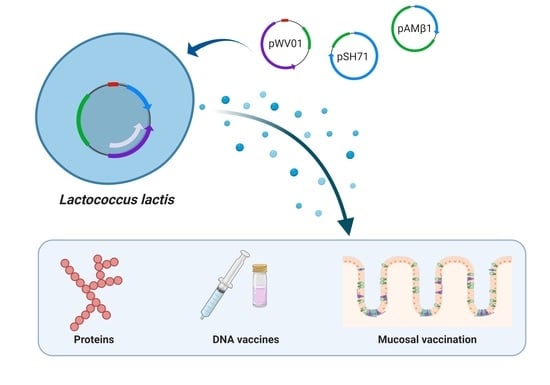
Plasmid Replicons for the Production of Pharmaceutical-Grade pDNA, Proteins and Antigens by Lactococcus lactis Cell Factories
Abstract
1. Introduction
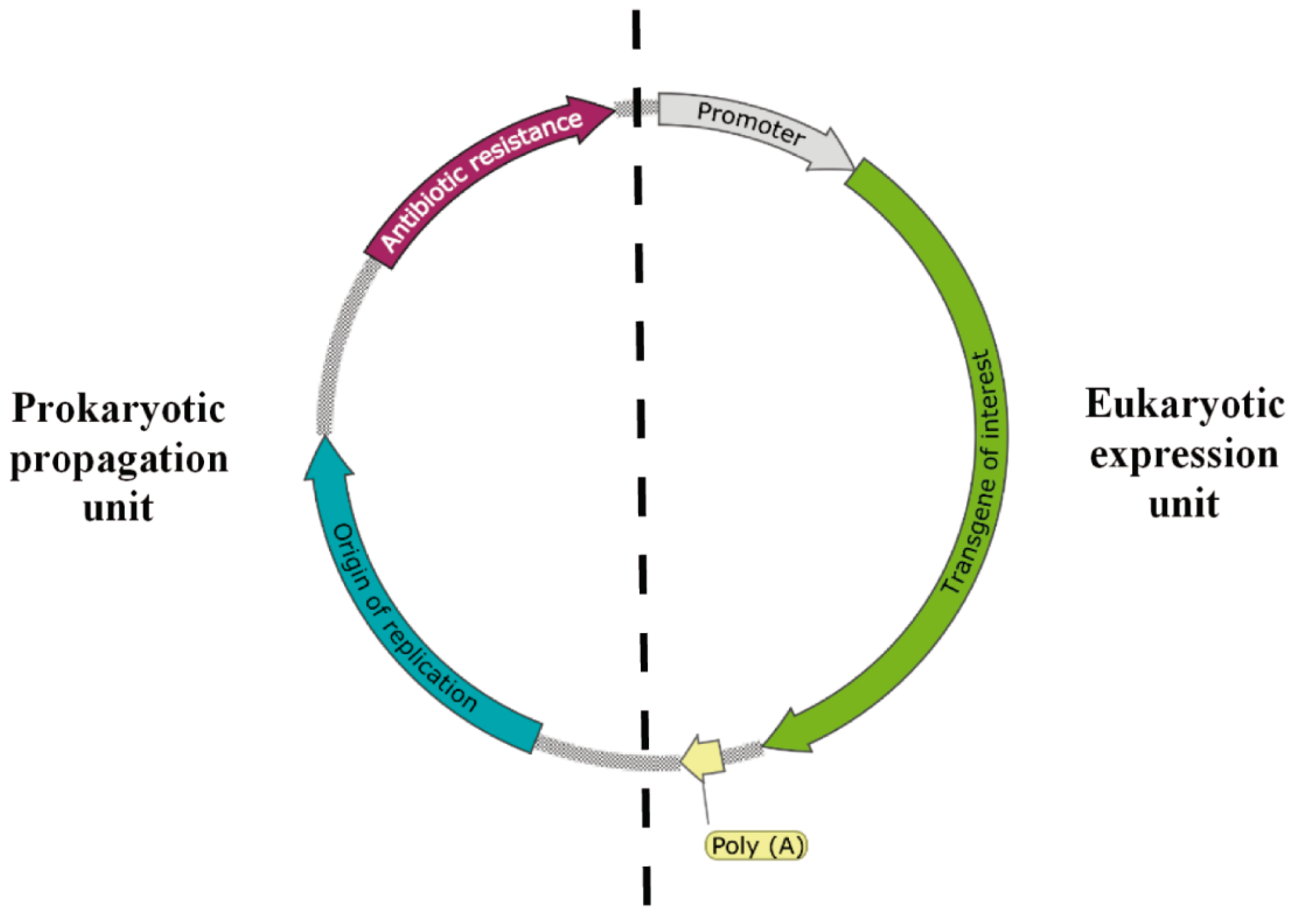
2. Plasmid Vectors Available for Plasmid DNA and Recombinant Protein Production by L. lactis
2.1. Production of Pharmaceutical- and Food-Grade Proteins and Metabolites
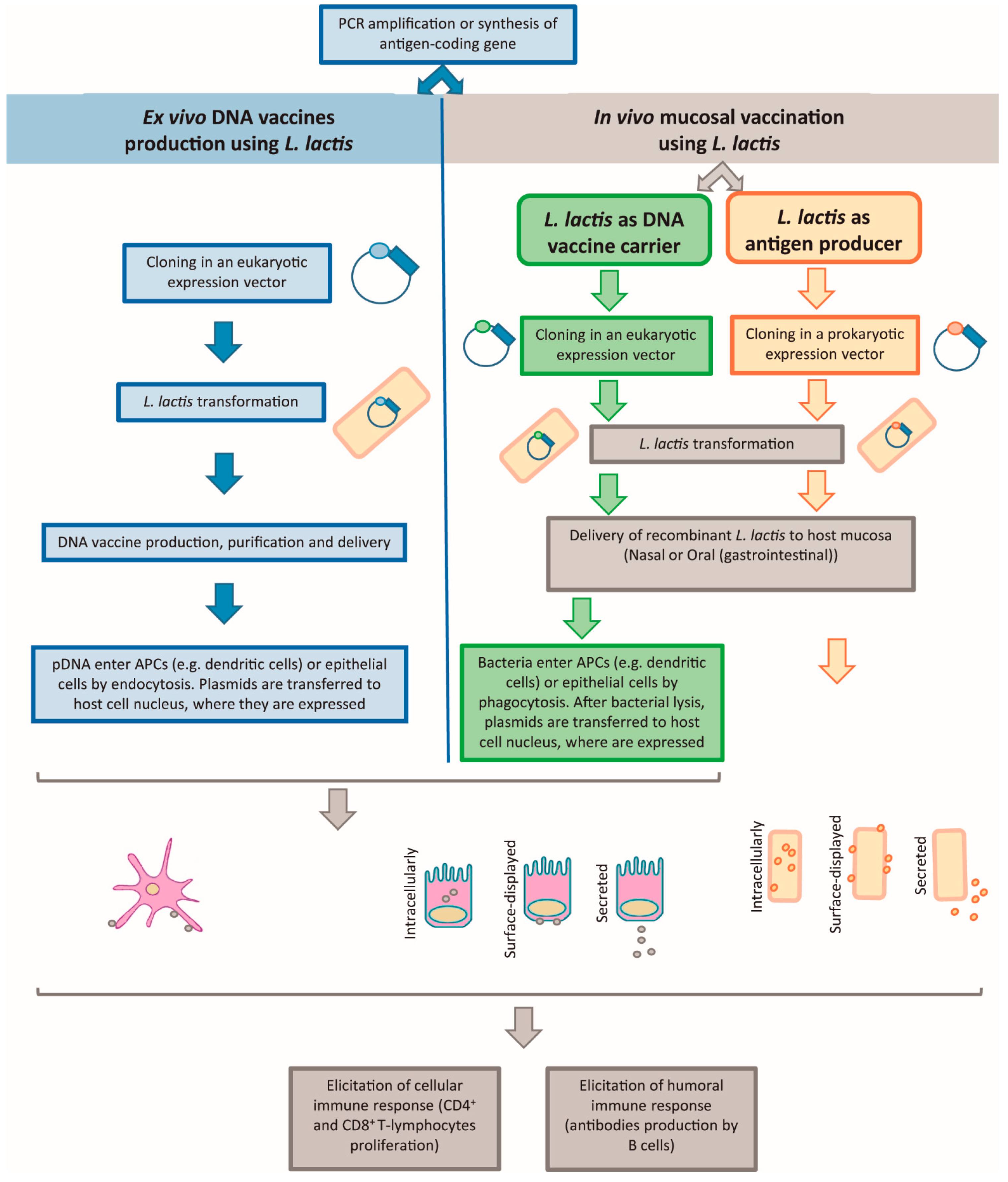
2.2. DNA Vaccination
2.2.1. Naked DNA Vaccination and L. lactis as pDNA Producer
2.2.2. Live Mucosal Vaccination and L. lactis as DNA Vaccine Carrier or as Antigen Producer
L. lactis as a DNA Vaccine Carrier
L. lactis as an Antigen Producer
Plasmids with a pWV01-Based Origin of Replication
Plasmids with a pSH71-Based Origin of Replication
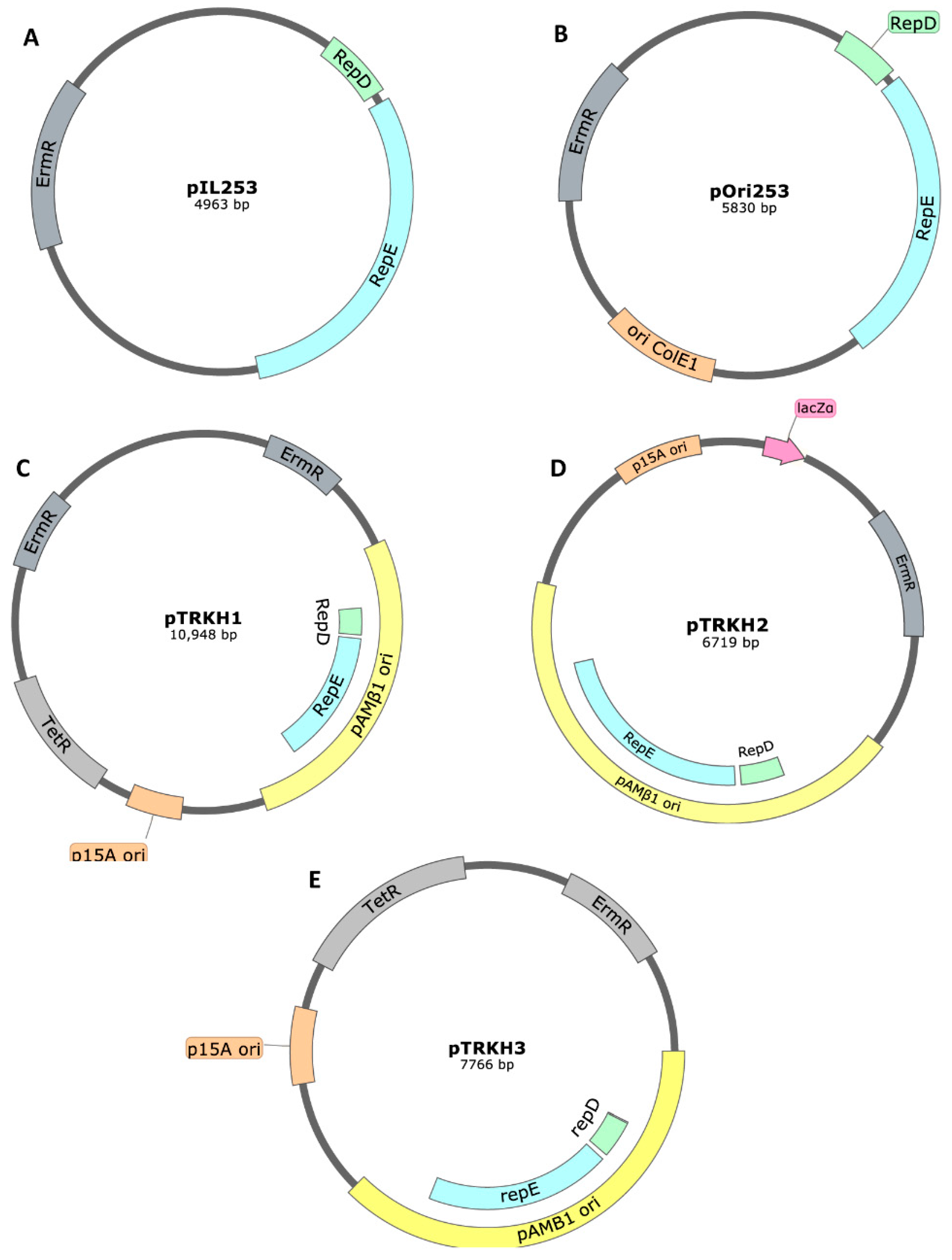
Plasmids with a pAMβ1-Based Origin of Replication
3. Conclusions: Choosing or Developing/Constructing the Best Vector for Each Application
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wood, B.J.B.; Holzapfel, W.H.N. The Genera of Lactic Acid Bacteria; Springer Science & Business Media: New York, NY, USA, 1992. [Google Scholar]
- Bolotin, A.; Wincker, P.; Mauger, S.; Jaillon, O.; Malarme, K.; Weissenbach, J.; Ehrlich, S.D.; Sorokin, A. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001, 11, 731–753. [Google Scholar] [CrossRef]
- Bermudez-Humaran, L.G.; Kharrat, P.; Chatel, J.-M.; Langella, P. Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb. Cell Fact. 2011, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.N.; Duarte, S.; Moreira, L.M.; Monteiro, G.A. Draft Genome Sequence of the Plasmid-Free Lactococcus lactis subsp. lactis Strain LMG 19460. Genome Announc. 2017, 5, e00210-17. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, R.; Pandeya, D.R.; Hong, S.T. Review: Lactococcus lactis: An efficient gram positive cell factory for the production and secretion of recombinant protein. Biomed. Res. 2012, 23, 1–7. [Google Scholar]
- Mamat, U.; Wilke, K.; Bramhill, D.; Schromm, A.B.; Lindner, B.; Kohl, T.A.; Corchero, J.L.; Villaverde, A.; Schaffer, L.; Head, S.R.; et al. Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb. Cell Fact. 2015, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Le Loir, Y.; Azevedo, V.; Oliveira, S.C.; Freitas, D.A.; Miyoshi, A.; Bermudez-Humaran, L.G.; Nouaille, S.; Ribeiro, L.A.; Leclercq, S.; Gabriel, J.E.; et al. Protein secretion in Lactococcus lactis: An efficient way to increase the overall heterologous protein production. Microb. Cell Fact. 2005, 4, 2. [Google Scholar] [CrossRef]
- Gram, G.J.; Fomsgaard, A.; Thorn, M.; Madsen, S.M.; Glenting, J. Immunological analysis of a Lactococcus lactis-based DNA vaccine expressing HIV gp120. Genet. Vaccines Ther. 2007, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.L.; Arnved, K.R.; Johansen, E. Genetic analysis of the minimal replicon of the Lactococcus lactis subsp. lactis biovar diacetylactis citrate plasmid. Mol. Gen. Genet. 1994, 244, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V.; Zurita-Turk, M.; Luerce-Saraiva, T.; Castro, C.; Souza, B.; Agresti, P.; Lima, F.; Pfeiffer, V.; Azevedo, M.; Rocha, C.; et al. DNA Vaccines Approach: From Concepts to Applications. World J. Vaccines 2014, 4, 50–71. [Google Scholar] [CrossRef]
- Singh, S.K.; Ahmed, S.U.; Pandey, A. Metabolic engineering approaches for lactic acid production. Process Biochem. 2006, 41, 991–1000. [Google Scholar] [CrossRef]
- Morello, E.; Bermudez-Humaran, L.G.; Llull, D.; Sole, V.; Miraglio, N.; Langella, P.; Poquet, I. Lactococcus lactis, an efficient cell factory for recombinant protein production and secretion. J. Mol. Microbiol. Biotechnol. 2008, 14, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Poquet, I.; Bolotin, A.; Gruss, A. Optimising the production of heterologous exported proteins in Lactococcus lactis by inactivation of HtrA, the unique housekeeping surface protease. Le Lait 2001, 81, 37–47. [Google Scholar] [CrossRef]
- Van Mellaert, L.; Anné, J. Gram-Positive Bacteria as Host Cells for Heterologous Production of Biopharmaceuticals. In Novel Frontiers in the Production of Compounds for Biomedical Use; Van Broekhoven, A., Shapiro, F., Anné, J., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 277–300. [Google Scholar] [CrossRef]
- Song, A.A.-L.; In, L.L.A.; Lim, S.H.E.; Rahim, R.A. A review on Lactococcus lactis: From food to factory. Microb. Cell Fact. 2017, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- Hols, P.; Kleerebezem, M.; Schanck, A.N.; Ferain, T.; Hugenholtz, J.; Delcour, J.; de Vos, W.M. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 1999, 17, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, I.; Molenaar, D.; Beekwilder, J.; Bouwmeester, H.; van Hylckama Vlieg, J.E.T. Expression of plant flavor genes in Lactococcus lactis. Appl. Environ. Microbiol. 2007, 73, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Song, A.A.L.; Abdullah, J.O.; Abdullah, M.P.; Shafee, N.; Rahim, R.A. Functional expression of an orchid fragrance gene in Lactococcus lactis. Int. J. Mol. Sci. 2012, 13, 1582–1597. [Google Scholar] [CrossRef] [PubMed]
