2. Bioengineered 2022, 13, 9662–9673. [Google Scholar] [CrossRef]
- Barrera-Ramirez, J.; Lavoie, J.R.; Maganti, H.B.; Stanford, W.L.; Ito, C.; Sabloff, M.; Brand, M.; Rosu-Myles, M.; Le, Y.; Allan, D.S. Micro-RNA Profiling of Exosomes from Marrow-Derived Mesenchymal Stromal Cells in Patients with Acute Myeloid Leukemia: Implications in Leukemogenesis. Stem Cell Rev. Rep. 2017, 13, 817–825. [Google Scholar] [CrossRef]
- Cheng, P.; Lu, P.; Guan, J.; Zhou, Y.; Zou, L.; Yi, X.; Cheng, H. LncRNA KCNQ1OT1 controls cell proliferation, differentiation and apoptosis by sponging miR-326 to regulate c-Myc expression in acute myeloid leukemia. Neoplasma 2020, 67, 238–248. [Google Scholar] [CrossRef]
- Gui, C.P.; Li, J.Y.; Fu, L.M.; Luo, C.G.; Zhang, C.; Tang, Y.M.; Zhang, L.Z.; Shu, G.N.; Wu, R.P.; Luo, J.H. Identification of mRNA vaccines and conserved ferroptosis related immune landscape for individual precision treatment in bladder cancer. J. Big Data 2022, 9, 88. [Google Scholar] [CrossRef]
- Yi, K.; Liu, J.; Rong, Y.; Wang, C.; Tang, X.; Zhang, X.; Xiong, Y.; Wang, F. Biological Functions and Prognostic Value of Ferroptosis-Related Genes in Bladder Cancer. Front. Mol. Biosci. 2021, 8, 631152. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.; Yin, H.; Yao, Z.Y.; Xing, X.L. Prognostic Signatures Based on Ferroptosis- and Immune-Related Genes for Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma. Front. Oncol. 2021, 11, 774558. [Google Scholar] [CrossRef] [PubMed]
- Tesfay, L.; Paul, B.T.; Konstorum, A.; Deng, Z.; Cox, A.O.; Lee, J.; Furdui, C.M.; Hegde, P.; Torti, F.M.; Torti, S.V. Stearoyl-CoA Desaturase 1 Protects Ovarian Cancer Cells from Ferroptotic Cell Death. Cancer Res. 2019, 79, 5355–5366. [Google Scholar] [CrossRef]
- Wohlhieter, C.A.; Richards, A.L.; Uddin, F.; Hulton, C.H.; Quintanal-Villalonga, À.; Martin, A.; de Stanchina, E.; Bhanot, U.; Asher, M.; Shah, N.S.; et al. Concurrent Mutations in STK11 and KEAP1 Promote Ferroptosis Protection and SCD1 Dependence in Lung Cancer. Cell Rep. 2020, 33, 108444. [Google Scholar] [CrossRef] [PubMed]
- Subedi, A.; Liu, Q.; Ayyathan, D.M.; Sharon, D.; Cathelin, S.; Hosseini, M.; Xu, C.; Voisin, V.; Bader, G.D.; D’Alessandro, A.; et al. Nicotinamide phosphoribosyltransferase inhibitors selectively induce apoptosis of AML stem cells by disrupting lipid homeostasis. Cell Stem Cell 2021, 28, 1851–1867.e1858. [Google Scholar] [CrossRef]

| Gene Names | Forward Primers (5′→3′) | Reverse Primers (5′→3′) |
|---|---|---|
| circZBTB46 divergent primer | GTTCGAGTACCTGCCCAGAG | GGTGTCGCCTCTTCTACAGA |
| circZBTB46 convergent primer | TTCAAGACGCTCTACTGCCA | GCGCTGAGTACATGAAGTCG |
| ZBTB46 | CGGGAAGAAGTTCACGCGG | CTGCACACCTTGCACACATAC |
| SCD | AAACCTGGCTTGCTGATG | GGGGGCTAATGTTCTTGTCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, F.; Lin, Z.; Long, Q.; Lu, Z.; Zhu, K.; Zhao, M.; Yang, M. CircZBTB46 Protects Acute Myeloid Leukemia Cells from Ferroptotic Cell Death by Upregulating SCD. Cancers 2023, 15, 459. https://doi.org/10.3390/cancers15020459
Long F, Lin Z, Long Q, Lu Z, Zhu K, Zhao M, Yang M. CircZBTB46 Protects Acute Myeloid Leukemia Cells from Ferroptotic Cell Death by Upregulating SCD. Cancers. 2023; 15(2):459. https://doi.org/10.3390/cancers15020459
Chicago/Turabian StyleLong, Fei, Zhi Lin, Qinpeng Long, Zhixing Lu, Kaiyu Zhu, Mingyi Zhao, and Minghua Yang. 2023. "CircZBTB46 Protects Acute Myeloid Leukemia Cells from Ferroptotic Cell Death by Upregulating SCD" Cancers 15, no. 2: 459. https://doi.org/10.3390/cancers15020459
APA StyleLong, F., Lin, Z., Long, Q., Lu, Z., Zhu, K., Zhao, M., & Yang, M. (2023). CircZBTB46 Protects Acute Myeloid Leukemia Cells from Ferroptotic Cell Death by Upregulating SCD. Cancers, 15(2), 459. https://doi.org/10.3390/cancers15020459




