Characterization and Biomarker Analyses of Post-COVID-19 Complications and Neurological Manifestations
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. SARS-CoV-2 IgG Antibody Test
2.3. APOE4 Genotyping
2.4. Plasma Multiplex Cytokine Analysis and NFL by MSD Assays
2.5. nEV Isolation
2.6. Characterization of nEVs by NTA and electron microscopy
2.7. Neuronal-Enriched EV Protein Cargo Analyses
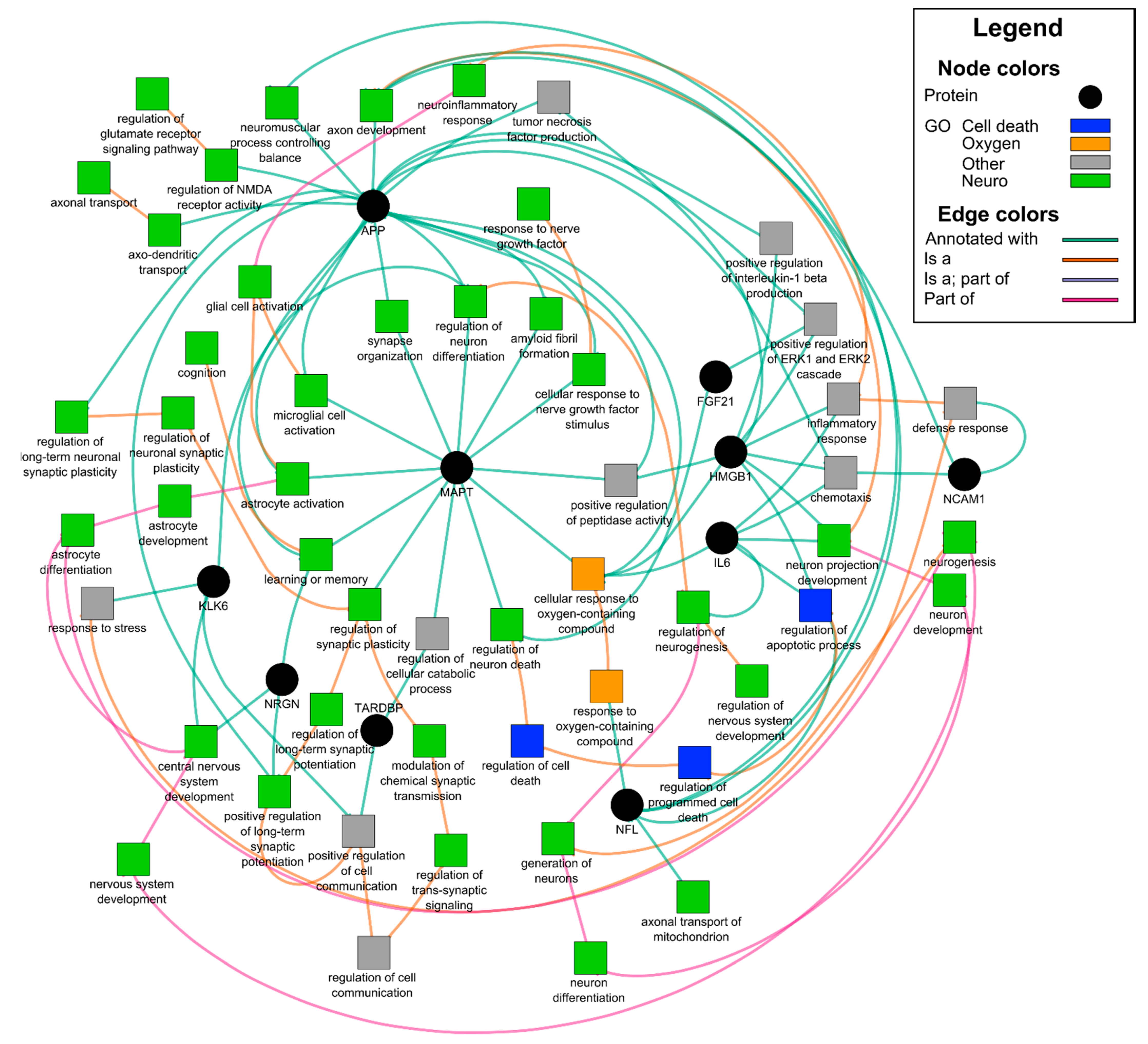
2.8. Statistics and Bioinformatics
3. Results
3.1. Participant Demographic and Clinical Data
3.2. Plasma Cytokines, NFL, and IgG Levels
3.3. Neuronal Enriched EV Characterization
3.4. Neuronal enriched EV protein cargo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paterson, R.W.; Brown, R.L.; Benjamin, L.; Nortley, R.; Wiethoff, S.; Bharucha, T.; Jayaseelan, D.L.; Kumar, G.; Raftopoulos, R.E.; Zambreanu, L.; et al. The emerging spectrum of COVID-19 neurology: Clinical, radiological and laboratory findings. Brain 2020, 143, 3104–3120. [Google Scholar] [CrossRef]
- Fotuhi, M.; Mian, A.; Meysami, S.; Raji, C.A. Neurobiology of COVID-19. J. Alzheimers Dis. 2020, 76, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Zubair, A.S.; McAlpine, L.S.; Gardin, T.; Farhadian, S.; Kuruvilla, D.E.; Spudich, S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: A review. JAMA Neurol. 2020, 77, 1018–1027. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef]
- Li, Y.C.; Bai, W.Z.; Hashikawa, T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020, 92, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Lazartigues, E. Expression of ACE2 in human neurons supports the neuro-invasive potential of COVID-19 virus. Cell. Mol. Neurobiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Espindola, O.M.; Brandao, C.O.; Gomes, Y.C.P.; Siqueira, M.; Soares, C.N.; Lima, M.; Leite, A.; Torezani, G.; Araujo, A.Q.C.; Silva, M.T.T. Cerebrospinal fluid findings in neurological diseases associated with COVID-19 and insights into mechanisms of disease development. Int. J. Infect. Dis. 2021, 102, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Z.; Chu, H.; Han, S.; Shuai, H.; Deng, J.; Hu, Y.F.; Gong, H.R.; Lee, A.C.; Zou, Z.; Yau, T.; et al. SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 2020, 30, 928–931. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Wong, S.Z.H.; Zhou, H.; Zhang, F.; Cubitt, B.; Chen, C.Z.; Xu, M.; Pradhan, M.; et al. Human pluripotent stem cell-derived neural cells and brain organoids reveal SARS-CoV-2 neurotropism. BioRxiv 2020. [Google Scholar] [CrossRef]