- Song, A.A.-L.; Abdullah, J.O.; Abdullah, M.P.; Shafee, N.; Othman, R.; Tan, E.-F.; Noor, N.M.; Raha, A.R. Overexpressing 3-hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) in the lactococcal mevalonate pathway for heterologous plant sesquiterpene production. PLoS ONE 2012, 7, e52444. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, V.; Puvendran, K.; Guru, B.R.; Jayaraman, G. Design of aqueous two-phase systems for purification of hyaluronic acid produced by metabolically engineered Lactococcus lactis. J. Sep. Sci. 2016, 39, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Sybesma, W.; Starrenburg, M.; Kleerebezem, M.; Mierau, I.; de Vos, W.M.; Hugenholtz, J. Increased production of folate by metabolic engineering of Lactococcus lactis. Appl. Environ. Microbiol. 2003, 69, 3069–3076. [Google Scholar] [CrossRef]
- Sybesma, W.; Van Den Born, E.; Starrenburg, M.; Mierau, I.; Kleerebezem, M.; De Vos, W.M.; Hugenholtz, J. Controlled modulation of folate polyglutamyl tail length by metabolic engineering of Lactococcus lactis. Appl. Environ. Microbiol. 2003, 69, 7101–7107. [Google Scholar] [CrossRef]
- Gu, W.; Xia, Q.; Yao, J.; Fu, S.; Guo, J.; Hu, X. Recombinant expressions of sweet plant protein mabinlin II in Escherichia coli and food-grade Lactococcus lactis. World J. Microbiol. Biotechnol. 2015, 31, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, M.; Baradaran, A.; Rosli, M.I.; Rosfarizan, M.; Khatijah, Y.; Raha, A.R. Effect of signal peptides on the secretion of beta-cyclodextrin glucanotransferase in Lactococcus lactis NZ9000. J. Mol. Microbiol. Biotechnol. 2012, 22, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Cuesta, M.C.; Gasson, M.J.; Narbad, A. Heterologous expression of the plant coumarate: CoA ligase in Lactococcus lactis. Lett. Appl. Microbiol. 2005, 40, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Zhang, J.; Li, H.; Du, G.; Chen, J.; Lee, B. Codon and Propeptide Optimizations to Improve the Food-grade Expression of Bile Salt Hydrolase in Lactococcus lactis. Protein Pept. Lett. 2015, 22, 727–735. [Google Scholar] [CrossRef]
- Yang, Y.; Kang, Z.; Zhou, J.; Chen, J.; Du, G. High-level expression and characterization of recombinant acid urease for enzymatic degradation of urea in rice wine. Appl. Microbiol. Biotechnol. 2015, 99, 301–308. [Google Scholar] [CrossRef]
- Garcia-Quintans, N.; Repizo, G.; Martin, M.; Magni, C.; Lopez, P. Activation of the diacetyl/acetoin pathway in Lactococcus lactis subsp. lactis bv. diacetylactis CRL264 by acidic growth. Appl. Environ. Microbiol. 2008, 74, 1988–1996. [Google Scholar] [CrossRef]
- Ramchandran, L.; Sanciolo, P.; Vasiljevic, T.; Broome, M.; Powell, I.; Duke, M. Improving cell yield and lactic acid production of Lactococcus lactis ssp. cremoris by a novel submerged membrane fermentation process. J. Memb. Sci. 2012, 403, 179–187. [Google Scholar] [CrossRef]
- Sybesma, W.; Burgess, C.; Starrenburg, M.; van Sinderen, D.; Hugenholtz, J. Multivitamin production in Lactococcus lactis using metabolic engineering. Metab. Eng. 2004, 6, 109–115. [Google Scholar] [CrossRef]
- Liu, J.; Dantoft, S.H.; Wurtz, A.; Jensen, P.R.; Solem, C. A novel cell factory for efficient production of ethanol from dairy waste. Biotechnol. Biofuels 2016, 9, 33. [Google Scholar] [CrossRef]
- Reyes-Sandoval, A.; Ertl, H.C. DNA vaccines. Curr. Mol. Med. 2001, 1, 217–243. [Google Scholar] [CrossRef]
- Fynan, E.F.; Lu, S.; Robinson, H.L. One Group’s Historical Reflections on DNA Vaccine Development. Hum. Gene Ther. 2018, 29, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Yurina, V. Live Bacterial Vectors-A Promising DNA Vaccine Delivery System. Med. Sci. 2018, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Hasson, S.S.A.A.; Al-Busaidi, J.K.Z.; Sallam, T.A. The past, current and future trends in DNA vaccine immunisations. Asian Pac. J. Trop. Biomed. 2015, 5, 344–353. [Google Scholar] [CrossRef]
- Gardlik, R.; Palffy, R.; Hodosy, J.; Lukacs, J.; Turna, J.; Celec, P. Vectors and delivery systems in gene therapy. Med. Sci. Monit. 2005, 11, RA110–RA121. [Google Scholar]
- Luo, D.; Saltzman, W.M. Synthetic DNA delivery systems. Nat. Biotechnol. 2000, 18, 33–37. [Google Scholar] [CrossRef]
- Williams, D.A.; Baum, C. Medicine. Gene therapy-new challenges ahead. Science 2003, 302, 400–401. [Google Scholar] [CrossRef]
- Ogra, P.L.; Faden, H.; Welliver, R.C. Vaccination strategies for mucosal immune responses. Clin. Microbiol. Rev. 2001, 14, 430–445. [Google Scholar] [CrossRef]
- Chatel, J.-M.; Pothelune, L.; Ah-Leung, S.; Corthier, G.; Wal, J.-M.; Langella, P. In vivo transfer of plasmid from food-grade transiting lactococci to murine epithelial cells. Gene Ther. 2008, 15, 1184–1190. [Google Scholar] [CrossRef]
- Hobernik, D.; Bros, M. DNA Vaccines-How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef]
- Praveen, B.; Golla, N. DNA Vaccines: Important Criteria Against Avian Viruses. Int. J. Virol. 2016, 13, 1–13. [Google Scholar] [CrossRef][Green Version]
- Pontes, D.S.; de Azevedo, M.S.P.; Chatel, J.-M.; Langella, P.; Azevedo, V.; Miyoshi, A. Lactococcus lactis as a live vector: Heterologous protein production and DNA delivery systems. Protein Expr. Purif. 2011, 79, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Zhu, L.; Kang, H.; Yang, Q. Mucosal Lactobacillus vectored vaccines. Hum. Vaccin. Immunother. 2013, 9, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Beard, C.W.; Mason, P.W. Out on the farm with DNA vaccines. Nat. Biotechnol. 1998, 16, 1325–1328. [Google Scholar] [CrossRef] [PubMed]
