Genetic Dissection of Tobacco (Nicotiana tabacum L.) Plant Height Using Single-Locus and Multi-Locus Genome-Wide Association Studies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Phenotyping
2.2. Statistical Analysis and Heritability Estimation for Plant Height
2.3. Genotyping and Genome-Wide Association Studies
2.4. Elite Allele Analysis for Plant Height
2.5. Potential Candidate Gene Analysis
2.6. Gene Ontology and KEGG Analysis
3. Results
3.1. Phenotypic Evaluation for Plant Height
3.2. Genome-Wide Association Mapping for Plant Height
3.3. Identification of Stable Quantitative Trait Nucleotides
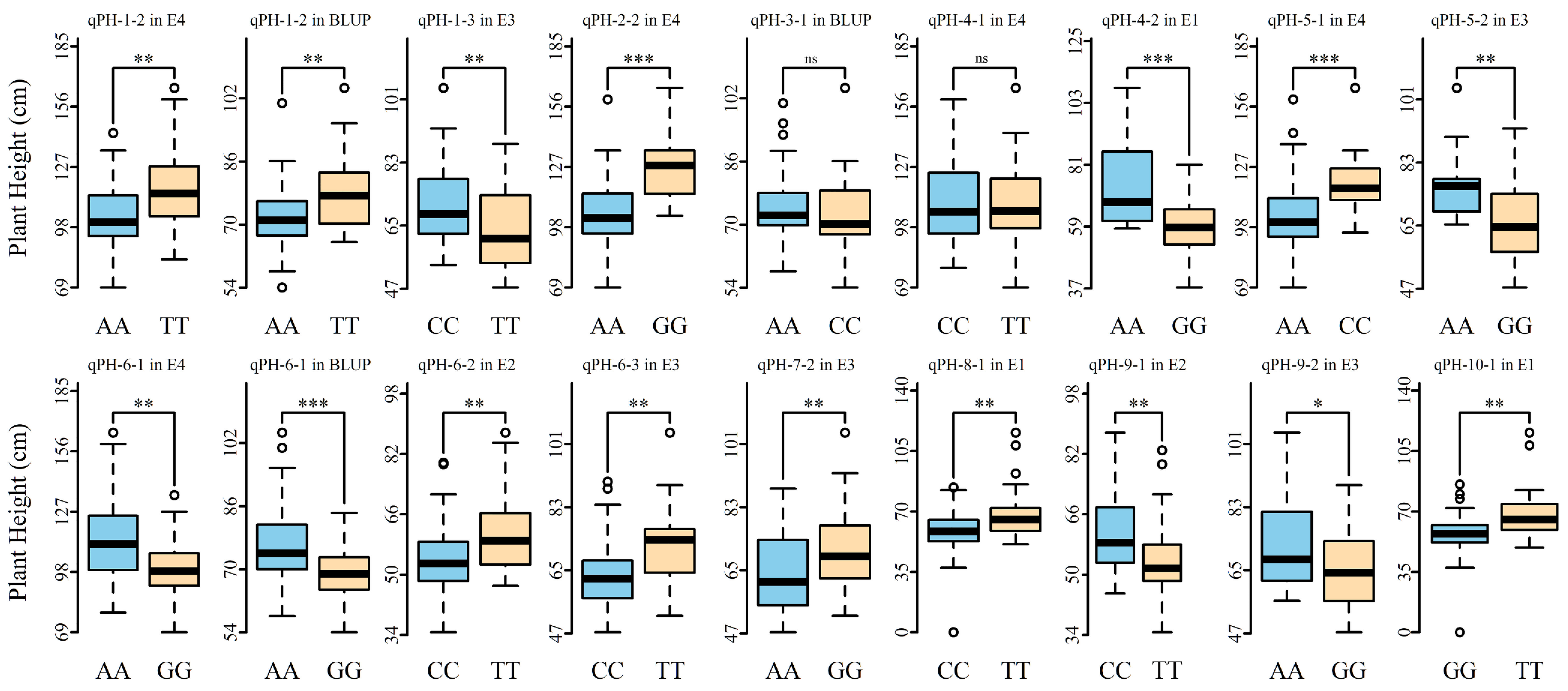
3.4. Allelic Effects of Stable QTNs for Plant Height in Multiple Environments
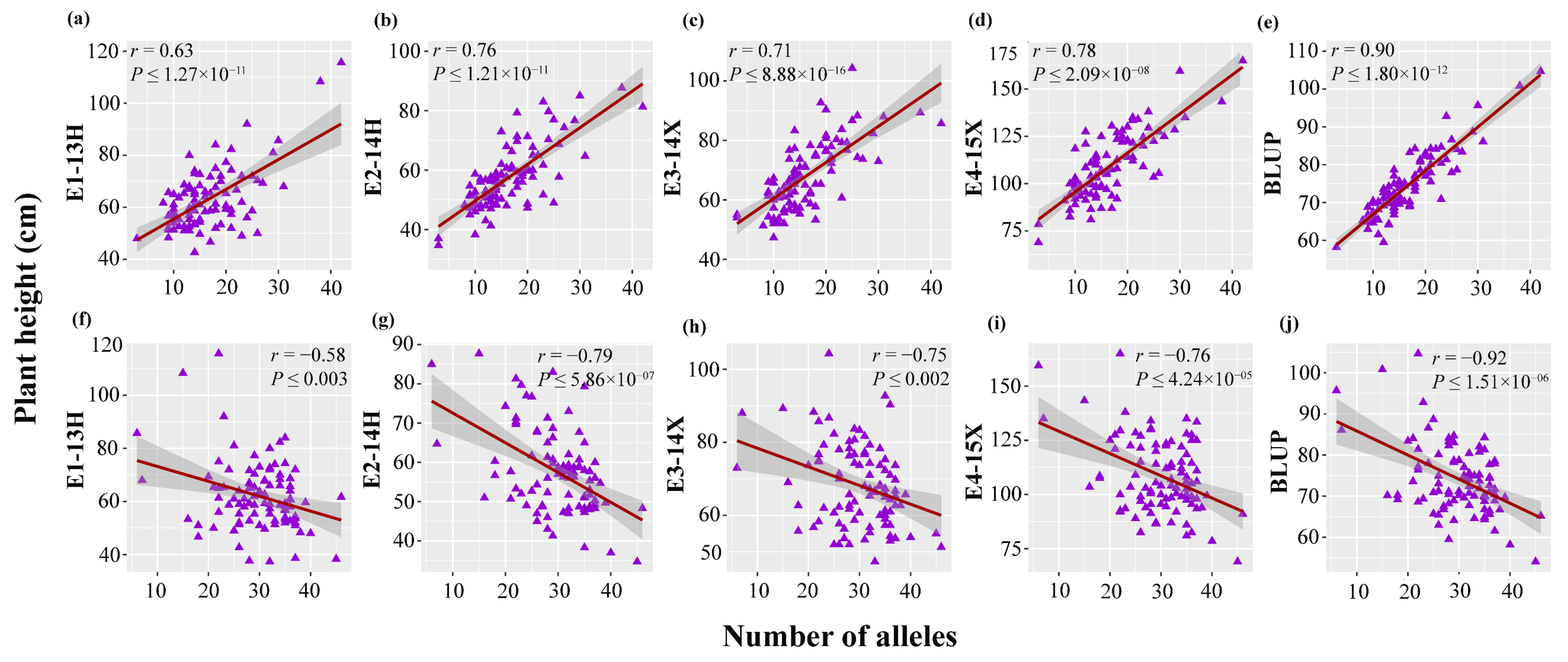
3.5. Distribution of Elite Alleles in Mapping Population and cross Combination in Two Directions
3.6. Identification of Candidate Genes Based on Stable QTNs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef] [PubMed]
- Nanda, C.; Sarala, K.; Nagesh, P.; Ramakrishnan, S. Heritability and genetic variability studies in the germplasm accessions of flue cured Virginia tobacco (Nicotiana tobaccum L.). Emergent Life Sci. Res. 2021, 07, 36–39. [Google Scholar] [CrossRef]
- Ahmed, S.; Mohammad, F. Heritability estimates and correlation analysis for production traits in fcv tobacco. Sarhad J. Agric. 2017, 33, 212–219. [Google Scholar] [CrossRef]
- Berbeć, A.K.; Matyka, M. Biomass characteristics and energy yields of tobacco (Nicotiana tabacum L.) cultivated in eastern Poland. Agriculture 2020, 10, 551. [Google Scholar] [CrossRef]
- Gong, D.; Huang, L.; Xu, X.; Wang, C.; Ren, M.; Wang, C.; Chen, M. Construction of a high-density SNP genetic map in flue-cured tobacco based on SLAF-seq. Mol. Breed. 2016, 36, 100. [Google Scholar] [CrossRef]
- Robinson, H.F.; Mann, T.; Comstock, R.E. An analysis of quantitative variability in Nicotiana tabacum. Heredity 1954, 8, 365–376. [Google Scholar] [CrossRef] [Green Version]
- Basit, A.; Farhan, M.; Mo, W.D.; Ding, H.X.; Ikram, M.; Farooq, T.; Ahmed, S.; Yang, Z.F.; Wang, Y.; Hashem, M.; et al. Enhancement of resistance by poultry manure and plant hormones (salicylic acid & citric acid) against tobacco mosaic virus. Saudi J. Biol. Sci. 2021, 28, 3526–3533. [Google Scholar]
- Cheng, L.; Yang, A.; Jiang, C.; Ren, M.; Zhang, Y.; Feng, Q.; Wang, S.; Gua, Y.; Luo, C. Quantitative trait loci mapping for plant height in tobacco using linkage and association mapping methods. Crop Sci. 2015, 55, 641–647. [Google Scholar] [CrossRef]
- Tong, Z.; Chen, X.; Fang, D.; Zeng, J.; Wu, X.; Xiao, B. SSR marker-based analyses on genetic diversity and relevant variations of agronomic traits and chemical composition of 231 flue-cured tobacco germplasm resources. Acta Tab. Sin. 2017, 23, 31–58. [Google Scholar]
- Tang, Z.; Chen, L.; Chen, Z.; Fu, Y.; Sun, X.; Wang, B.; Xia, T. Climatic factors determine the yield and quality of Honghe flue-cured tobacco. Sci. Rep. 2020, 10, 19868. [Google Scholar] [CrossRef]
- Lai, R.; Ikram, M.; Li, R.; Xia, Y.; Yuan, Q.; Zhao, W.; Zhang, Z.; Siddique, K.H.M.; Guo, P. Identification of novel quantitative trait nucleotides and candidate genes for bacterial wilt resistance in tobacco (Nicotiana tabacum L.) using genotyping-by-sequencing and multi-locus genome-wide association studies. Front. Plant Sci. 2021, 12, 744175. [Google Scholar] [CrossRef] [PubMed]