- Nordvig, A.S.; Rimmer, K.T.; Willey, J.Z.; Thakur, K.T.; Boehme, A.K.; Vargas, W.S.; Smith, C.J.; Elkind, M.S.V. Potential neurological manifestations of COVID-19. Neurology 2020. [Google Scholar] [CrossRef]
- Garrigues, E.; Janvier, P.; Kherabi, Y.; Le Bot, A.; Hamon, A.; Gouze, H.; Doucet, L.; Berkani, S.; Oliosi, E.; Mallart, E.; et al. Post-discharge persistent symptoms and health-related quality of life after hospitalization for COVID-19. J. Infect. 2020, 81, e4–e6. [Google Scholar] [CrossRef] [PubMed]
- Klok, F.A.; Boon, G.; Barco, S.; Endres, M.; Geelhoed, J.J.M.; Knauss, S.; Rezek, S.A.; Spruit, M.A.; Vehreschild, J.; Siegerink, B. The Post-COVID-19 Functional Status scale: A tool to measure functional status over time after COVID-19. Eur. Respir. J. 2020, 56. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; Assaf, G.S.; McCorkell, L.; Wei, H.; Low, R.J.; Re’em, Y.; Redfield, S.; Austin, J.P.; Akrami, A. Characterizing long COVID in an International Cohort: 7 months of symptoms and their impact. medRxiv 2020. [Google Scholar] [CrossRef]
- Gupta, A.; Pulliam, L. Exosomes as mediators of neuroinflammation. J. Neuroinflammation 2014, 11, 68. [Google Scholar] [CrossRef]
- Pulliam, L.; Gupta, A. Modulation of cellular function through immune-activated exosomes. DNA Cell Biol. 2015, 34, 459–463. [Google Scholar] [CrossRef]
- Dalvi, P.; Sun, B.; Tang, N.; Pulliam, L. Immune activated monocyte exosomes alter microRNAs in brain endothelial cells and initiate an inflammatory response through the TLR4/MyD88 pathway. Sci. Rep. 2017, 7, 9954. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Sun, B.; Gupta, A.; Rempel, H.; Pulliam, L. Monocyte exosomes induce adhesion molecules and cytokines via activation of NF-kappaB in endothelial cells. FASEB J. 2016, 30, 3097–3106. [Google Scholar] [CrossRef] [PubMed]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimers Dement. 2015, 11, 600–607 e601. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Carlson, O.D.; Mustapic, M.; Kapogiannis, D. Low neural exosomal levels of cellular survival factors in Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 769–773. [Google Scholar] [CrossRef]