- Gulce-Iz, S.; Saglam-Metiner, P. Current State of the Art in DNA Vaccine Delivery and Molecular Adjuvants: Bcl-xL Anti-Apoptotic Protein as a Molecular Adjuvant. Immune Response Activ. Immunomodulation 2019, 1, 1–16. [Google Scholar] [CrossRef]
- da Silva, A.J.; Zangirolami, T.C.; Novo-Mansur, M.T.M.; de Campos Giordano, R.; Martins, E.A.L. Live bacterial vaccine vectors: An overview. Braz. J. Microbiol. 2014, 45, 1117–1129. [Google Scholar] [CrossRef]
- Wells, J.M.; Mercenier, A. Mucosal delivery of therapeutic and prophylactic molecules using lactic acid bacteria. Nat. Rev. Microbiol. 2008, 6, 349–362. [Google Scholar] [CrossRef]
- Klijn, N.; Weerkamp, A.H.; De Vos, W.M. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 1995, 61, 2771–2774. [Google Scholar] [CrossRef]
- Kimoto, H.; Kurisaki, J.; Tsuji, N.M.; Ohmomo, S.; Okamoto, T. Lactococci as probiotic strains: Adhesion to human enterocyte-like Caco-2 cells and tolerance to low pH and bile. Lett. Appl. Microbiol. 1999, 29, 313–316. [Google Scholar] [CrossRef]
- Guimarães, V.; Innocentin, S.; Chatel, J.-M.; Lefèvre, F.; Langella, P.; Azevedo, V.; Miyoshi, A. A new plasmid vector for DNA delivery using lactococci. Genet. Vaccines Ther. 2009, 7, 4. [Google Scholar] [CrossRef]
- Tao, L.; Pavlova, S.I.; Ji, X.; Jin, L.; Spear, G. A novel plasmid for delivering genes into mammalian cells with noninvasive food and commensal lactic acid bacteria. Plasmid 2011, 65, 8–14. [Google Scholar] [CrossRef]
- Yagnik, B.; Padh, H.; Desai, P. Construction of a new shuttle vector for DNA delivery into mammalian cells using non-invasive Lactococcus lactis. Microbes Infect. 2016, 18, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Yagnik, B.; Sharma, D.; Padh, H.; Desai, P. Dual recombinant Lactococcus lactis for enhanced delivery of DNA vaccine reporter plasmid pPERDBY. Microbiol. Immunol. 2017, 61, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Dean, D.A.; Dean, B.S.; Muller, S.; Smith, L.C. Sequence requirements for plasmid nuclear import. Exp. Cell Res. 1999, 253, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Bakari, S.; Lembrouk, M.; Sourd, L.; Ousalem, F.; Andre, F.; Orlowski, S.; Delaforge, M.; Frelet-Barrand, A. Lactococcus lactis is an Efficient Expression System for Mammalian Membrane Proteins Involved in Liver Detoxification, CYP3A4, and MGST1. Mol. Biotechnol. 2016, 58, 299–310. [Google Scholar] [CrossRef]
- Mao, R.; Zhou, K.; Han, Z.; Wang, Y. Subtilisin QK-2: Secretory expression in Lactococcus lactis and surface display onto gram-positive enhancer matrix (GEM) particles. Microb. Cell Fact. 2016, 15, 80. [Google Scholar] [CrossRef][Green Version]
- Mao, R.; Wu, D.; Hu, S.; Zhou, K.; Wang, M.; Wang, Y. Secretory expression and surface display of a new and biologically active single-chain insulin (SCI-59) analog by lactic acid bacteria. Appl. Microbiol. Biotechnol. 2017, 101, 3259–3271. [Google Scholar] [CrossRef]
- Ma, C.; Zhang, L.; Gao, M.; Ma, D. Construction of Lactococcus lactis expressing secreted and anchored Eimeria tenella 3-1E protein and comparison of protective immunity against homologous challenge. Exp. Parasitol. 2017, 178, 14–20. [Google Scholar] [CrossRef]
- Ribeiro, L.A.; Azevedo, V.; Le Loir, Y.; Oliveira, S.C.; Dieye, Y.; Piard, J.-C.; Gruss, A.; Langella, P. Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis: A first step towards food-grade live vaccines against brucellosis. Appl. Environ. Microbiol. 2002, 68, 910–916. [Google Scholar] [CrossRef]
- Michon, C.; Langella, P.; Eijsink, V.G.H.; Mathiesen, G.; Chatel, J.M. Display of recombinant proteins at the surface of lactic acid bacteria: Strategies and applications. Microb. Cell Fact. 2016, 15, 70. [Google Scholar] [CrossRef]
- Seegers, J.F.; Zhao, A.C.; Meijer, W.J.; Khan, S.A.; Venema, G.; Bron, S. Structural and functional analysis of the single-strand origin of replication from the lactococcal plasmid pWV01. Mol. Gen. Genet. 1995, 249, 43–50. [Google Scholar] [CrossRef]
- Bryksin, A.V.; Matsumura, I. Rational design of a plasmid origin that replicates efficiently in both gram-positive and gram-negative bacteria. PLoS ONE 2010, 5, e13244. [Google Scholar] [CrossRef]
- Gasson, M.J.; de Vos, W. Genetics and Biotechnology of Lactic Acid Bacteria; Springer Science & Business Media: New York, NY, USA, 2012. [Google Scholar]
- Kok, J.; van der Vossen, J.M.; Venema, G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 1984, 48, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Leenhouts, K.J.; Kok, J.; Venema, G. Lactococcal plasmid pWV01 as an integration vector for lactococci. Appl. Environ. Microbiol. 1991, 57, 2562–2567. [Google Scholar] [CrossRef] [PubMed]
- Leenhouts, K.J.; Tolner, B.; Bron, S.; Kok, J.; Venema, G.; Seegers, J.F. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 1991, 26, 55–66. [Google Scholar] [CrossRef]
- Pereira, V.B.; da Cunha, V.P.; Preisser, T.M.; Souza, B.M.; Turk, M.Z.; De Castro, C.P.; Azevedo, M.S.P.; Miyoshi, A. Lactococcus lactis carrying a DNA vaccine coding for the ESAT-6 antigen increases IL-17 cytokine secretion and boosts the BCG vaccine immune response. J. Appl. Microbiol. 2017, 122, 1657–1662. [Google Scholar] [CrossRef]
- Mancha-Agresti, P.; de Castro, C.P.; Dos Santos, J.S.C.; Araujo, M.A.; Pereira, V.B.; LeBlanc, J.G.; Leclercq, S.Y.; Azevedo, V. Recombinant Invasive Lactococcus lactis Carrying a DNA Vaccine Coding the Ag85A Antigen Increases INF-gamma, IL-6, and TNF-alpha Cytokines after Intranasal Immunization. Front. Microbiol. 2017, 8, 1263. [Google Scholar] [CrossRef]
