Screening for a Novel Gene, OsPSLSq6, Using QTL Analysis for Lodging Resistance in Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Experiment Design
2.2. Phenotype Evaluation
2.3. QTL Analysis for PSLSA and PSLSB
2.4. Gene Information Analysis and Statistical Analysis
3. Results
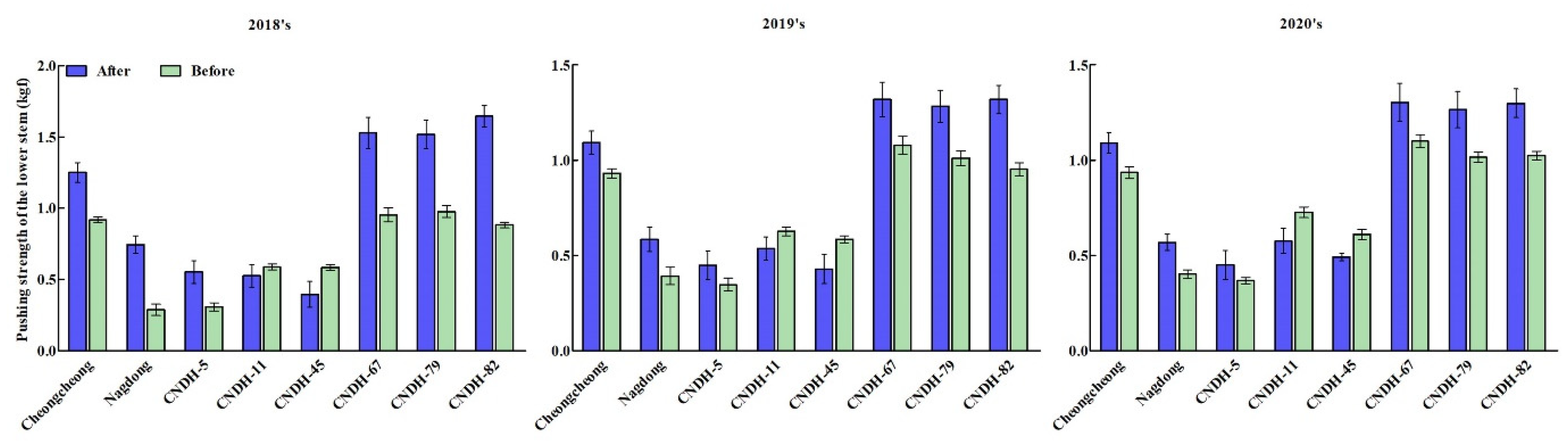
3.1. Phenotypic Evaluation and Comparison to Agricultural Characteristics
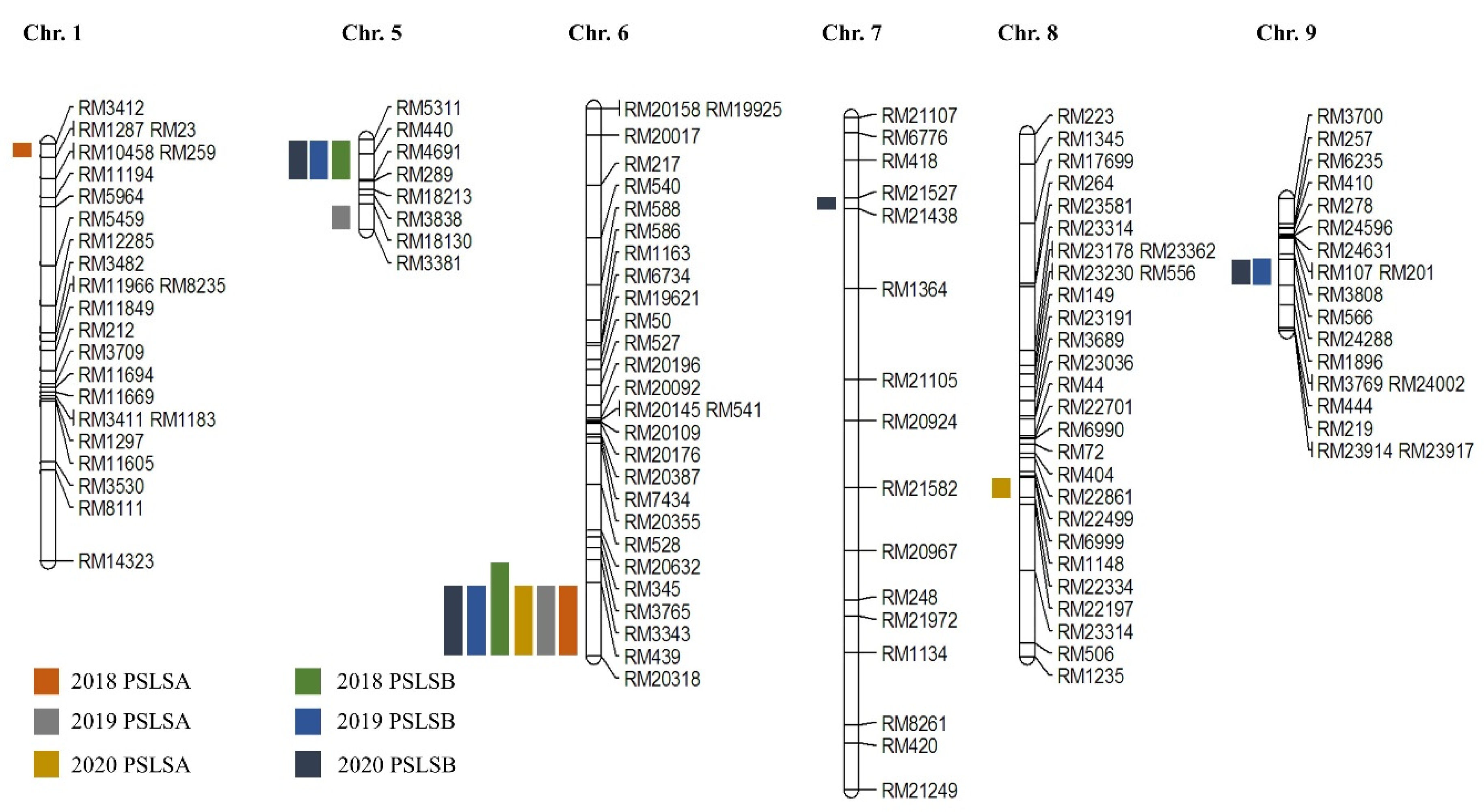
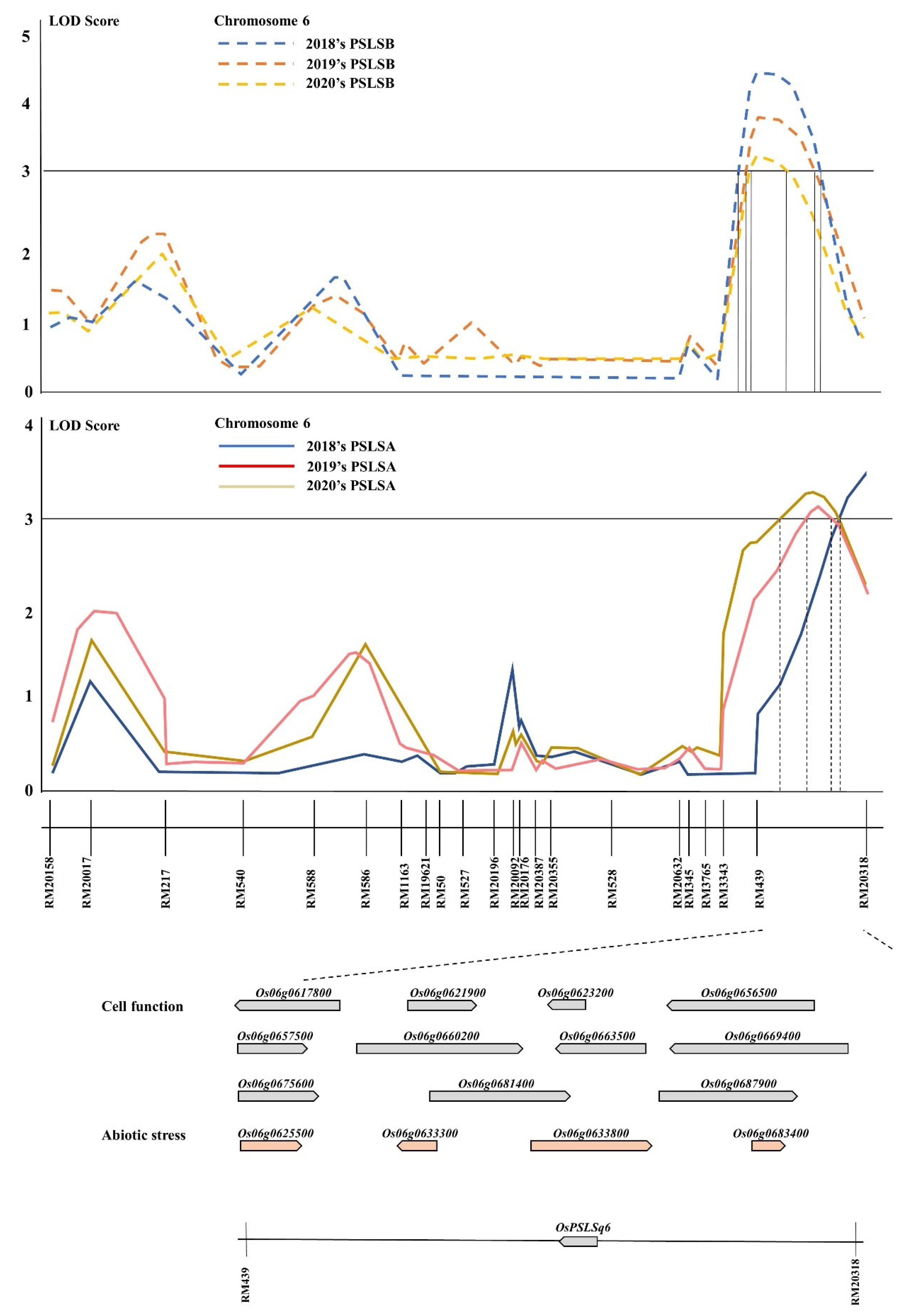
3.2. QTL Analysis Associated with PSLSA and PSLSB
3.3. Search Candidate Genes Associated with PSLSA and PSLSB
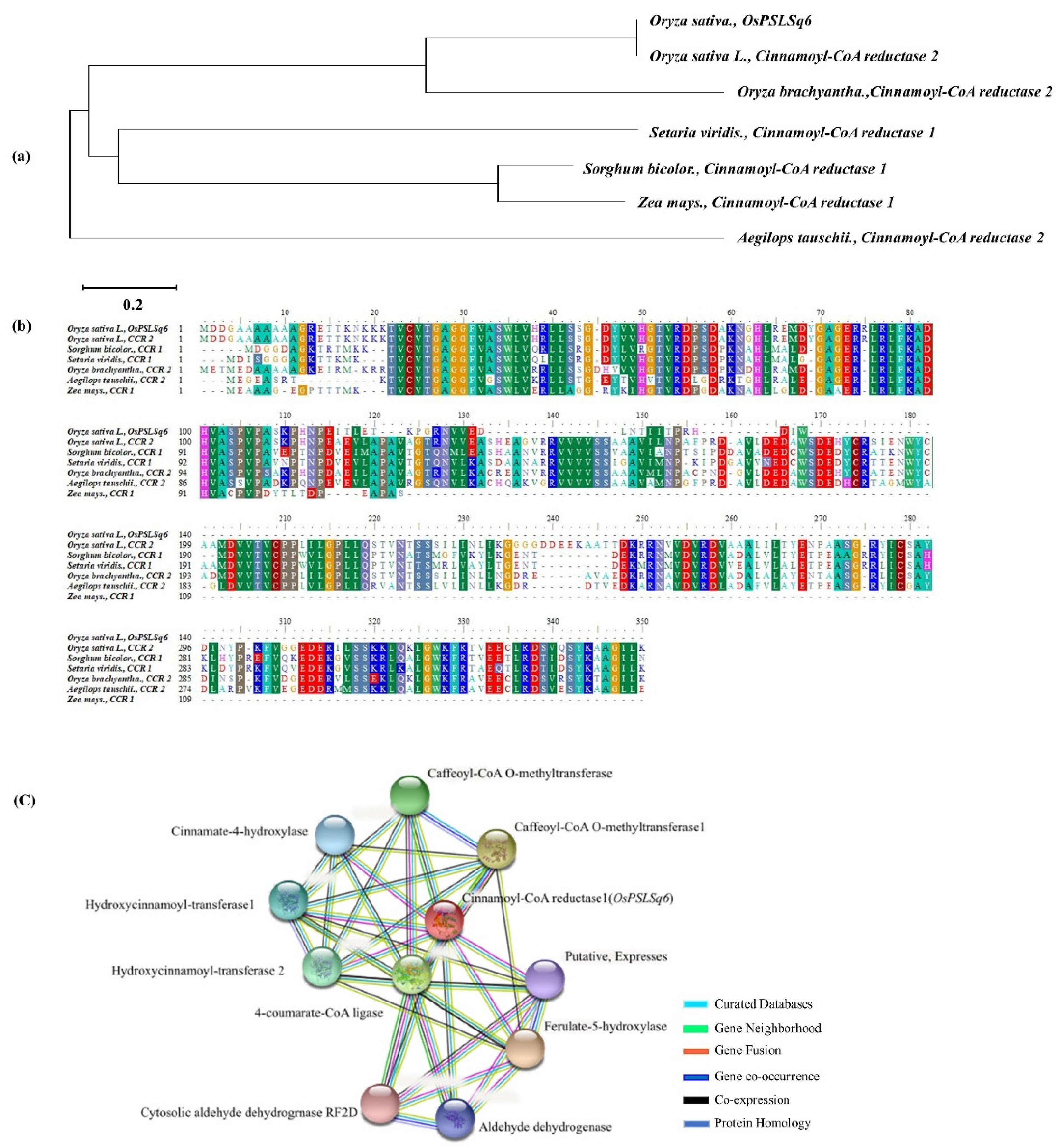
3.4. Phylogenetic Tree Analysis and Homology Sequence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kashiwagi, T.; Ishimaru, K. Identification and functional analysis of a locus for improvement of lodging resistance in rice. Plant Physiol. 2004, 134, 676–683. [Google Scholar] [CrossRef] [Green Version]
- Shah, L.; Yahya, M.; Shah, S.M.A.; Nadeem, M.; Ali, A.; Ali, A.; Wang, J.; Riaz, M.W.; Rehman, S.; Wu, W. Improving lodging resistance: Using wheat and rice as classical examples. Int. J. Mol. Sci. 2019, 20, 4211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.-J.; Li, G.-H.; Yang, Y.-M.; Quan, L.; Zhang, J.; Liu, J.-Y.; Shaohua, W.; She, T.; Ding, Y.-F. Effects of nitrogen application rate and ratio on lodging resistance of super rice with different genotypes. J. Integr. Agric. 2014, 13, 63–72. [Google Scholar] [CrossRef]
- Kuai, J.; Yang, Y.; Sun, Y.; Zhou, G.; Zuo, Q.; Wu, J.; Ling, X. Paclobutrazol increases canola seed yield by enhancing lodging and pod shatter resistance in Brassica napus L. Field Crops Res. 2015, 180, 10–20. [Google Scholar] [CrossRef]
- Zuo, Q.; Kuai, J.; Zhao, L.; Hu, Z.; Wu, J.; Zhou, G. The effect of sowing depth and soil compaction on the growth and yield of rapeseed in rice straw returning field. Field Crops Res. 2017, 203, 47–54. [Google Scholar] [CrossRef]