- Sun, B.; Dalvi, P.; Abadjian, L.; Tang, N.; Pulliam, L. Blood neuron-derived exosomes as biomarkers of cognitive impairment in HIV. AIDS 2017, 31, F9–F17. [Google Scholar] [CrossRef]
- Sun, B.; Fernandes, N.; Pulliam, L. Profile of neuronal exosomes in HIV cognitive impairment exposes sex differences. AIDS 2019, 33, 1683–1692. [Google Scholar] [CrossRef]
- Winston, C.N.; Goetzl, E.J.; Akers, J.C.; Carter, B.S.; Rockenstein, E.M.; Galasko, D.; Masliah, E.; Rissman, R.A. Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimers Dement. 2016, 3, 63–72. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Ledreux, A.; Granholm, A.C.; Elahi, F.M.; Goetzl, L.; Hiramoto, J.; Kapogiannis, D. Neuron-derived exosome proteins may contribute to progression from repetitive mild traumatic brain injuries to chronic traumatic encephalopathy. Front. Neurosci. 2019, 13, 452. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Peltz, C.B.; Mustapic, M.; Kapogiannis, D.; Yaffe, K. Neuron-derived plasma exosome proteins after remote traumatic brain injury. J. Neurotrauma 2020, 37, 382–388. [Google Scholar] [CrossRef]
- Winston, C.N.; Romero, H.K.; Ellisman, M.; Nauss, S.; Julovich, D.A.; Conger, T.; Hall, J.R.; Campana, W.; O’Bryant, S.E.; Nievergelt, C.M.; et al. Assessing neuronal and astrocyte derived exosomes from individuals with mild traumatic brain injury for markers of neurodegeneration and cytotoxic activity. Front. Neurosci. 2019, 13, 1005. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Abner, E.L.; Jicha, G.A.; Kapogiannis, D.; Schwartz, J.B. Declining levels of functionally specialized synaptic proteins in plasma neuronal exosomes with progression of Alzheimer’s disease. FASEB J. 2018, 32, 888–893. [Google Scholar] [CrossRef]
- Kapogiannis, D.; Mustapic, M.; Shardell, M.D.; Berkowitz, S.T.; Diehl, T.C.; Spangler, R.D.; Tran, J.; Lazaropoulos, M.P.; Chawla, S.; Gulyani, S.; et al. Association of extracellular vesicle biomarkers with alzheimer disease in the baltimore longitudinal study of aging. JAMA Neurol. 2019. [Google Scholar] [CrossRef]
- Pulliam, L.; Sun, B.; Mustapic, M.; Chawla, S.; Kapogiannis, D. Plasma neuronal exosomes serve as biomarkers of cognitive impairment in HIV infection and Alzheimer’s disease. J. Neurovirology 2019, 25, 702–709. [Google Scholar] [CrossRef] [PubMed]