- Souza, B.M.; Preisser, T.M.; Pereira, V.B.; Zurita-Turk, M.; de Castro, C.P.; da Cunha, V.P.; de Oliveira, R.P.; Gomes-Santos, A.C.; de Faria, A.M.C.; Machado, D.C.C.; et al. Lactococcus lactis carrying the pValac eukaryotic expression vector coding for IL-4 reduces chemically-induced intestinal inflammation by increasing the levels of IL-10-producing regulatory cells. Microb. Cell Fact. 2016, 15, 150. [Google Scholar] [CrossRef]
- Chiabai, M.J.; Almeida, J.F.; De Azevedo, M.G.D.; Fernandes, S.S.; Pereira, V.B.; De Castro, R.J.A.; Jerônimo, M.S.; Sousa, I.G.; De Souza Vianna, L.M.; Miyoshi, A.; et al. Mucosal delivery of Lactococcus lactis carrying an anti-TNF scFv expression vector ameliorates experimental colitis in mice. BMC Biotechnol. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Zurita-Turk, M.; Souza, B.M.; De Castro, C.P.; Pereira, V.B.; Da Cunha, V.P.; Preisser, T.M.; De Faria, A.M.C.; Machado, D.C.C.; Miyoshi, A. Attenuation of intestinal inflammation in IL-10 deficient mice by a plasmid carrying Lactococcus lactis strain. BMC Biotechnol. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Del Carmen, S.; de Moreno de LeBlanc, A.; Levit, R.; Azevedo, V.; Langella, P.; Bermudez-Humaran, L.G.; LeBlanc, J.G. Anti-cancer effect of lactic acid bacteria expressing antioxidant enzymes or IL-10 in a colorectal cancer mouse model. Int. Immunopharmacol. 2017, 42, 122–129. [Google Scholar] [CrossRef]
- Yagnik, B.; Sharma, D.; Padh, H.; Desai, P. Immunization with r-Lactococcus lactis expressing outer membrane protein A of Shigella dysenteriae type-1: Evaluation of oral and intranasal route of administration. J. Appl. Microbiol. 2017, 122, 493–505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, S.; Du, X.; Li, T.; Han, L.; Kong, J. Heterologous expression of carcinoembryonic antigen in Lactococcus lactis via LcsB-mediated surface displaying system for oral vaccine development. J. Microbiol. Immunol. Infect. 2016, 49, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Santos, A.C.; de Oliveira, R.P.; Moreira, T.G.; Castro-Junior, A.B.; Horta, B.C.; Lemos, L.; de Almeida, L.A.; Rezende, R.M.; Cara, D.C.; Oliveira, S.C.; et al. Hsp65-Producing Lactococcus lactis Prevents Inflammatory Intestinal Disease in Mice by IL-10- and TLR2-Dependent Pathways. Front. Immunol. 2017, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Herrera Ramirez, J.C.; De la Mora, A.C.; De la Mora Valle, A.; Lopez-Valencia, G.; Hurtado, R.M.B.; Renteria Evangelista, T.B.; Rodriguez Castillo, J.L.; Rodriguez Gardea, A.; Gomez Gomez, S.D.; Medina-Basulto, G.E. Immunopathological evaluation of recombinant mycobacterial antigen Hsp65 expressed in Lactococcus lactis as a novel vaccine candidate. Iran. J. Vet. Res. 2017, 18, 197–202. [Google Scholar] [PubMed]
- Gao, X.Y.; Zhou, X.F.; Wang, H.; Lv, N.; Liu, Y.; Guo, J.R. Effects of heme oxygenase-1 recombinant Lactococcus lactis on the intestinal barrier of hemorrhagic shock rats. Braz. J. Med. Biol. Res. 2017, 50, e5601. [Google Scholar] [CrossRef]
- Carvalho, R.D.; Breyner, N.; Menezes-Garcia, Z.; Rodrigues, N.M.; Lemos, L.; Maioli, T.U.; da Gloria Souza, D.; Carmona, D.; de Faria, A.M.C.; Langella, P.; et al. Secretion of biologically active pancreatitis-associated protein I (PAP) by genetically modified dairy Lactococcus lactis NZ9000 in the prevention of intestinal mucositis. Microb. Cell Fact. 2017, 16, 1–11. [Google Scholar] [CrossRef]
- Carvalho, R.; Vaz, A.; Pereira, F.L.; Dorella, F.; Aguiar, E.; Chatel, J.M.; Bermudez, L.; Langella, P.; Fernandes, G.; Figueiredo, H.; et al. Gut microbiome modulation during treatment of mucositis with the dairy bacterium Lactococcus lactis and recombinant strain secreting human antimicrobial PAP. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Takahashi, K.; Yano, A.; Watanabe, S.; Langella, P.; Bermúdez-Humarán, L.G.; Inoue, N. M cell–targeting strategy enhances systemic and mucosal immune responses induced by oral administration of nuclease-producing L lactis. Appl. Microbiol. Biotechnol. 2018, 102, 10703–10711. [Google Scholar] [CrossRef]
- Gusmao-Silva, G.; Aguiar, S.L.F.; Miranda, M.C.G.; Guimarães, M.A.; Alves, J.L.; Vieira, A.T.; Cara, D.C.; Miyoshi, A.; Azevedo, V.A.; Oliveira, R.P.; et al. Hsp65-Producing Lactococcocus lactis Prevents Antigen-Induced Arthritis in Mice. Front. Immunol. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- de Lacerda, L.B.; Rios, W.M.; Masson, A.P.; Brandão, I.T.; Milani, T.M.; de Carvalho Borges, M.; Ramalho, L.N.Z.; Barbosa, M.C.R.; Miyoshi, A.; Silva, C.L. Oral Administration of Hsp65-producing Lactococcus lactis attenuates allergic asthma in a murine model. J. Appl. Microbiol. 2020. [Google Scholar] [CrossRef]
- Yagnik, B.; Sharma, D.; Padh, H.; Desai, P. Oral immunization with LacVax® OmpA induces protective immune response against Shigella flexneri 2a ATCC 12022 in a murine model. Vaccine 2019, 37, 3097–3105. [Google Scholar] [CrossRef]
- Kim, D.; Beck, B.R.; Lee, S.M.; Jeon, J.; Lee, D.W.; Lee, J.I.; Song, S.K. Pellet feed adsorbed with the recombinant Lactococcus lactis BFE920 expressing SiMA antigen induced strong recall vaccine effects against Streptococcus iniae infection in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2016, 55, 374–383. [Google Scholar] [CrossRef]