- Kashiwagi, T. Identification of quantitative trait loci for resistance to bending-type lodging in rice (Oryza sativa L.). Euphytica 2014, 198, 353–367. [Google Scholar] [CrossRef]
- Mulsanti, I.W.; Yamamoto, T.; Ueda, T.; Samadi, A.F.; Kamahora, E.; Rumanti, I.A.; Thanh, V.C.; Adachi, S.; Suzuki, S.; Kanekatsu, M. Finding the superior allele of japonica-type for increasing stem lodging resistance in indica rice varieties using chromosome segment substitution lines. Rice 2018, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Feng, S.; Ding, W.; Li, G. Influence of speed and rainfall on large-scale wheat lodging from 2007 to 2014 in China. PLoS ONE 2016, 11, e0157677. [Google Scholar] [CrossRef]
- Setter, T.; Laureles, E.; Mazaredo, A. Lodging reduces yield of rice by self-shading and reductions in canopy photosynthesis. Field Crops Res. 1997, 49, 95–106. [Google Scholar] [CrossRef]
- Samadi, A.F.; Suzuki, H.; Ueda, T.; Yamamoto, T.; Adachi, S.; Ookawa, T. Identification of quantitative trait loci for breaking and bending types lodging resistance in rice, using recombinant inbred lines derived from Koshihikari and a strong culm variety, Leaf Star. Plant Growth Regul. 2019, 89, 83–98. [Google Scholar] [CrossRef]
- Berry, P.; Berry, S. Understanding the genetic control of lodging-associated plant characters in winter wheat (Triticum aestivum L.). Euphytica 2015, 205, 671–689. [Google Scholar] [CrossRef]
- Ookawa, T.; Aoba, R.; Yamamoto, T.; Ueda, T.; Takai, T.; Fukuoka, S.; Ando, T.; Adachi, S.; Matsuoka, M.; Ebitani, T. Precise estimation of genomic regions controlling lodging resistance using a set of reciprocal chromosome segment substitution lines in rice. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Upadhya, D.; Dhakal, R.; Khadka, K.; Rana, S.; Acharya, P.; Rana, R.; Chaudhary, P. Local knowledge on climate-induced traits in rice for improving crop yield, food security and climate resilience. Int. Agric. Innov. Res. J. 2016, 5, 385–396. [Google Scholar]