- LIINC. Long-term Impact of Infection with Novel Coronavirus (LIINC). Available online: https://www.liincstudy.org/en/study-information (accessed on 4 February 2021).
- Rabin, R.; de Charro, F. EQ-5D: A measure of health status from the EuroQol Group. Ann. Med. 2001, 33, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, R183. [Google Scholar] [CrossRef] [PubMed]
- Pomaznoy, M.; Ha, B.; Peters, B. GOnet: A tool for interactive Gene Ontology analysis. BMC Bioinform. 2018, 19, 470. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Aureli, A.; Sebastiani, P.; Del Beato, T.; Marimpietri, A.E.; Graziani, A.; Sechi, E.; Di Loreto, S. Involvement of IL-6 and IL-1 receptor antagonist on intellectual disability. Immunol. Lett. 2014, 162, 124–131. [Google Scholar] [CrossRef]
- Bettcher, B.M.; Watson, C.L.; Walsh, C.M.; Lobach, I.V.; Neuhaus, J.; Miller, J.W.; Green, R.; Patel, N.; Dutt, S.; Busovaca, E.; et al. Interleukin-6, age, and corpus callosum integrity. PLoS ONE 2014, 9, e106521. [Google Scholar] [CrossRef]
- Ferrucci, L.; Harris, T.B.; Guralnik, J.M.; Tracy, R.P.; Corti, M.C.; Cohen, H.J.; Penninx, B.; Pahor, M.; Wallace, R.; Havlik, R.J. Serum IL-6 level and the development of disability in older persons. J. Am. Geriatr. Soc. 1999, 47, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Antoine, D.J.; Andersson, U.; Tracey, K.J. The many faces of HMGB1: Molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J. Leukoc. Biol. 2013, 93, 865–873. [Google Scholar] [CrossRef]
- Mella, C.; Figueroa, C.D.; Otth, C.; Ehrenfeld, P. Involvement of kallikrein-related peptidases in nervous system disorders. Front. Cell. Neurosci. 2020, 14, 166. [Google Scholar] [CrossRef]
- Vogt, M.A.; Ehsaei, Z.; Knuckles, P.; Higginbottom, A.; Helmbrecht, M.S.; Kunath, T.; Eggan, K.; Williams, L.A.; Shaw, P.J.; Wurst, W.; et al. TDP-43 induces p53-mediated cell death of cortical progenitors and immature neurons. Sci. Rep. 2018, 8, 8097. [Google Scholar] [CrossRef]
- Restelli, L.M.; Oettinghaus, B.; Halliday, M.; Agca, C.; Licci, M.; Sironi, L.; Savoia, C.; Hench, J.; Tolnay, M.; Neutzner, A.; et al. Neuronal mitochondrial dysfunction activates the integrated stress response to induce fibroblast growth factor 21. Cell Rep. 2018, 24, 1407–1414. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, C.; Li, J.; Yuan, J.; Wei, J.; Huang, F.; Wang, F.; Li, G.; Li, Y.; Xing, L.; et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin. Immunol. 2020, 146, 119e4–127e4 e114. [Google Scholar] [CrossRef]