- Liu, K.-F.; Liu, X.-R.; Li, G.-L.; Lu, S.-P.; Jin, L.; Wu, J. Oral administration of Lactococcus lactis-expressing heat shock protein 65 and tandemly repeated IA2P2 prevents type 1 diabetes in NOD mice. Immunol. Lett. 2016, 174, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.; Wang, X.; Liu, K.; He, D.; Niu, P.; Cao, R.; Jin, L.; Wu, J. Oral delivery of staphylococcal nuclease by Lactococcus lactis prevents type 1 diabetes mellitus in NOD mice. Appl. Microbiol. Biotechnol. 2017, 101, 7653–7662. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.R.; Lee, S.H.; Kim, D.; Park, J.H.; Lee, H.K.; Kwon, S.-S.; Lee, K.H.; Lee, J.I.; Song, S.K. A Lactococcus lactis BFE920 feed vaccine expressing a fusion protein composed of the OmpA and FlgD antigens from Edwardsiella tarda was significantly better at protecting olive flounder (Paralichthys olivaceus) from edwardsiellosis than single antigen vaccines. Fish Shellfish Immunol. 2017, 68, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Breyner, N.M.; Vilas Boas, P.B.; Fernandes, G.; de Carvalho, R.D.; Rochat, T.; Michel, M.L.; Chain, F.; Sokol, H.; de Azevedo, M.; Myioshi, A.; et al. Oral delivery of pancreatitis-associated protein by Lactococcus lactis displays protective effects in dinitro-benzenesulfonic-acid-induced colitis model and is able to modulate the composition of the microbiota. Environ. Microbiol. 2019, 21, 4020–4031. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Zhang, C.; Liu, D.; Bai, W.; Zhang, Q.; Xiang, Q.; Huang, Y.; Su, Z. Prevention of gastrointestinal lead poisoning using recombinant Lactococcus lactis expressing human metallothionein-I fusion protein. Sci. Rep. 2016, 6, 23716. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.Y.; Sun, J.C.; Li, X.; Zhang, Y.; Luo, B.; Jiang, N.; Liu, M.C. Analysis of immune responses in mice orally immunized with recombinant pMG36e-SP-TSOL18/lactococcus lactis and pMG36e-TSOL18/lactococcus lactis vaccines of taenia solium. J. Immunol. Res. 2018, 2018, 9262631. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, Y.; Tao, L.; Wu, X.; Wu, W. Recombinant lactococcus lactis secreting viral protein 1 of enterovirus 71 and its immunogenicity in mice. Biotechnol. Lett. 2019, 41, 867–872. [Google Scholar] [CrossRef]
- Fang, X.; Zhou, X.; Miao, Y.; Han, Y.; Wei, J.; Chen, T. Therapeutic effect of GLP-1 engineered strain on mice model of Alzheimer’s disease and Parkinson’s disease. AMB Express 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Enouf, V.; Langella, P.; Commissaire, J.; Cohen, J.; Corthier, G. Bovine rotavirus nonstructural protein 4 produced by Lactococcus lactis is antigenic and immunogenic. Appl. Environ. Microbiol. 2001, 67, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M. Gene cloning and expression in lactic streptococci. FEMS Microbiol. Rev. 1987, 3, 281–295. [Google Scholar] [CrossRef]
- Tauer, C.; Heinl, S.; Egger, E.; Heiss, S.; Grabherr, R. Tuning constitutive recombinant gene expression in Lactobacillus plantarum. Microb. Cell Fact. 2014, 13, 150. [Google Scholar] [CrossRef] [PubMed]
- Frelet-Barrand, A.; Boutigny, S.; Moyet, L.; Deniaud, A.; Seigneurin-Berny, D.; Salvi, D.; Bernaudat, F.; Richaud, P.; Pebay-Peyroula, E.; Joyard, J.; et al. Lactococcus lactis, an alternative system for functional expression of peripheral and intrinsic Arabidopsis membrane proteins. PLoS ONE 2010, 5, e8746. [Google Scholar] [CrossRef] [PubMed]
- Mierau, I.; Kleerebezem, M. 10 Years of the nisin-controlled gene expression system (NICE) in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2005, 68, 705–717. [Google Scholar] [CrossRef]
- Mierau, I.; Leij, P.; van Swam, I.; Blommestein, B.; Floris, E.; Mond, J.; Smid, E.J. Industrial-scale production and purification of a heterologous protein in Lactococcus lactis using the nisin-controlled gene expression system NICE: The case of lysostaphin. Microb. Cell Fact. 2005, 4, 1–9. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, C.; Cheng, W.; Duan, G.; Shi, Q.; Chen, S.; Fan, Q. Delivery of Helicobacter pylori HpaA to gastrointestinal mucosal immune sites using Lactococcus lactis and its immune efficacy in mice. Biotechnol. Lett. 2018, 40, 585–590. [Google Scholar] [CrossRef]
- Peng, X.; Zhang, R.; Duan, G.; Wang, C.; Sun, N.; Zhang, L.; Chen, S.; Fan, Q.; Xi, Y. Production and delivery of Helicobacter pylori NapA in Lactococcus lactis and its protective efficacy and immune modulatory activity. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, R.; Duan, G.; Peng, X.; Wang, C.; Fan, Q.; Chen, S.; Xi, Y. An engineered food-grade Lactococcus lactis strain for production and delivery of heat-labile enterotoxin B subunit to mucosal sites. BMC Biotechnol. 2017, 17, 25. [Google Scholar] [CrossRef]
- Sun, N.; Zhang, R.; Duan, G.; Peng, X.; Wang, C.; Chen, S.; Fan, Q. A food-grade engineered Lactococcus lactis strain delivering Helicobacter pylori Lpp20 alleviates bacterial infection in H. pylori-challenged mice. Biotechnol. Lett. 2019, 41, 1415–1421. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, W.; Song, Y.; Wang, W.; Zhang, Y.; Wang, T.; Li, K.; Pan, Q.; Qi, X.; Gao, Y.; et al. Recombinant Lactococcus lactis co-expressing OmpH of an M cell-targeting ligand and IBDV-VP2 protein provide immunological protection in chickens. Vaccine 2018, 36, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Song, Y.; Liu, L.; Zhang, Y.; Wang, T.; Zhang, W.; Li, K.; Qi, X.; Gao, Y.; Gao, L.; et al. Neutralizing-antibody-mediated protection of chickens against infectious bursal disease via one-time vaccination with inactivated recombinant Lactococcus lactis expressing a fusion protein constructed from the RCK protein of Salmonella enterica and VP2 of infectious bursal disease virus. Microb. Cell Fact. 2019, 18, 1–12. [Google Scholar] [CrossRef]