- Ookawa, T.; Hobo, T.; Yano, M.; Murata, K.; Ando, T.; Miura, H.; Asano, K.; Ochiai, Y.; Ikeda, M.; Nishitani, R. New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 2010, 1, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.; Singh, U.M.; Naik, S.M.; Venkateshwarlu, C.; Ramayya, P.J.; Raman, K.A.; Sandhu, N.; Kumar, A. Molecular mapping of QTLs associated with lodging resistance in dry direct-seeded rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 1431. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Togawa, E.; Hirotsu, N.; Ishimaru, K. Improvement of lodging resistance with QTLs for stem diameter in rice (Oryza sativa L.). Theor. Appl. Genet. 2008, 117, 749–757. [Google Scholar] [CrossRef]
- Kong, E.; Liu, D.; Guo, X.; Yang, W.; Sun, J.; Li, X.; Zhan, K.; Cui, D.; Lin, J.; Zhang, A. Anatomical and chemical characteristics associated with lodging resistance in wheat. Crop J. 2013, 1, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.; Kim, K.; Manigbas, N.L.; Yi, G.; Sohn, J. Identification of quantitative trait loci for resistance to white-backed planthopper (Sogatella furcifera) in rice with Milyang 46 (Cheongcheongbyeo) background. Philipp. J. Crop Sci. 2013, 38, 30–36. [Google Scholar]
- Lee, G.-H.; Yun, B.-W.; Kim, K.-M. Analysis of QTLs associated with the rice quality related gene by double haploid populations. Int. J. Genom. 2014, 2014, 781832. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chung, I.K.; Kim, H.Y.; Kim, K.-M. Plant development of new ecological model related to yield using QTL analysis. Euphytica 2018, 214, 1–11. [Google Scholar] [CrossRef]
- Kim, Y.; Choi, M.; Lee, K.; Baek, M.; Ku, B.; Kang, S.; Park, H.; Kim, B. Growth and yield of rice in levels of nitrogen and water management of reclaimed saline soil in southwestern area. Korean J. Crop Sci./Hanguk Jakmul Hakhoe Chi 2012, 57, 203–208. [Google Scholar] [CrossRef]
- Yoshida, S. Fundamentals of Rice Crop Science; International Rice Research Institute: Los Baños, Philippines, 1981. [Google Scholar]