- Gadani, S.P.; Cronk, J.C.; Norris, G.T.; Kipnis, J. IL-4 in the brain: A cytokine to remember. J. Immunol. 2012, 189, 4213–4219. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, A.; Salvati, L.; Maggi, L.; Capone, M.; Vanni, A.; Spinicci, M.; Mencarini, J.; Caporale, R.; Peruzzi, B.; Antonelli, A.; et al. Impaired immune cell cytotoxicity in severe COVID-19 is IL-6 dependent. J. Clin. Investig. 2020, 130, 4694–4703. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef] [PubMed]
- Gisslen, M.; Price, R.W.; Andreasson, U.; Norgren, N.; Nilsson, S.; Hagberg, L.; Fuchs, D.; Spudich, S.; Blennow, K.; Zetterberg, H. Plasma concentration of the neurofilament light protein (NFL) is a biomarker of CNS injury in HIV infection: A Cross-sectional study. EBioMedicine 2016, 3, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Li, C.H.; Yang, K.C.; Lin, F.J.; Wu, C.C.; Chieh, J.J.; Chiu, M.J. Blood NfL: A biomarker for disease severity and progression in Parkinson disease. Neurology 2019, 93, e1104–e1111. [Google Scholar] [CrossRef]
- Preische, O.; Schultz, S.A.; Apel, A.; Kuhle, J.; Kaeser, S.A.; Barro, C.; Graber, S.; Kuder-Buletta, E.; LaFougere, C.; Laske, C.; et al. Serum neurofilament dynamics predicts neurodegeneration and clinical progression in presymptomatic Alzheimer’s disease. Nat. Med. 2019, 25, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, M.; Garcia, G., Jr.; Tian, E.; Cui, Q.; Chen, X.; Sun, G.; Wang, J.; Arumugaswami, V.; Shi, Y. ApoE-isoform-dependent SARS-CoV-2 neurotropism and cellular response. Cell Stem Cell 2021, 28, 331–342. [Google Scholar] [CrossRef]
- Kuo, C.L.; Pilling, L.C.; Atkins, J.L.; Masoli, J.A.H.; Delgado, J.; Kuchel, G.A.; Melzer, D. APOE e4 genotype predicts severe COVID-19 in the UK biobank community cohort. J. Gerontol. 2020, 75, 2231–2232. [Google Scholar] [CrossRef]
- Sharma, P.; Mesci, P.; Carromeu, C.; McClatchy, D.R.; Schiapparelli, L.; Yates, J.R., 3rd; Muotri, A.R.; Cline, H.T. Exosomes regulate neurogenesis and circuit assembly. Proc. Nat. Acad. Sci. USA 2019, 116, 16086–16094. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Nat. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef]