- Badu-Apraku, B.; Adewale, S.; Paterne, A.; Gedil, M.; Asiedu, R. Identification of QTLs controlling resistance/tolerance to striga hermonthica in an extra-early maturing yellow maize population. Agronomy 2020, 10, 1168. [Google Scholar] [CrossRef]
- Nishi, T.; Tajima, T.; Noguchi, S.; Ajisaka, H.; Negishi, H. Identification of DNA markers of tobacco linked to bacterial wilt resistance. Theor. Appl. Genet. 2003, 106, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Kizil, S.; Basak, M.; Guden, B.; Tosun, H.S.; Uzun, B.; Yol, E. Genome-wide discovery of indel markers in sesame (Sesamum indicum L.) using ddradseq. Plants 2020, 9, 1262. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Chhapekar, S.S.; Rameneni, J.J.; Kim, S.; Gan, T.H.; Choi, S.R.; Lim, Y.P. Identification of qtls and candidate genes related to flower traits and bolting time in radish (Raphanus sativus L.). Agronomy 2021, 11, 1623. [Google Scholar] [CrossRef]
- Chao, W.S.; Horvath, D.P.; Stamm, M.J.; Anderson, J.V. Genome-wide association mapping of freezing tolerance loci in canola (Brassica napus L.). Agronomy 2021, 11, 233. [Google Scholar] [CrossRef]
- Tong, Z.-J.; Jiao, F.-C.; Wu, X.-F.; Wang, F.-Q.; Chen, X.-J.; Li, X.-Y.; Gao, Y.-L.; Zhang, Y.-H.; Xiao, B.-G.; Wu, W.-R. Mapping of quantitative trait loci underlying six agronomic traits in flue- cured tobacco (Nicotiana tabacum L.). Acta Agron. Sin. 2013, 38, 1407–1415. [Google Scholar] [CrossRef]
- Li, R.H.; Carfi, L.; Lv, Y.H.; Xia, Y.S.; Wu, C.; Yu, Y.W.; Qiu, M.W.; Zhao, W.C.; Guo, P.G. Association analysis of MFLP markers with bacterial wilt resistance in tobacco. Proc. Environ. Sci. Biol. Eng. 2014, 1, 303–310. [Google Scholar]
- Tong, Z.; Xiu, Z.; Ming, Y.; Fang, D.; Chen, X.; Hu, Y.; Zhou, J.; He, W.; Jiao, F.; Zhang, C.; et al. Quantitative trait locus mapping and genomic selection of tobacco (Nicotiana tabacum L.) based on high-density genetic map. Plant Biotechnol. Rep. 2021, 15, 845–854. [Google Scholar] [CrossRef]
- Guo, P.; Baum, M.; Varshney, R.K.; Graner, A.; Grando, S.; Ceccarelli, S. QTLs for chlorophyll and chlorophyll fluorescence parameters in barley under post-flowering drought. Euphytica 2008, 163, 203–214. [Google Scholar] [CrossRef]
- Buckler IV, E.S.; Thornsberry, J.M. Plant molecular diversity and applications to genomics. Curr. Opin. Plant Biol. 2002, 5, 107–111. [Google Scholar] [CrossRef]
- Zhang, J.-S.; Wang, R.-G.; Yang, C.-Y.; Wu, C.; Shi, Y.-W.; Wang, Z.-H.; Wang, Y.; Ren, X.-L. Genetic diversity of agronomic traits and association analysis with SRAP markers in flue-cured tobacco (Nicotiana tabacum L) varieties from China and Abroad. Acta Agron. Sin. 2013, 38, 1029–1041. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, H.; Xiang, X.; Yang, A.; Feng, Q.; Dai, P.; Li, Y.; Jiang, X.; Liu, G.; Zhang, X. Construction of a SNP fingerprinting database and population genetic analysis of cigar tobacco germplasm resources in China. Front. Plant Sci. 2021, 12, 618133. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Luo, J.; Qi, C.; Ruan, Y.; Li, J.; Zhang, A.; Yang, X.; He, Y. Genome-wide association study (GWAS) reveals the genetic architecture of four husk traits in maize. BMC Genom. 2016, 17, 946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safdar, L.B.; Almas, F.; Sarfraz, S.; Ejaz, M.; Ali, Z.; Mahmood, Z.; Yang, L.; Tehseen, M.M.; Ikram, M.; Liu, S.; et al. Genome-wide association study identifies five new cadmium uptake loci in wheat. Plant Genome 2020, 13, e20030. [Google Scholar] [CrossRef]