- Lincoln, S.E.; Daly, M.J.; Lander, E.S. Constructing genetic linkage maps with MAPMAKER/EXP Version 3.0: A tutorial and reference manual. Whitehead Inst. Biomed. Res. Tech. Rep. 1993, 3, 6–40. [Google Scholar]
- Yun, B.-W.; Kim, M.-G.; Handoyo, T.; Kim, K.-M. Analysis of rice grain quality-associated quantitative trait loci by using genetic mapping. Am. J. Plant Sci. 2014, 2014, 1125. [Google Scholar] [CrossRef] [Green Version]
- McCough, S.R.; Doerge, R.W. QTL mapping in rice. Trends Genet. 1995, 11, 482–487. [Google Scholar] [CrossRef]
- Zeng, Z.-B. Precision mapping of quantitative trait loci. Genetics 1994, 136, 1457–1468. [Google Scholar] [CrossRef]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.-C.; Iwamoto, M.; Abe, T. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Sato, Y.; Takehisa, H.; Kamatsuki, K.; Minami, H.; Namiki, N.; Ikawa, H.; Ohyanagi, H.; Sugimoto, K.; Antonio, B.A.; Nagamura, Y. RiceXPro version 3.0: Expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013, 41, D1206–D1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007, 35, D61–D65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T. BioEdit Version 7.0.0. Distributed by the Author. 2004. Available online: www.mbio.ncsu.edu/BioEdit/bioedit.html (accessed on 27 December 2020).
- Park, J.-R.; Yang, W.-T.; Kim, D.-H.; Kim, K.-M. Identification of a Novel Gene, Osbht, in Response to High Temperature Tolerance at Booting Stage in Rice. Int. J. Mol. Sci. 2020, 21, 5862. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-F.; Shen, L.-S.; Tan, Z.; Xu, Y.; He, P.; Chen, Y.; Zhu, L. Comparative mapping of QTLs for agronomic traits of rice across environments using a doubled haploid population. Theor. Appl. Genet. 1996, 93, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Mun, B.-G.; Imran, Q.M.; Lee, S.-U.; Adamu, T.A.; Shahid, M.; Kim, K.-M.; Yun, B.-W. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebbs, S.D.; Kochian, L.V. Phytoextraction of zinc by oat (Avena sativa), barley (Hordeum vulgare), and Indian mustard (Brassica juncea). Environ. Sci. Technol. 1998, 32, 802–806. [Google Scholar] [CrossRef]