- Budnik, V.; Ruiz-Canada, C.; Wendler, F. Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 2016, 17, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Olsson, B.; Lautner, R.; Andreasson, U.; Ohrfelt, A.; Portelius, E.; Bjerke, M.; Holtta, M.; Rosen, C.; Olsson, C.; Strobel, G.; et al. CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: A systematic review and meta-analysis. Lancet Neurol. 2016, 15, 673–684. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Elahi, F.M.; Mustapic, M.; Kapogiannis, D.; Pryhoda, M.; Gilmore, A.; Gorgens, K.A.; Davidson, B.; Granholm, A.C.; Ledreux, A. Altered levels of plasma neuron-derived exosomes and their cargo proteins characterize acute and chronic mild traumatic brain injury. FASEB J. 2019, 33, 5082–5088. [Google Scholar] [CrossRef]
- Shi, M.; Kovac, A.; Korff, A.; Cook, T.J.; Ginghina, C.; Bullock, K.M.; Yang, L.; Stewart, T.; Zheng, D.; Aro, P.; et al. CNS tau efflux via exosomes is likely increased in Parkinson’s disease but not in Alzheimer’s disease. Alzheimers Dement. 2016, 12, 1125–1131. [Google Scholar] [CrossRef] [PubMed]
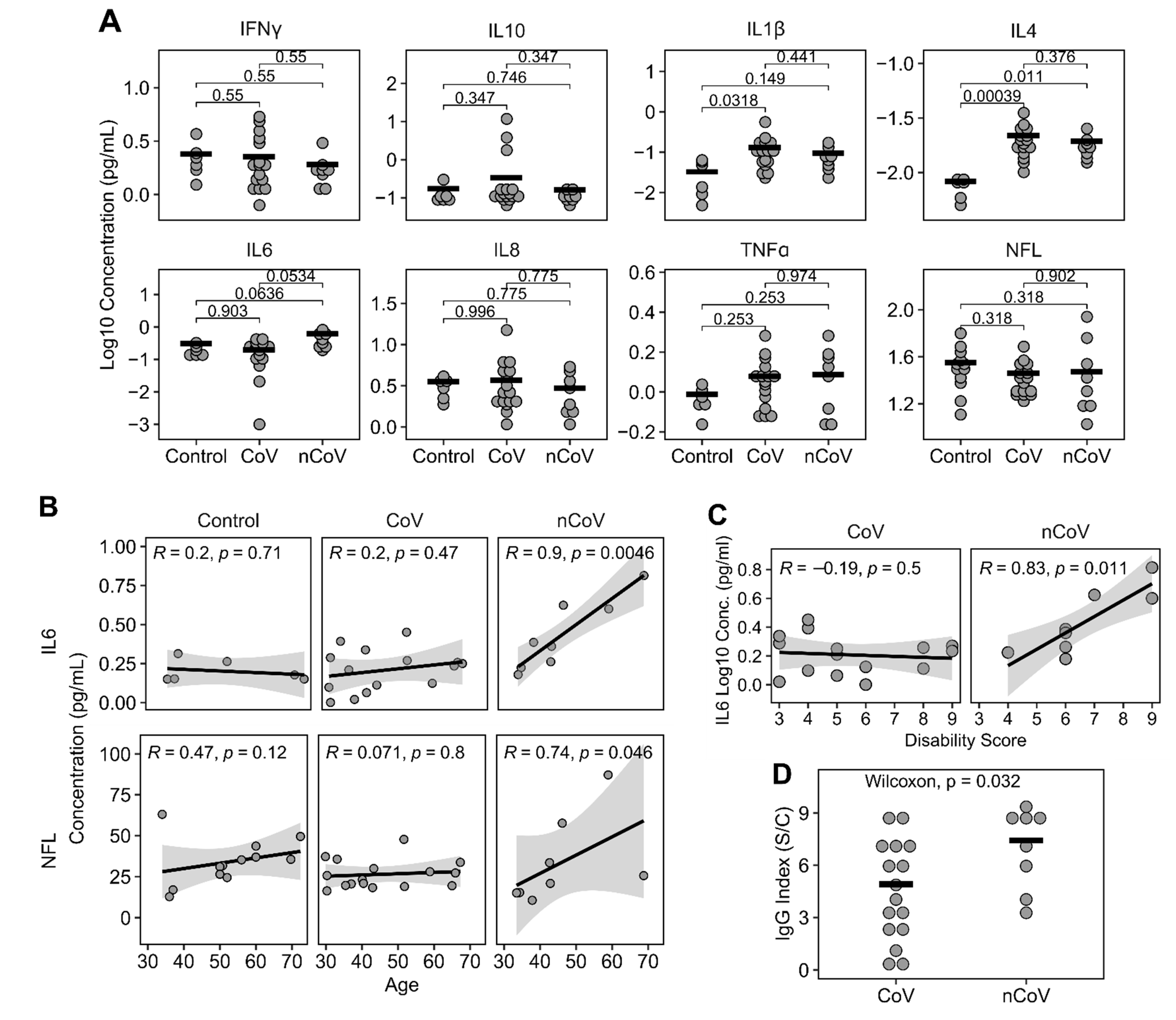
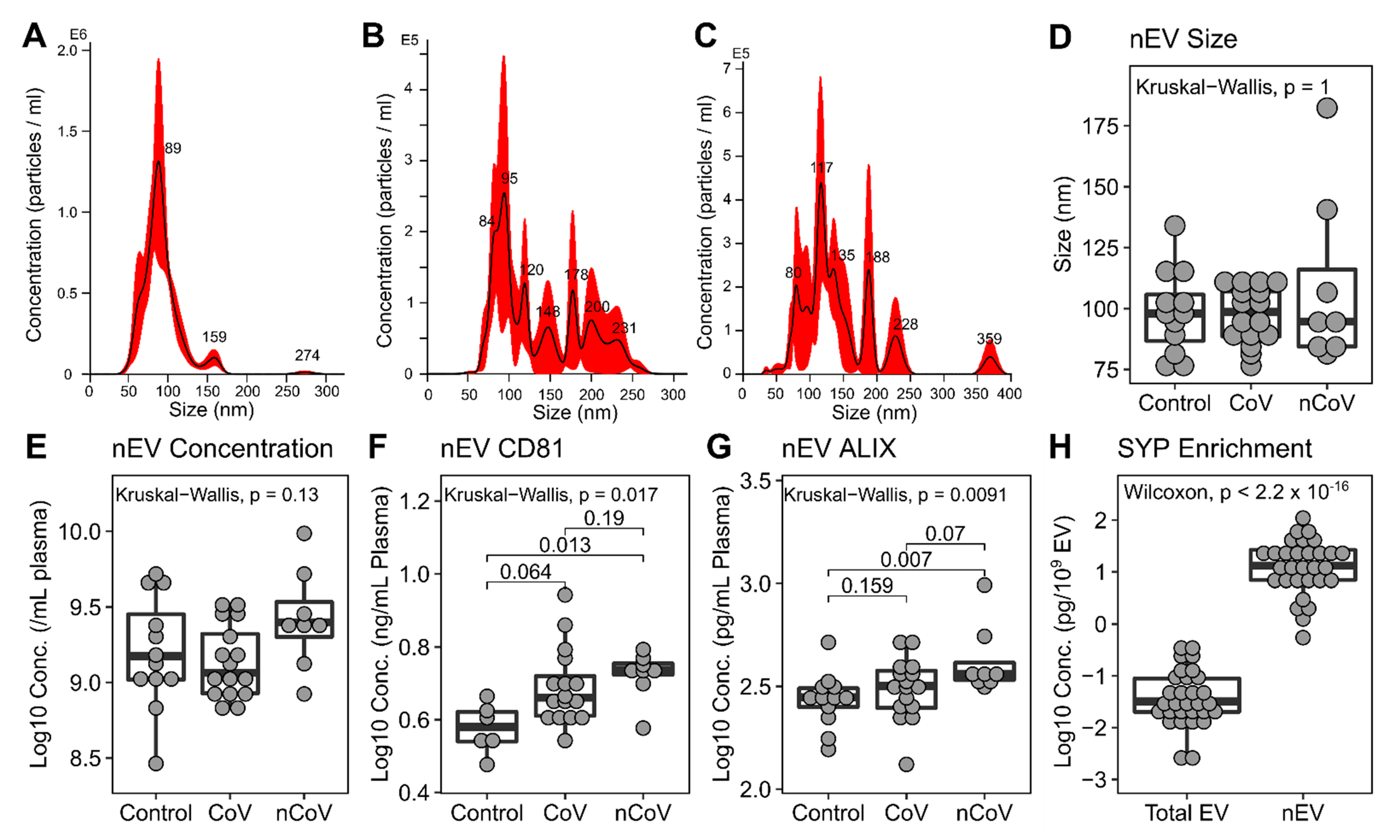
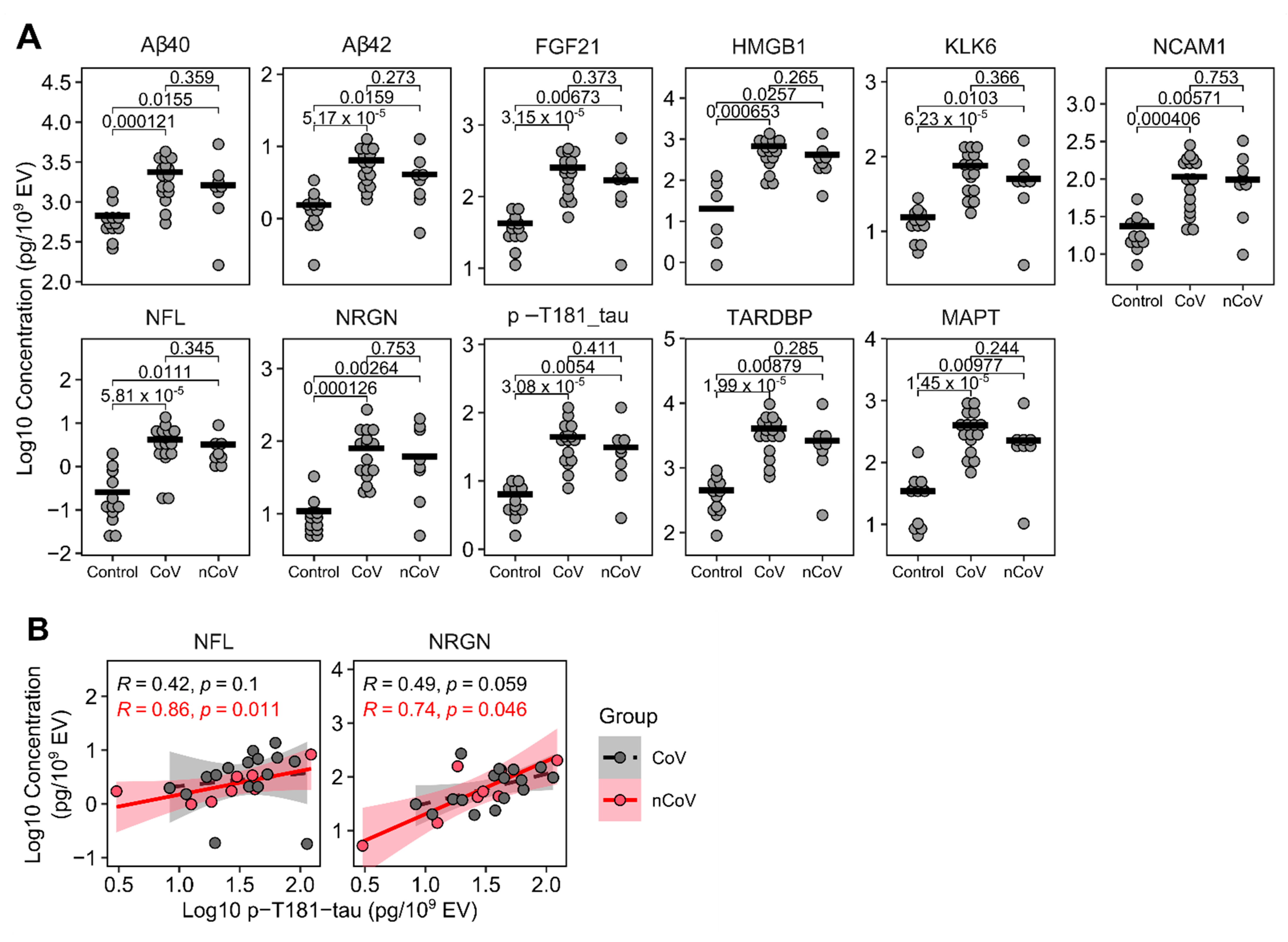
| Control (%) † | CoV (%) | nCoV (%) | p-Value | |||||
|---|---|---|---|---|---|---|---|---|
| Sex | Female (N = 22) | 4 | (33.3) | 12 | (75.0) | 6 | (75.0) | 0.053 |
| Male (N = 14) | 8 | (66.7) | 4 | (25.0) | 2 | (25.0) | ||
| Race | Asian | 2 | (16.7) | 0 | (0.0) | 1 | (12.5) | 0.154 |
| Black | 4 | (33.3) | 2 | (12.5) | 1 | (12.5) | ||
| White | 6 | (50.0) | 11 | (68.8) | 5 | (62.5) | ||
| American Indian | 0 | (0.0) | 0 | (0.0) | 1 | (12.5) | ||
| Undisclosed | 0 | (0.0) | 3 | (18.8) | 0 | (0.0) | ||
| Ethnicity | Hispanic | 1 | (8.3) | 3 | (18.8) | 4 | (50.0) | 0.081 |
| Non-Hispanic | 11 | (91.7) | 13 | (81.3) | 4 | (50.0) | ||
| APOE genotype | ε2, ε3 | NA | NA | 2 | (12.5) | 0 | (0.0) | 0.615 |
| ε3, ε3 | NA | NA | 9 | (56.3) | 6 | (75.0) | ||
| ε3, ε4 | NA | NA | 4 | (25.0) | 2 | (25.0) | ||
| ε4, ε4 | NA | NA | 1 | (6.3) | 0 | (0.0) | ||
| Comorbidity | NA | NA | 7 | (43.8) | 8 | (100) | 0.0095 | |
| Days till visit, Mean (SD) | NA | 56.6 (20.3) | 76.5 (27.2) | 0.094 | ||||
| Age in years, Mean (SD) | 52.3 (12.4) | 45.2 (13.2) | 45.6 (12.3) | 0.316 | ||||
| ID | Age | Sex | Race/Ethnicity | Mobility Issues ‡ | Selfcare Issues ‡ | Activity Issues ‡ | Hospitalized | Days Till visit § | IgG | ApoE Genotype | Neuro Symptoms | Comorbidity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 34 | F | AI/L | 2 | 1 | 1 | No | 36 | 4.2 | ε3,ε3 | DV | LD |