- Elhadi, G.M.I.; Kamal, N.M.; Gorafi, Y.S.A.; Yamasaki, Y.; Ban, Y.; Kato, K.; Tahir, I.S.A.; Ishii, T.; Tanaka, H.; Tsujimoto, H. Novel loci for kernel hardness appeared as a response to heat and combined heat-drought conditions in wheat harboring Aegilops tauschii diversity. Agronomy 2021, 11, 1061. [Google Scholar] [CrossRef]
- Reddy, V.R.P.; Das, S.; Dikshit, H.K.; Mishra, G.P.; Aski, M.S.; Singh, A.; Tripathi, K.; Pandey, R.; Bansal, R.; Singh, M.P.; et al. Genetic dissection of phosphorous uptake and utilization efficiency traits using gwas in mungbean. Agronomy 2021, 11, 1401. [Google Scholar] [CrossRef]
- Xia, Y.; Li, R.; Bai, G.; Siddique, K.H.M.; Varshney, R.K.; Baum, M.; Yan, G.; Guo, P. Genetic variations of HvP5CS1 and their association with drought tolerance related traits in barley (Hordeum vulgare L.). Sci. Rep. 2017, 7, 7870. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Li, R.; Ning, Z.; Bai, G.; Siddique, K.H.M.; Yan, G.; Baum, M.; Varshney, R.K.; Guo, P. Single nucleotide polymorphisms in HSP17.8 and their association with agronomic traits in barley. PLoS ONE 2013, 8, e56816. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Ning, Z.; Bai, G.; Li, R.; Yan, G.; Siddique, K.H.M.; Baum, M.; Guo, P. Allelic variations of a light harvesting chlorophyll a/b-binding protein gene (Lhcb1) associated with agronomic traits in barley. PLoS ONE 2012, 7, e37573. [Google Scholar] [CrossRef] [Green Version]
- Faccini, N.; Delbono, S.; Oǧuz, A.Ç.; Cattivelli, L.; Vale, G.; Tondelli, A. Resistance of european spring 2-row barley cultivars to pyrenophora graminea and detection of associated loci. Agronomy 2021, 11, 374. [Google Scholar] [CrossRef]
- Jain, M.; Moharana, K.C.; Shankar, R.; Kumari, R.; Garg, R. Genomewide discovery of DNA polymorphisms in rice cultivars with contrasting drought and salinity stress response and their functional relevance. Plant Biotechnol. J. 2014, 12, 253–264. [Google Scholar] [CrossRef]
- Wang, K.; Zhuang, J.Y.; Huang, D.R.; Ying, J.Z.; Fan, Y.Y. Genome-wide polymorphisms between the parents of an elite hybrid rice and the development of a novel set of PCR-based InDel markers. Genet. Mol. Res. 2015, 14, 3209–3222. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Kong, C.; Han, F.; Yang, L.; Zhuang, M.; Zhang, Y.; Wang, Y.; Ji, J.; Li, Z.; Fang, Z.; et al. Two user-friendly molecular markers developed for the identification of hybrid lethality genes in brassica oleracea. Agronomy 2021, 11, 982. [Google Scholar] [CrossRef]
- Lee, G.J.; Lee, S.; Carter, T.E.; Shannon, G.; Boerma, H.R. Identification of soybean yield QTL in irrigated and rain-fed environments. Agronomy 2021, 11, 2207. [Google Scholar] [CrossRef]
- Bus, A.; Hecht, J.; Huettel, B.; Reinhardt, R.; Stich, B. High-throughput polymorphism detection and genotyping in Brassica napus using next-generation RAD sequencing. BMC Genom. 2012, 13, 281. [Google Scholar] [CrossRef] [Green Version]
- Barchi, L.; Lanteri, S.; Portis, E.; Acquadro, A.; Valè, G.; Toppino, L.; Rotino, G.L. Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genom. 2011, 12, 304. [Google Scholar] [CrossRef] [Green Version]
- Rebetzke, G.J.; Richards, R.A. Gibberellic acid-sensitive dwarfing genes reduce plant height to increase kernel number and grain yield of wheat. Aust. J. Agric. Res. 2000, 51, 235–245. [Google Scholar] [CrossRef]