- Zhou, J.; Wei, K.; Wang, C.; Dong, W.; Ma, N.; Zhu, L.; Hu, L.P.; Huang, H.; Zhu, R. Oral immunisation with Taishan Pinus massoniana pollen polysaccharide adjuvant with recombinant Lactococcus lactis -expressing Proteus mirabilis ompA confers optimal protection in mice. Allergol. Immunopathol. 2017, 45, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Maghvan, M.A.; Jafari, P.; Hoseini, S.D.; Behrozikhah, A.M. Cloning and expression of B. Mellitensis bp26 gene in Lactococcus lactis as a food grade vaccine. Avicenna J. Med. Biotechnol. 2019, 11, 264–267. [Google Scholar]
- Wozniak, A.; Scioscia, N.; García, P.C.; Dale, J.B.; Paillavil, B.A.; Legarraga, P.; Salazar-Echegarai, F.J.; Bueno, S.M.; Kalergis, A.M. Protective immunity induced by an intranasal multivalent vaccine comprising 10 Lactococcus lactis strains expressing highly prevalent M-protein antigens derived from Group A Streptococcus. Microbiol. Immunol. 2018, 62, 395–404. [Google Scholar] [CrossRef]
- Torkashvand, A.; Bahrami, F.; Adib, M.; Ajdary, S. Mucosal and systemic immune responses elicited by recombinant Lactococcus lactis expressing a fusion protein composed of pertussis toxin and filamentous hemagglutinin from Bordetella pertussis. Microb. Pathog. 2018, 120, 155–160. [Google Scholar] [CrossRef]
- Torkashvand, A.; Bahrami, F.; Adib, M.; Ajdary, S. Subcutaneous immunization with recombinant Lactococcus lactis expressing F1S1 fusion protein induces systemic and mucosal immune responses in BALB/C mice. Rep. Biochem. Mol. Biol. 2019, 7, 196–203. [Google Scholar]
- Guo, M.; Yi, S.; Guo, Y.; Zhang, S.; Niu, J.; Wang, K.; Hu, G. Construction of a Recombinant Lactococcus lactis Strain Expressing a Variant Porcine Epidemic Diarrhea Virus S1 Gene and Its Immunogenicity Analysis in Mice. Viral Immunol. 2019, 32, 144–150. [Google Scholar] [CrossRef]
- Song, S.; Li, P.; Zhang, R.; Chen, J.; Lan, J.; Lin, S.; Guo, G.; Xie, Z.; Jiang, S. Oral vaccine of recombinant Lactococcus lactis expressing the VP1 protein of duck hepatitis A virus type 3 induces mucosal and systemic immune responses. Vaccine 2019, 37, 4364–4369. [Google Scholar] [CrossRef]
- Yildiz, M.; Askin, H. Heterologous expression of azurin from Pseudomonas aeruginosa in food-grade Lactococcus lactis. Prep. Biochem. Biotechnol. 2019, 49, 800–806. [Google Scholar] [CrossRef]
- Wang, C.; Zhou, H.; Guo, F.; Yang, B.; Su, X.; Lin, J.; Xu, F. Oral Immunization of Chickens with Lactococcus lactis Expressing cjaA Temporarily Reduces Campylobacter jejuni Colonization. Foodborne Pathog. Dis. 2020, 17, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.Y.; Dong, M.; Hu, Z.Y.; Wu, J.; Li, Y.C.; Xu, H.D. Recombinant Lactococcus lactis NZ3900 expressing bioactive human FGF21 reduced body weight of Db/Db mice through the activity of brown adipose tissue. Benef. Microbes 2020, 11, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.F.; Chiang, C.H.; Lin, S.J.; Song, P.P.; Liu, H.C.; Wu, T.J.; Lin, W.W. Recombinant Lactococcus lactis Expressing Ling Zhi 8 Protein Ameliorates Nonalcoholic Fatty Liver and Early Atherogenesis in Cholesterol-Fed Rabbits. Biomed Res. Int. 2020, 2020, 3495682. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Zhao, L.; Song, M. A Lactococcus lactis-vectored oral vaccine induces protective immunity of mice against enterotoxigenic Escherichia coli lethal challenge. Immunol. Lett. 2020, 225, 57–63. [Google Scholar] [CrossRef]
- Szczepankowska, A.K.; Szatraj, K.; Salanski, P.; Rozga, A.; Gorecki, R.K.; Bardowski, J.K. Recombinant Lactococcus lactis Expressing Haemagglutinin from a Polish Avian H5N1 Isolate and Its Immunological Effect in Preliminary Animal Trials. Biomed Res. Int. 2017, 2017, 6747482. [Google Scholar] [CrossRef]
- Joan, S.S.X.; Pui-Fong, J.; Song, A.A.-L.; Chang, L.-Y.; Yusoff, K.; AbuBakar, S.; Rahim, R.A. Oral vaccine of Lactococcus lactis harbouring pandemic H1N1 2009 haemagglutinin1 and nisP anchor fusion protein elevates anti-HA1 sIgA levels in mice. Biotechnol. Lett. 2016, 38, 793–799. [Google Scholar] [CrossRef]
- Zeng, Z.; Yu, R.; Zuo, F.; Zhang, B.; Ma, H.; Chen, S. Recombinant Lactococcus lactis expressing bioactive exendin-4 to promote insulin secretion and beta-cell proliferation in vitro. Appl. Microbiol. Biotechnol. 2017, 101, 7177–7186. [Google Scholar] [CrossRef]
- Lahiri, A.; Sharif, S.; Mallick, A.I. Intragastric delivery of recombinant Lactococcus lactis displaying ectodomain of influenza matrix protein 2 (M2e) and neuraminidase (NA) induced focused mucosal and systemic immune responses in chickens. Mol. Immunol. 2019, 114, 497–512. [Google Scholar] [CrossRef]
- Stedman, A.; Chambers, M.A.; Gutierrez-Merino, J. Secretion and functional expression of Mycobacterium bovis antigens MPB70 and MPB83 in lactic acid bacteria. Tuberculosis 2019, 117, 24–30. [Google Scholar] [CrossRef]