| 2 † | 69 | M | W | 3 | 3 | 3 | No | 62 | 8.3 | ε3,ε3 | M/C | HT,LD,O |
| 3 | 38 | F | Asian | 2 | 2 | 2 | Yes | 103 | 8.6 | ε3,ε3 | H | AID, DM,O |
| 4 † | 59 | M | W | 3 | 3 | 3 | Yes | 80 | 7.5 | ε3,ε3 | M/C | HT,LD |
| 5 | 33 | F | W/L | 2 | 1 | 3 | No | 97 | 3.3 | ε3,ε3 | M/C | HT |
| 6 | 43 | F | B/L | 2 | 1 | 3 | Yes | 99 | 9.4 | ε3,ε4 | M/C | HT,LD,O |
| 7 | 46 | F | W/L | 2 | 2 | 3 | Yes | 95 | 5.9 | ε3,ε4 | M/C | DM,O |
| 8 | 43 | F | W | 2 | 2 | 2 | No | 40 | 9.1 | ε3,ε3 | M/C | O |
| 9 | 30 | F | W | 1 | 2 | 3 | No | 38 | 2 | ε2,ε3 | None | |
| 10 | 37 | F | W | 1 | 1 | 1 | No | 30 | 0.4 | ε3,ε3 | None | |
| 11 | 30 | F | W | 1 | 1 | 1 | No | 31 | 3.9 | ε3,ε3 | None | |
| 12 | 43 | F | W | 3 | 2 | 3 | No | 52 | 1.1 | ε3,ε4 | None | |
| 13 | 35 | F | W | 2 | 1 | 2 | No | 41 | 8.8 | ε3,ε4 | None | |
| 14 | 30 | F | W | 2 | 1 | 1 | No | 44 | 6.1 | ε3,ε4 | None | |
| 15 | 66 | F | W | 3 | 2 | 3 | No | 49 | 8.4 | ε3,ε3 | None | LD |
| 16 | 40 | F | B | 1 | 1 | 1 | No | 38 | 4.9 | ε3,ε4 | None | O |
| 17 | 33 | F | W | 1 | 2 | 1 | No | 60 | 3.5 | ε3,ε3 | None | O |
| 18 | 67 | M | W | 2 | 1 | 2 | No | 64 | 2.6 | ε4,ε4 | None | O |
| 19 | 43 | F | B | 1 | 3 | 2 | No | 70 | 7.3 | ε3,ε3 | None | HT |
| 20 | 59 | F | W | 2 | 2 | 2 | No | 60 | 0.3 | ε2,ε3 | None | |
| 21 | 40 | F | W | 2 | 1 | 2 | No | 60 | 7.2 | ε3,ε3 | None | |
| 22 | 52 | M | Dc/L | 3 | 3 | 3 | Yes | 88 | 5.8 | ε3,ε3 | None | DM |
| 23 | 65 | M | Dc/L | 3 | 3 | 3 | Yes | 84 | 6.7 | ε3,ε3 | None | DM,O |
| 24 | 52 | M | Dc/L | 2 | 1 | 1 | Yes | 97 | 4.2 | ε3,ε3 | None |
| Term | p Value | Genes |
|---|---|---|
| Gene Ontology | ||
| GO:0030182~neuron differentiation | 0.0002 | APP, IL6, NCAM1, MAPT, HMGB1, KLK6 |
| GO:0050803~regulation of synapse structure or activity | 0.0002 | APP, NCAM1, SYP, NRGN |
| GO:0007399~nervous system development | 0.0003 | APP, IL6, NCAM1, MAPT, HMGB1, NRGN, KLK6 |
| GO:0070997~neuron death | 0.0003 | APP, IL6, FGF21, KLK6 |
| GO:0022008~neurogenesis | 0.0004 | APP, IL6, NCAM1, MAPT, HMGB1, KLK6 |
| GO:0031175~neuron projection development | 0.0006 | APP, IL6, NCAM1, MAPT, HMGB1 |
| GO:0008219~cell death | 0.0018 | APP, IL6, HMGB1, TARDBP, FGF21, KLK6 |
| GO:0048167~regulation of synaptic plasticity | 0.0024 | NCAM1, SYP, NRGN |