- Noor, R.B.M.; Caviness, C. Influence of Induced Lodging on Pod Distribution and Seed Yield in Soybeans 1. Agron. J. 1980, 72, 904–906. [Google Scholar] [CrossRef]
- Evans, J.R. Nitrogen and photosynthesis in the flag leaf of wheat (Triticum aestivum L.). Plant Physiol. 1983, 72, 297–302. [Google Scholar] [CrossRef] [Green Version]
- Kashiwagi, T.; Sasaki, H.; Ishimaru, K. Factors responsible for decreasing sturdiness of the lower part in lodging of rice (Oryza sativa L.). Plant Prod. Sci. 2005, 8, 166–172. [Google Scholar] [CrossRef]
- Weber, C.; Fehr, W. Seed yield losses from lodging and combine harvesting in soybeans 1. Agron. J. 1966, 58, 287–289. [Google Scholar] [CrossRef]
- Lang, Y.-Z.; Yang, X.-D.; Wang, M.-E.; Zhu, Q.-S. Effects of lodging at different filling stages on rice yield and grain quality. Rice Sci. 2012, 19, 315–319. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Yang, L.; Li, P.; Zhu, J.; Kazuhiko, K.; Wang, Y. Ozone stress increases lodging risk of rice cultivar Liangyoupeijiu: A FACE study. Shengtai Xuebao/Acta Ecol. Sin. 2011, 31, 6098–6107. [Google Scholar]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, S.C.; Sayre, K.; Kaul, J.; Narang, R. Growth and morphology of spring wheat (Triticum aestivum L.) culms and their association with lodging: Effects of genotypes, N levels and ethephon. Field Crops Res. 2003, 84, 271–290. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Ann. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Kawasaki, T.; Koita, H.; Nakatsubo, T.; Hasegawa, K.; Wakabayashi, K.; Takahashi, H.; Umemura, K.; Umezawa, T.; Shimamoto, K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 230–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, D.; Chen, X.; Yin, Y.; Lu, K.; Yang, W.; Tang, Y.; Wang, Z. Lodging resistance of winter wheat (Triticum aestivum L.): Lignin accumulation and its related enzymes activities due to the application of paclobutrazol or gibberellin acid. Field Crops Res. 2014, 157, 1–7. [Google Scholar] [CrossRef]