- Gorain, C.; Singh, A.; Bhattacharyya, S.; Kundu, A.; Lahiri, A.; Gupta, S.; Mallick, A.I. Mucosal delivery of live Lactococcus lactis expressing functionally active JlpA antigen induces potent local immune response and prevent enteric colonization of Campylobacter jejuni in chickens. Vaccine 2020, 38, 1630–1642. [Google Scholar] [CrossRef]
- Ma, D.; Gao, M.; Dalloul, R.A.; Ge, J.; Ma, C.; Li, J. Protective effects of oral immunization with live Lactococcus lactis expressing Eimeria tenella 3-1E protein. Parasitol. Res. 2013, 112, 4161–4167. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, F.; Ma, C.; Huang, Y.; Wang, D.; Ma, D. Recombinant lactococcus lactis expressing Eimeria tenella AMA1 protein and its immunological effects against homologous challenge. Exp. Parasitol. 2018, 191, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Ma, C.; Ma, D.; Zhao, Q.; Wang, F.; Huang, Y.; Li, J.; Zhang, L.; Zhou, E.M. Live recombinant Lactococcus lactis expressing avian hepatitis virus ORF2 protein: Immunoprotection against homologous virus challenge in chickens. Vaccine 2018, 36, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, A.; Zuo, F.; Yu, R.; Zeng, Z.; Ma, H.; Chen, S. Recombinant Lactococcus lactis NZ9000 secretes a bioactive kisspeptin that inhibits proliferation and migration of human colon carcinoma HT-29 cells. Microb. Cell Fact. 2016, 15, 102. [Google Scholar] [CrossRef] [PubMed]
- Zadravec, P.; Mareckova, L.; Petrokova, H.; Hodnik, V.; Perisic Nanut, M.; Anderluh, G.; Strukelj, B.; Maly, P.; Berlec, A. Development of Recombinant Lactococcus lactis Displaying Albumin-Binding Domain Variants against Shiga Toxin 1 B Subunit. PLoS ONE 2016, 11, e0162625. [Google Scholar] [CrossRef] [PubMed]
- Shigemori, S.; Ihara, M.; Sato, T.; Yamamoto, Y.; Nigar, S.; Ogita, T.; Shimosato, T. Secretion of an immunoreactive single-chain variable fragment antibody against mouse interleukin 6 by Lactococcus lactis. Appl. Microbiol. Biotechnol. 2017, 101, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Y.; Deng, B.; Xu, Z. Recombinant Lactococcus lactis expressing porcine insulin-like growth factor I ameliorates DSS-induced colitis in mice. BMC Biotechnol. 2016, 16, 25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Ai, C. Development of House Dust Mite Vaccine. Methods Mol. Biol. 2016, 1403, 739–751. [Google Scholar] [CrossRef]
- Van Hoang, V.; Ochi, T.; Kurata, K.; Arita, Y.; Ogasahara, Y.; Enomoto, K. Nisin-induced expression of recombinant T cell epitopes of major Japanese cedar pollen allergens in Lactococcus lactis. Appl. Microbiol. Biotechnol. 2018, 102, 261–268. [Google Scholar] [CrossRef]
- Mustopa, A.Z.; Mariyah, S.; Fatimah; Budiarti, S.; Murtiyaningsih, H.; Alfisyahrin, W.N. Construction, heterologous expression, partial purification, and in vitro cytotoxicity of the recombinant plantaricin E produced by Lactococcus lactis against Enteropathogenic Escherichia coli K.1.1 and human cervical carcinoma (HeLa) cells. Mol. Biol. Rep. 2018, 45, 1235–1244. [Google Scholar] [CrossRef]
- Škrlec, K.; Ručman, R.; Jarc, E.; Sikirić, P.; Švajger, U.; Petan, T.; Perišić Nanut, M.; Štrukelj, B.; Berlec, A. Engineering recombinant Lactococcus lactis as a delivery vehicle for BPC-157 peptide with antioxidant activities. Appl. Microbiol. Biotechnol. 2018, 102, 10103–10117. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tian, M.; Li, W.; Hao, F. Preventative delivery of IL-35 by Lactococcus lactis ameliorates DSS-induced colitis in mice. Appl. Microbiol. Biotechnol. 2019, 103, 7931–7941. [Google Scholar] [CrossRef] [PubMed]
- Plavec, T.V.; Kuchař, M.; Benko, A.; Lišková, V.; Černý, J.; Berlec, A.; Malý, P. Engineered Lactococcus lactis secreting IL-23 receptor-targeted REX protein blockers for modulation of IL-23/Th17-mediated inflammation. Microorganisms 2019, 7, 152. [Google Scholar] [CrossRef] [PubMed]
- Sreerohini, S.; Balakrishna, K.; Parida, M. Oral immunization of mice with Lactococcus lactis expressing Shiga toxin truncate confers enhanced protection against Shiga toxins of Escherichia coli O157:H7 and Shigella dysenteriae. Apmis 2019, 127, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Sagi, S.; Konduru, B.; Parida, M. Heterologous expression of Intimin and IpaB fusion protein in Lactococcus lactis and its mucosal delivery elicit protection against pathogenicity of Escherichia coli O157 and Shigella flexneri in a murine model. Int. Immunopharmacol. 2020, 85, 106617. [Google Scholar] [CrossRef] [PubMed]
- Namai, F.; Murakami, A.; Ueda, A.; Tsukagoshi, M.; Shigemori, S.; Ogita, T.; Sato, T.; Shimosato, T. Construction of Genetically Modified Lactococcus lactis Producing Anti-human-CTLA-4 Single-Chain Fragment Variable. Mol. Biotechnol. 2020, 62, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Sha, Z.; Shang, H.; Miao, Y.; Huang, J.; Niu, X.; Chen, R.; Hu, L.; Huang, H.; Wei, K.; Zhu, R. Recombinant Lactococcus Lactis Expressing M1-HA2 Fusion Protein Provides Protective Mucosal Immunity Against H9N2 Avian Influenza Virus in Chickens. Front. Vet. Sci. 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Zeng, L.; Tan, J.; Xue, M.; Liu, L.; Wang, M.; Liang, L.; Deng, J.; Chen, W.; Chen, Y. An engineering probiotic producing defensin-5 ameliorating dextran sodium sulfate-induced mice colitis via Inhibiting NF-kB pathway. J. Transl. Med. 2020, 18, 1–15. [Google Scholar] [CrossRef]