- Li, Z.F.; Guo, Y.; Ou, L.; Hong, H.; Wang, J.; Liu, Z.X.; Guo, B.; Zhang, L.; Qiu, L. Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet. 2018, 131, 1001–1016. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Watanabe, S.; Uchiyama, T.; Kong, F.; Kanazawa, A.; Xia, Z.; Nagamatsu, A.; Arai, M.; Yamada, T.; Kitamura, K.; et al. The soybean stem growth habit gene Dt1 is an ortholog of arabidopsis Terminal Flower1. Plant Physiol. 2010, 153, 198–210. [Google Scholar] [CrossRef] [Green Version]
- Salas Fernandez, M.G.; Becraft, P.W.; Yin, Y.; Lübberstedt, T. From dwarves to giants? Plant height manipulation for biomass yield. Trends Plant Sci. 2009, 14, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Xu, H.; Wang, Y.; Wang, H.; Li, Z.; Liu, X.; Shu, Y.; Li, G.; Liu, W.; Ying, J.; et al. Osabf1 represses gibberellin biosynthesis to regulate plant height and seed germination in rice (Oryza sativa L.). Int. J. Mol. Sci. 2021, 22, 12220. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Peng, Z.; Peng, Q.; Cai, Q.; Peng, W.; Chen, M.; Yao, J. Regulation of plant height in rice by the Polycomb group genes OsEMF2b, OsFIE2 and OsCLF. Plant Sci. 2018, 267, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, C.; Lin, J.; Liu, J.; Liu, B.; Wang, J.; Huang, A.; Li, H.; Zhao, T. OsMPH1 regulates plant height and improves grain yield in rice. PLoS ONE 2017, 12, e0180825. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Wu, T.; Huang, K.; Jin, Y.M.; Li, Z.; Chen, M.; Yun, S.; Zhang, H.; Yang, X.; Chen, H.; et al. A Novel AP2/ERF transcription factor, OsRPH1, negatively regulates plant height in rice. Front. Plant Sci. 2020, 11, 709. [Google Scholar] [CrossRef]
- Teng, F.; Zhai, L.; Liu, R.; Bai, W.; Wang, L.; Huo, D.; Tao, Y.; Zheng, Y.; Zhang, Z. ZmGA3ox2, a candidate gene for a major QTL, qPH3.1, for plant height in maize. Plant J. 2013, 73, 405–416. [Google Scholar] [CrossRef]
- Li, H.; Wang, L.; Liu, M.; Dong, Z.; Li, Q.; Fei, S.; Xiang, H.; Liu, B.; Jin, W. Maize plant architecture is regulated by the ethylene biosynthetic gene ZmACS7. Plant Physiol. 2020, 183, 1184–1199. [Google Scholar] [CrossRef]
- Visscher, A.M.; Belfield, E.J.; Vlad, D.; Irani, N.; Moore, I.; Harberd, N.P. Overexpressing the multiple-stress responsive gene At1g74450 reduces plant height and male fertility in Arabidopsis Thaliana. PLoS ONE 2015, 10, e0140368. [Google Scholar]
- Zheng, M.; Zhang, L.; Tang, M.; Liu, J.; Liu, H.; Yang, H.; Fan, S.; Terzaghi, W.; Wang, H.; Hua, W. Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 2020, 18, 644–654. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; He, J.; Wan, S.; Li, W.; Chen, W.; Wang, Y.; Jiang, X.; Cheng, P.; Chu, P.; Shen, W.; et al. Fine mapping of the BnaC04.BIL1 gene controlling plant height in Brassica napus L. BMC Plant Biol. 2021, 21, 359. [Google Scholar] [CrossRef]
- Benavente, E.; Giménez, E. Modern approaches for the genetic improvement of rice, wheat and maize for abiotic constraints-related traits: A comparative overview. Agronomy 2021, 11, 376. [Google Scholar] [CrossRef]
- Ikram, M.; Han, X.; Zuo, J.F.; Song, J.; Han, C.Y.; Zhang, Y.W.; Zhang, Y.M. Identification of QTNs and their candidate genes for 100-seed weight in soybean (Glycine max L.) using multi-locus genome-wide association studies. Genes 2020, 11, 714. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Xu, Z.R.; Zhou, L.; Han, C.Y.; Zhang, Y.M. Identification of QTNs and their candidate genes for flowering time and plant height in soybean using multi-locus genome-wide association studies. Mol. Breed. 2021, 41, 39. [Google Scholar] [CrossRef]