- Ookawa, T.; Inoue, K.; Matsuoka, M.; Ebitani, T.; Takarada, T.; Yamamoto, T.; Ueda, T.; Yokoyama, T.; Sugiyama, C.; Nakaba, S. Increased lodging resistance in long-culm, low-lignin gh2 rice for improved feed and bioenergy production. Sci. Rep. 2014, 4, 6567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lüderitz, T.; Grisebach, H. Enzymic Synthesis of Lignin Precursors Comparison of Cinnamoyl-CoA Reductase and Cinnamyl Alcohol: NADP+ Dehydrogenase from Spruce (Picea abies L.) and Soybean (Glycine max L.). Eur. J. Biochem. 1981, 119, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Zhou, R.; Louie, G.V.; Mühlemann, J.K.; Bomati, E.K.; Bowman, M.E.; Dudareva, N.; Dixon, R.A.; Noel, J.P.; Wang, X. Structural studies of cinnamoyl-CoA reductase and cinnamyl-alcohol dehydrogenase, key enzymes of monolignol biosynthesis. Plant Cell 2014, 26, 3709–3727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, A.; Tobimatsu, Y.; Goeminne, G.; Phillips, L.; Flint, H.; Steward, D.; Torr, K.; Donaldson, L.; Boerjan, W.; Ralph, J. Suppression of CCR impacts metabolite profile and cell wall composition in Pinus radiata tracheary elements. Plant Mol. Biol. 2013, 81, 105–117. [Google Scholar] [CrossRef]
- Zhou, R.; Jackson, L.; Shadle, G.; Nakashima, J.; Temple, S.; Chen, F.; Dixon, R.A. Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc. Natl. Acad. Sci. USA 2010, 107, 17803–17808. [Google Scholar] [CrossRef] [Green Version]
- Thévenin, J.; Pollet, B.; Letarnec, B.; Saulnier, L.; Gissot, L.; Maia-Grondard, A.; Lapierre, C.; Jouanin, L. The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Mol. Plant 2011, 4, 70–82. [Google Scholar] [CrossRef]