| GO:0006935~chemotaxis | 0.0026 | APP, IL6, NCAM1, HMGB1 |
| GO:0030424~axon | 0.0050 | APP, MAPT, NRGN |
| Uniprotein: Neurodegeneration | 0.0068 | APP, MAPT, TARDBP |
| GO:0006915~apoptotic process | 0.0096 | APP, IL6, HMGB1, TARDBP, FGF21 |
| GO:0048812~neuron projection morphogenesis | 0.0323 | APP, NCAM1, MAPT |
| GO:0099536~synaptic signaling | 0.0393 | NCAM1, SYP, NRGN |
| Genetic Association Database | ||
| Dementia | 0.0009 | APP, IL6, MAPT |
| Alzheimer’s Disease | 0.0013 | APP, IL6, MAPT, TARDBP |
| Schizophrenia | 0.0141 | IL6, NCAM1, MAPT, NRGN |
| Neuroinflammation Signaling Pathway † | 0.0000 | APP, IL6, HMGB1, MAPT |
| Neuroprotective Role of THOP1 in Alzheimer’s Disease † | 0.0000 | APP, KLK6, MAPT |
| Transcription factor binding site | ||
| NFKB | 0.0298 | APP, IL6, NCAM1, SYP, MAPT, TARDBP, NRGN, KLK6 |
| STAT3 | 0.0217 | APP, NCAM1, SYP, MAPT, TARDBP, FGF21, NRGN, KLK6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, B.; Tang, N.; Peluso, M.J.; Iyer, N.S.; Torres, L.; Donatelli, J.L.; Munter, S.E.; Nixon, C.C.; Rutishauser, R.L.; Rodriguez-Barraquer, I.; et al. Characterization and Biomarker Analyses of Post-COVID-19 Complications and Neurological Manifestations. Cells 2021, 10, 386. https://doi.org/10.3390/cells10020386
Sun B, Tang N, Peluso MJ, Iyer NS, Torres L, Donatelli JL, Munter SE, Nixon CC, Rutishauser RL, Rodriguez-Barraquer I, et al. Characterization and Biomarker Analyses of Post-COVID-19 Complications and Neurological Manifestations. Cells. 2021; 10(2):386. https://doi.org/10.3390/cells10020386
Chicago/Turabian StyleSun, Bing, Norina Tang, Michael J. Peluso, Nikita S. Iyer, Leonel Torres, Joanna L. Donatelli, Sadie E. Munter, Christopher C. Nixon, Rachel L. Rutishauser, Isabel Rodriguez-Barraquer, and et al. 2021. "Characterization and Biomarker Analyses of Post-COVID-19 Complications and Neurological Manifestations" Cells 10, no. 2: 386. https://doi.org/10.3390/cells10020386
APA StyleSun, B., Tang, N., Peluso, M. J., Iyer, N. S., Torres, L., Donatelli, J. L., Munter, S. E., Nixon, C. C., Rutishauser, R. L., Rodriguez-Barraquer, I., Greenhouse, B., Kelly, J. D., Martin, J. N., Deeks, S. G., Henrich, T. J., & Pulliam, L. (2021). Characterization and Biomarker Analyses of Post-COVID-19 Complications and Neurological Manifestations. Cells, 10(2), 386. https://doi.org/10.3390/cells10020386