- Hou, S.; Zhu, G.; Li, Y.; Li, W.; Fu, J.; Niu, E.; Li, L.; Zhang, D.; Guo, W. Genome-wide association studies reveal genetic variation and candidate genes of drought stress related traits in cotton (Gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 1276. [Google Scholar] [CrossRef]
- Ma, L.; Liu, M.; Yan, Y.; Qing, C.; Zhang, X.; Zhang, Y.; Long, Y.; Wang, L.; Pan, L.; Zou, C.; et al. Genetic dissection of maize embryonic callus regenerative capacity using multi-locus genome-wide association studies. Front. Plant Sci. 2018, 9, 561. [Google Scholar] [CrossRef] [Green Version]
- Butorac, J.; Beljo, J.; Gunjača, J. Study of inheritance of some agronomic and morphological traits in burley tobacco by graphic analysis of diallel cross. Plant, Soil Environ. 2004, 50, 162–167. [Google Scholar] [CrossRef] [Green Version]
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Tamba, C.L.; Wen, Y.J.; Li, P.; Ren, W.L.; Ni, Y.L.; Gao, J.; Zhang, Y.M. mrMLM v4.0: An R platform for multi-locus genome-wide association studies. Genom. Proteom. Bioinform. 2020, 18, 481–487. [Google Scholar] [CrossRef]
- Wang, S.B.; Feng, J.Y.; Ren, W.L.; Huang, B.; Zhou, L.; Wen, Y.J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Feng, J.Y.; Ni, Y.L.; Wen, Y.J.; Niu, Y.; Tamba, C.L.; Yue, C.; Song, Q.; Zhang, Y.M. PLARmEB: Integration of least angle regression with empirical Bayes for multilocus genome-wide association studies. Heredity 2017, 118, 517–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamba, C.L.; Ni, Y.L.; Zhang, Y.M. Iterative sure independence screening EM-Bayesian LASSO algorithm for multi-locus genome-wide association studies. PLoS Comput. Biol. 2017, 13, e1005357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Jia, Z.; Dunwell, J.M. Editorial: The applications of new multi-locus gwas methodologies in the genetic dissection of complex traits. Front. Plant Sci. 2019, 10, 100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [Green Version]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, J. Biosynthesis, catabolism, and signal transduction of brassinosteroids. Plant Physiol. J. 2017, 53, 291–307. [Google Scholar]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar] [CrossRef]
- Lewis, R.S.; Milla, S.R.; Kernodle, S.P. Analysis of an introgressed Nicotiana tomentosa genomic region affecting leaf number and correlated traits in Nicotiana tabacum. Theor. Appl. Genet. 2007, 114, 841–854. [Google Scholar] [CrossRef]
- Vontimitta, V.; Lewis, R.S. Mapping of quantitative trait loci affecting resistance to Phytophthora nicotianae in tobacco (Nicotiana tabacum L.) line Beinhart-1000. Mol. Breed. 2012, 29, 89–98. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, T.; Zhou, Y.; Yin, S.; Li, P.; Liu, J.; Xu, S.; Yang, Z.; Xu, C. Genome-wide association mapping of starch pasting properties in maize using single-locus and multi-locus models. Front. Plant Sci. 2018, 9, 1311. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, F.; Zhou, Y. The application of multi-locus GWAS for the detection of salt-tolerance loci in rice. Front. Plant Sci. 2018, 9, 1464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Fu, Y.; Sun, R.; Wang, Y.; Wang, Q. Single-locus and multi-locus genome-wide association studies in the genetic dissection of fiber quality traits in upland cotton (gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 1083. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, S.; Shah, T.; Xie, C.; Hao, Z.; Li, X.; Farkhari, M.; Ribaut, J.M.; Cao, M.; Rong, T.; et al. Joint linkage-linkage disequilibrium mapping is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. Proc. Natl. Acad. Sci. USA 2010, 107, 19585–19590. [Google Scholar] [CrossRef] [Green Version]