- Tu, Y.; Rochfort, S.; Liu, Z.; Ran, Y.; Griffith, M.; Badenhorst, P.; Louie, G.V.; Bowman, M.E.; Smith, K.F.; Noel, J.P. Functional analyses of caffeic acid O-methyltransferase and cinnamoyl-CoA-reductase genes from perennial ryegrass (Lolium perenne). Plant Cell 2010, 22, 3357–3373. [Google Scholar] [CrossRef] [Green Version]
- Liang, M.; Davis, E.; Gardner, D.; Cai, X.; Wu, Y. Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 2006, 224, 1185. [Google Scholar] [CrossRef]
- Vanholme, R.; Storme, V.; Vanholme, B.; Sundin, L.; Christensen, J.H.; Goeminne, G.; Halpin, C.; Rohde, A.; Morreel, K.; Boerjan, W. A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis. Plant Cell 2012, 24, 3506–3529. [Google Scholar] [CrossRef] [Green Version]
- Derikvand, M.M.; Sierra, J.B.; Ruel, K.; Pollet, B.; Do, C.-T.; Thévenin, J.; Buffard, D.; Jouanin, L.; Lapierre, C. Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 2008, 227, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Cass, C.L.; Mazaheri, M.; Sekhon, R.S.; Heckwolf, M.; Kaeppler, H.; de Leon, N.; Mansfield, S.D.; Kaeppler, S.M.; Sedbrook, J.C. Suppression of CINNAMOYL-CoA REDUCTASE increases the level of monolignol ferulates incorporated into maize lignins. Biotechnol. Biofuels 2017, 10, 109. [Google Scholar] [CrossRef]
- Piquemal, J.; Lapierre, C.; Myton, K.; O’connell, A.; Schuch, W.; Grima-Pettenati, J.; Boudet, A.-M. Down-regulation of cinnamoyl-CoA reductase induces significant changes of lignin profiles in transgenic tobacco plants. Plant J. 1998, 13, 71–83. [Google Scholar] [CrossRef]
- Chabannes, M.; Barakate, A.; Lapierre, C.; Marita, J.M.; Ralph, J.; Pean, M.; Danoun, S.; Halpin, C.; Grima-Pettenati, J.; Boudet, A.M. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 2001, 28, 257–270. [Google Scholar] [CrossRef]
- Giordano, A.; Liu, Z.; Panter, S.N.; Dimech, A.M.; Shang, Y.; Wijesinghe, H.; Fulgueras, K.; Ran, Y.; Mouradov, A.; Rochfort, S. Reduced lignin content and altered lignin composition in the warm season forage grass Paspalum dilatatum by down-regulation of a Cinnamoyl CoA reductase gene. Transgenic Res. 2014, 23, 503–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Li, J.; Yuan, L.; Tanksley, S. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor. Appl. Genet. 1996, 92, 230–244. [Google Scholar] [CrossRef]