- Tian, Z.; Qian, Q.; Liu, Q.; Yan, M.; Liu, X.; Yan, C.; Liu, G.; Gao, Z.; Tang, S.; Zeng, D.; et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. USA 2009, 106, 21760–21765. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wan, X.; Crossa, J.; Crouch, J.; Weng, J.; Zhai, H.; Wan, J. QTL mapping of grain length in rice (Oryza sativa L.) using chromosome segment substitution lines. Genet. Res. 2006, 88, 93–104. [Google Scholar] [CrossRef]
- Luo, H.; Chen, S.; Jiang, J.; Teng, N.; Chen, Y.; Chen, F. The AP2-like gene NsAP2 from water lily is involved in floral organogenesis and plant height. J. Plant Physiol. 2012, 169, 992–998. [Google Scholar] [CrossRef]
- Ma, X.; Song, L.; Yang, Y.; Liu, D. A gain-of-function mutation in the ROC1 gene alters plant architecture in Arabidopsis. New Phytol. 2013, 197, 751–762. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, J.; Wu, Y. Arabidopsis ATAF1 enhances the tolerance to salt stress and ABA in transgenic rice. J. Plant Res. 2016, 129, 955–962. [Google Scholar] [CrossRef]
- Waterworth, W.M.; Kozak, J.; Provost, C.M.; Bray, C.M.; Angelis, K.J.; West, C.E. DNA ligase 1 deficient plants display severe growth defects and delayed repair of both DNA single and double strand breaks. BMC Plant Biol. 2009, 9, 79. [Google Scholar] [CrossRef] [Green Version]
- Abercrombie, J.M.; Halfhill, M.D.; Ranjan, P.; Rao, M.R.; Saxton, A.M.; Yuan, J.S.; Stewart, C.N. Transcriptional responses of Arabidopsis thaliana plants to As (V) stress. BMC Plant Biol. 2008, 8, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Pujol, G.; Ferrer, A.; Ariño, J. Protein phosphatase 2A and protein phosphatase X genes in Arabidopsis thaliana. In Protein Phosphatase Protocols; Humana Press: Totowa, NJ, USA, 1998; Volume 93, pp. 201–212. [Google Scholar]
- Wen, F.; Wang, J.; Xing, D. A protein phosphatase 2A catalytic subunit modulates blue light-induced chloroplast avoidance movements through regulating actin cytoskeleton in arabidopsis. Plant Cell Physiol. 2012, 53, 1366–1379. [Google Scholar] [CrossRef]
- Visioli, G.; Maestri, E.; Polverini, E.; Pavesi, A.; Marmiroli, N. L1 a Non-LTR retrotrasposon fragment in the genome of Arabidopsis thaliana with homology to plants and animals. Am. J. Plant Sci. 2013, 04, 806–816. [Google Scholar] [CrossRef] [Green Version]
- Serrano, I.; Gu, Y.; Qi, D.; Dubiella, U.; Innes, R.W. The arabidopsis EDR1 protein kinase negatively regulates the ATL1 e3 ubiquitin ligase to suppress cell death. Plant Cell 2014, 26, 4532–4546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwager, K.M.; Calderon-Villalobos, L.I.A.; Dohmann, E.M.N.; Willige, B.C.; Knierer, S.; Nill, C.; Schwechheimer, C. Characterization of the VIER F-BOX PROTEINE genes from arabidopsis reveals their importance for plant growth and development. Plant Cell 2007, 19, 1163–1178. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Wu, J.; Zhao, M.; Ma, X.; Zhang, W.; Xia, G.; Wang, M. A novel wheat cysteine-rich receptor-like kinase gene CRK41 is involved in the regulation of seed germination under osmotic stress in Arabidopsis thaliana. J. Plant Biol. 2017, 60, 571–581. [Google Scholar] [CrossRef]
- Zhu, S.H.; Xue, F.; Li, Y.J.; Liu, F.; Zhang, X.Y.; Zhao, L.J.; Sun, Y.Q.; Zhu, Q.H.; Sun, J. Identification and functional characterization of a microtubule-associated protein, GhCLASP2, from upland cotton (Gossypium hirsutum L.). Front. Plant Sci. 2018, 9, 882. [Google Scholar] [CrossRef]
- Eng, R.C.; Schneider, R.; Matz, T.W.; Carter, R.; Ehrhardt, D.W.; Jönsson, H.; Nikoloski, Z.; Sampathkumar, A. KATANIN and CLASP function at different spatial scales to mediate microtubule response to mechanical stress in Arabidopsis cotyledons. Curr. Biol. 2021, 31, 3262–3274.e6. [Google Scholar] [CrossRef]
- Fan, W.; Lu, J.; Pan, C.; Tan, M.; Lin, Q.; Liu, W.; Li, D.; Wang, L.; Hu, L.; Wang, L.; et al. Sequencing of Chinese castor lines reveals genetic signatures of selection and yield-associated loci. Nat. Commun. 2019, 10, 3418. [Google Scholar] [CrossRef]
- Knutson, B.A. Emergence and expansion of TFIIB-like factors in the plant kingdom. Gene 2013, 526, 30–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, Q.K.; Liang, Y.; Zhou, J.J.; Dou, X.Y.; Gao, S.C.; Chen, L.Q.; Zhang, X.Q.; Ye, D. Pollen-expressed transcription factor 2 encodes a novel plant-specific TFIIB-related protein that is required for pollen germination and embryogenesis in arabidopsis. Mol. Plant 2013, 6, 1091–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osakabe, Y.; Arinaga, N.; Umezawa, T.; Katsura, S.; Nagamachi, K.; Tanaka, H.; Ohiraki, H.; Yamada, K.; Seo, S.U.; Abo, M.; et al. Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 2013, 25, 609–624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobbie, L.; McGovern, M.; Hurwitz, L.R.; Pierro, A.; Liu, N.Y.; Bandyopadhyay, A.; Estelle, M. The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development 2000, 127, 23–32. [Google Scholar] [CrossRef]
- Furutani, M.; Sakamoto, N.; Yoshida, S.; Kajiwara, T.; Robert, H.S.; Friml, J.; Tasaka, M. Polar-localized NPH3-like proteins regulate polarity and endocytosis of PIN-FORMED auxin efflux carriers. Development 2011, 138, 2069–2078. [Google Scholar] [CrossRef] [Green Version]
- Mehmood, S.; Ahmed, W.; Ikram, M.; Imtiaz, M.; Mahmood, S.; Tu, S.; Chen, D. Chitosan modified biochar increases soybean (Glycine max L.) resistance to salt-stress by augmenting root morphology, antioxidant defense mechanisms and the expression of stress-responsive genes. Plants 2020, 9, 1173. [Google Scholar] [CrossRef]
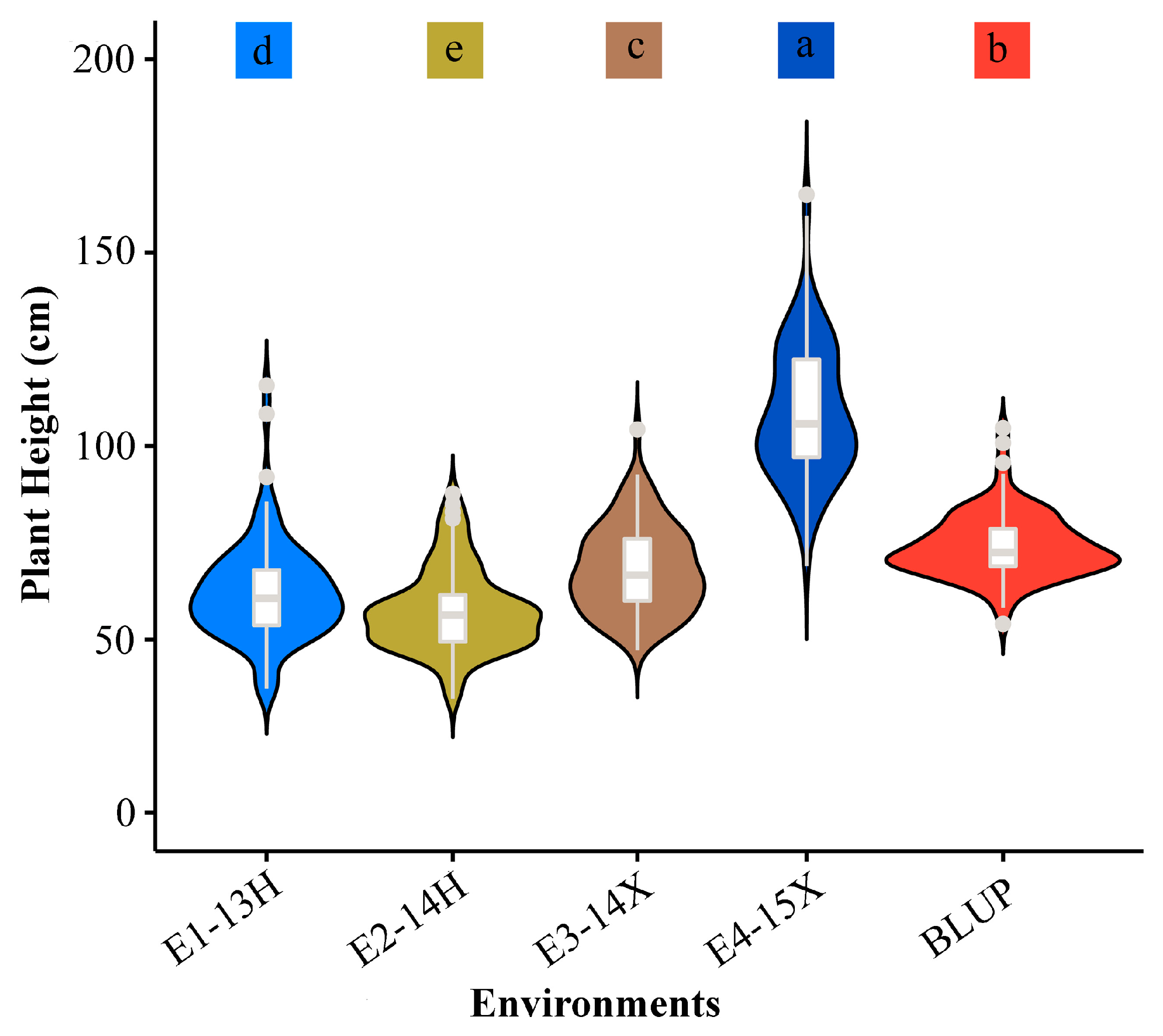



| Environment | Mean | Min | Max | SD | CV (%) | Kurtosis | Skewness | FG | FE | FG×E | h2B (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E1-13H | 62.20 | 37.33 | 115.67 | 12.95 | 23.39 | 3.70 | 1.25 | 506.64 ** | 158.39 ** | 24.81 ** | 80.31 |
| E2-14H | 57.55 | 34.70 | 87.70 | 10.73 | 18.64 | 0.44 | 0.73 | 87.09 | |||
| E3-14X | 68.25 | 47.30 | 104.30 | 11.23 | 16.46 | 0.01 | 0.53 | 83.37 | |||
| E4-15X | 108.80 | 69.00 | 165.00 | 17.37 | 15.96 | 0.58 | 0.58 | 78.93 |
| QTN name | Position (bp) | LOD score | ‘−log10(p)’ | r2 (%) a | Environments b | Method c |
|---|---|---|---|---|---|---|
| qPH-1-1 | Nt01_39983559 | 3.01–5.00 | 3.71–5.79 | 4.69–6.84 | E3-14X, BLUP | M2, M3, M4 |
| qPH-1-2 | Nt01_102574685 | 3.31–6.45 | 4.03–7.29 | 4.9–12.84 | E4-15X, BLUP | M2, M3 |
| qPH-6-1 | Nt06_34417817 | 4.43–5.59 | 5.2–6.41 | 4.43–9.85 | E4-15X, BLUP | M1, M2, M4 |
| qPH-10-1 | Nt10_26491005 | 4.13–13.45 | 4.89–14.45 | 7.12–25.67 | E1-13H, E2-14H | M1, M2, M3 |
| qPH-10-2 | Nt10_62768687 | 4.12–9.48 | 4.88–10.41 | 9.5–21.48 | E1-13H, E2-14H | M2, M3, M4 |