| Year | Trait | PSLSA (kgf) | PSLSB (kgf) | Yield (kg/10a) |
|---|---|---|---|---|
| 2018 | PSLSA | 1.000 | ||
| PSLSB | 0.261 ** | 1.000 | ||
| Yield | 0.425 ** | 0.103 | 1.000 | |
| 2019 | PSLSA | 1.000 | ||
| PSLSB | 0.530 ** | 1.000 | ||
| Yield | 0.544 ** | 0.297 ** | 1.000 | |
| 2020 | PSLSA | 1.000 | ||
| PSLSB | 0.643 ** | 1.000 | ||
| Yield | 0.553 ** | 0.423 ** | 1.000 |
| Gene | No. of Genes |
|---|---|
| Cell function | 11 |
| Ribose-phosphate pyrophosphokinase 2 | |
| Similar to Alpha-expansin OsEXPA16 | |
| Similar to Cinnamoyl-CoA reductase | |
| Similar to 4-coumarate--CoA ligase 1 | |
| Similar to ANT (Ovule development protein aintegumenta) | |
| Similar to Auxin efflux carrier protein | |
| SBP domain-containing protein | |
| Similar to FtsH protease (VAR2) (Zinc-dependent protease) | |
| Similar to GRAB2 protein | |
| Polyubiquitin | |
| Glycosyl transferase, family 43 protein | |
| Abiotic stress | 4 |
| Similar to Thioredoxin peroxidase | |
| Phytosulfokines 1 precursor | |
| Amino acid/polyamine transporter II family protein | |
| Similar to EF-hand Ca2+-binding protein CCD1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, D.-D.; Son, J.H.; Lee, G.-S.; Kim, K.-M. Screening for a Novel Gene, OsPSLSq6, Using QTL Analysis for Lodging Resistance in Rice. Agronomy 2021, 11, 334. https://doi.org/10.3390/agronomy11020334
Zhao D-D, Son JH, Lee G-S, Kim K-M. Screening for a Novel Gene, OsPSLSq6, Using QTL Analysis for Lodging Resistance in Rice. Agronomy. 2021; 11(2):334. https://doi.org/10.3390/agronomy11020334
Chicago/Turabian StyleZhao, Dan-Dan, Ju Hyeong Son, Gang-Seob Lee, and Kyung-Min Kim. 2021. "Screening for a Novel Gene, OsPSLSq6, Using QTL Analysis for Lodging Resistance in Rice" Agronomy 11, no. 2: 334. https://doi.org/10.3390/agronomy11020334