| qPH-11-1 | Nt11_5419687 | 3.52–4.06 | 4.24–4.81 | 3.03–7.58 | E3-14X, BLUP | M1, M2 |
| qPH-13-2 | Nt13_62198057 | 3.27–8.33 | 3.98–9.23 | 6.43–11.84 | E2-14H, BLUP | M2, M3, SL2 |
| qPH-14-1 | Nt14_88069397 | 3.53–11.64 | 4.26–12.61 | 4.25–11.48 | E4-15X, BLUP | M1, M2, M3, M4, SL1, SL2 |
| qPH-17-2 | Nt17_140487212 | 3.38–5.45 | 4.1–6.27 | 0.64–2.09 | E1-13H, E2-14H | M2, M4 |
| qPH-17-3 | Nt17_200407178 | 4.62–5.02 | 5.4–5.82 | 6.31–7.08 | E2-14H, BLUP | M2, M4, SL1 |
| qPH-18-1 | Nt18_10034331 | 3.38–8.4 | 4.1–9.31 | 6.63–15.68 | E4-15X, BLUP | M1, M3, M4 |
| qPH-18-3 | Nt18_37042785 | 4.3–10.55 | 5.06–11.5 | 3.95–13.32 | E1-13H, E2-14H, BLUP | M2, M4, SL2 |
| qPH-18-4 | Nt18_61973096 | 4.06–6.09 | 4.81–6.93 | 3.65–6.43 | E3-14X, BLUP | M2, M4, SL1, SL2 |
| qPH-20-1 | Nt20_3868276 | 3.13–5.87 | 3.83–6.7 | 1.57–6.54 | E4-15X, BLUP | M2, M3 |
| qPH-22-1 | Nt22_19764404 | 3.67–9.86 | 4.4–10.8 | 7.29–24.78 | E3-14X, BLUP | M2, M3, M4 |
| qPH-22-2 | Nt22_102807360 | 3.15–8.63 | 3.86–9.54 | 1.39–7.8 | E1-13H, E2-14H | M3, M4 |
| qPH-23-3 | Nt23_56966891 | 3.09–4.55 | 3.79–5.33 | 4.16–6.4 | E4-15X, BLUP | M2, M3, M4 |
| qPH-23-4 | Nt23_116192791 | 3.18–3.95 | 3.89–4.69 | 2.63–7.86 | E2-14H, BLUP | M2, M3, M4 |
| qPH-24-2 | Nt24_34952292 | 2.48E-08 | 8.513637 | E1-13H, E2-14H, BLUP | SL1, SL2 |
| Direction | P1 | P2 | P1-Phenotype (cm) | P2-Phenotype (cm) | P1-Elite Alleles | P2-Elite Alleles | Offspring Alleles |
|---|---|---|---|---|---|---|---|
| Taller plants | Qinggeng | KY14 | 111.91 | 88.92 | 42 | 31 | 47 |
| Japan 4 | Qinggeng | 78.86 | 111.91 | 25 | 42 | 46 | |
| ROX28 | 81-26 | 89.55 | 107.16 | 25 | 38 | 46 | |
| H66B | KY14 | 100.79 | 88.92 | 30 | 31 | 45 | |
| 81-26 | KY14 | 107.16 | 88.92 | 38 | 31 | 45 | |
| Hicks 187 | Qinggeng | 79.7 | 111.91 | 26 | 42 | 44 | |
| Japan 4 | 81-26 | 78.86 | 107.16 | 25 | 38 | 44 | |
| Shorter plants | K399 | C319 | 49.25 | 54.37 | 45 | 46 | 49 |
| NC82 | C319 | 60.28 | 54.37 | 40 | 46 | 48 | |
| G33 | RG112 | 72.29 | 63.67 | 37 | 37 | 44 | |
| K394 | VA116 | 69.03 | 65.92 | 39 | 37 | 46 | |
| K399 | RG112 | 49.25 | 63.67 | 45 | 37 | 49 | |
| Nanxuan No. 1 | K394 | 69.03 | 63.33 | 37 | 39 | 45 | |
| RG112 | VA115 | 63.67 | 63.67 | 37 | 37 | 42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ikram, M.; Lai, R.; Xia, Y.; Li, R.; Zhao, W.; Siddique, K.H.M.; Chen, J.; Guo, P. Genetic Dissection of Tobacco (Nicotiana tabacum L.) Plant Height Using Single-Locus and Multi-Locus Genome-Wide Association Studies. Agronomy 2022, 12, 1047. https://doi.org/10.3390/agronomy12051047
Ikram M, Lai R, Xia Y, Li R, Zhao W, Siddique KHM, Chen J, Guo P. Genetic Dissection of Tobacco (Nicotiana tabacum L.) Plant Height Using Single-Locus and Multi-Locus Genome-Wide Association Studies. Agronomy. 2022; 12(5):1047. https://doi.org/10.3390/agronomy12051047
Chicago/Turabian StyleIkram, Muhammad, Ruiqiang Lai, Yanshi Xia, Ronghua Li, Weicai Zhao, Kadambot H. M. Siddique, Jianjun Chen, and Peiguo Guo. 2022. "Genetic Dissection of Tobacco (Nicotiana tabacum L.) Plant Height Using Single-Locus and Multi-Locus Genome-Wide Association Studies" Agronomy 12, no. 5: 1047. https://doi.org/10.3390/agronomy12051047
APA StyleIkram, M., Lai, R., Xia, Y., Li, R., Zhao, W., Siddique, K. H. M., Chen, J., & Guo, P. (2022). Genetic Dissection of Tobacco (Nicotiana tabacum L.) Plant Height Using Single-Locus and Multi-Locus Genome-Wide Association Studies. Agronomy, 12(5), 1047. https://doi.org/10.3390/agronomy12051047