- Liu, X.; Qi, L.; Lv, J.; Zhang, Z.; Zhou, P.; Ma, Z.; Wang, Y.; Zhang, Y.; Pan, L. The immune response to a recombinant Lactococcus lactis oral vaccine against foot-and-mouth disease virus in mice. Biotechnol. Lett. 2020, 42, 1907–1917. [Google Scholar] [CrossRef]
- Naderi-Samani, M.; Soltani, M.; Dadar, M.; Taheri-Mirghaed, A.; Zargar, A.; Ahmadivand, S.; Hassanzadeh, R.; Goudarzi, L.M. Oral immunization of trout fry with recombinant Lactococcus lactis NZ3900 expressing G gene of viral hemorrhagic septicaemia virus (VHSV). Fish Shellfish Immunol. 2020, 105, 62–70. [Google Scholar] [CrossRef]
- Lei, H.; Gao, T.; Cen, Q.; Peng, X. Haemagglutinin displayed on the surface of Lactococcus lactis confers broad cross-clade protection against different H5N1 viruses in chickens. Microb. Cell Fact. 2020, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Guo, S.; Zhao, Z.; Guo, Z.R.; Ma, R.; Wang, G.X.; Zhu, B. Surface display of spring viremia of carp virus glycoprotein on Lactococcus lactis and its protection efficacy in common carp (Cyprinus carpio L.). Fish Shellfish Immunol. 2020, 104, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Namai, F.; Shigemori, S.; Ogita, T.; Sato, T.; Shimosato, T. Construction of genetically modified Lactococcus lactis that produces bioactive anti-interleukin-4 single-chain fragment variable. Mol. Biol. Rep. 2020, 47, 7039–7047. [Google Scholar] [CrossRef] [PubMed]
- Shigemori, S.; Namai, F.; Yamamoto, Y.; Nigar, S.; Sato, T.; Ogita, T.; Shimosato, T. Genetically modified Lactococcus lactis producing a green fluorescent protein-bovine lactoferrin fusion protein suppresses proinflammatory cytokine expression in lipopolysaccharide-stimulated RAW 264.7 cells. J. Dairy Sci. 2017, 100, 7007–7015. [Google Scholar] [CrossRef] [PubMed]
- Yumoto, K.; Sato, T.; Nakashima, K.; Namai, F.; Shigemori, S.; Shimosato, T.; Kaneko, T. Nasally administered lactococcus lactis secreting heme oxygenase-1 attenuates murine emphysema. Antioxidants 2020, 9, 1049. [Google Scholar] [CrossRef] [PubMed]
- Namai, F.; Shigemori, S.; Ogita, T.; Sato, T.; Shimosato, T. Microbial therapeutics for acute colitis based on genetically modified Lactococcus lactis hypersecreting IL-1Ra in mice. Exp. Mol. Med. 2020, 52, 1627–1636. [Google Scholar] [CrossRef]
- Zhao, L.; Tang, X.; Sheng, X.; Xing, J.; Zhan, W. Surface display of hirame novirhabdovirus (HIRRV) G protein in Lactococcus lactis and its immune protection in flounder (Paralichthys olivaceus). Microb. Cell Fact. 2019, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.M.S.; Lorenz, N.; Tsai, C.J.-Y.; Khemlani, A.H.J.; Proft, T. Mucosal vaccination with pili from Group A Streptococcus expressed on Lactococcus lactis generates protective immune responses. Sci. Rep. 2017, 7, 7174. [Google Scholar] [CrossRef]
- Wagachchi, D.; Tsai, J.-Y.C.; Chalmers, C.; Blanchett, S.; Loh, J.M.S.; Proft, T. PilVax—A novel peptide delivery platform for the development of mucosal vaccines. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef]
- Clow, F.; Peterken, K.; Pearson, V.; Proft, T.; Radcliff, F.J. PilVax, a novel Lactococcus lactis-based mucosal vaccine platform, stimulates systemic and mucosal immune responses to Staphylococcus aureus. Immunol. Cell Biol. 2020, 98, 369–381. [Google Scholar] [CrossRef]
- Taghinezhad-S, S.; Mohseni, A.H.; Keyvani, H.; Razavi, M.R. Phase 1 Safety and Immunogenicity Trial of Recombinant Lactococcus lactis Expressing Human Papillomavirus Type 16 E6 Oncoprotein Vaccine. Mol. Ther. Methods Clin. Dev. 2019, 15, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Taghinezhad-S, S.; Mohseni, A.H.; Keyvani, H.; Razavilar, V. Protection against human papillomavirus type 16-induced tumors in C57BL/6 mice by mucosal vaccination with Lactococcus lactis NZ9000 expressing E6 oncoprotein. Microb. Pathog. 2019, 126, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, A.H.; Razavilar, V.; Keyvani, H.; Razavi, M.R.; Nejad, R.A.K. Codon Usage Optimization and Construction of Plasmid Encoding Iranian Human Papillomavirus Type 16 E7 Oncogene for Lactococcus Lactis Subsp. Cremoris MG1363. Asian Pac. J. Cancer Prev. 2017, 18, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, A.H.; Razavilar, V.; Keyvani, H.; Razavi, M.R.; Khavari-Nejad, R.A. Efficient production and optimization of E7 oncoprotein from Iranian human papillomavirus type 16 in Lactococcus lactis using nisin-controlled gene expression (NICE) system. Microb. Pathog. 2017, 110, 554–560. [Google Scholar] [CrossRef]
- Mohseni, A.H.; Razavilar, V.; Keyvani, H.; Razavi, M.R.; Khavari-Nejad, R.A. Oral immunization with recombinant Lactococcus lactis NZ9000 expressing human papillomavirus type 16 E7 antigen and evaluation of its immune effects in female C57BL/6 mice. J. Med. Virol. 2019, 91, 296–307. [Google Scholar] [CrossRef]
- Mohseni, A.H.; Taghinezhad-S, S.; Keyvani, H.; Razavilar, V. Extracellular overproduction of E7 oncoprotein of Iranian human papillomavirus type 16 by genetically engineered Lactococcus lactis. BMC Biotechnol. 2019, 19, 1–13. [Google Scholar] [CrossRef]
- Mohseni, A.H.; Taghinezhad-S, S.; Keyvani, H. The First Clinical Use of a Recombinant Lactococcus lactis Expressing Human Papillomavirus Type 16 E7 Oncogene Oral Vaccine: A phase I safety and immunogenicity trial in healthy women volunteers. Mol. Cancer Ther. 2020, 19, 717–727. [Google Scholar] [CrossRef]
- Davarpanah, E.; Seyed, N.; Bahrami, F.; Rafati, S.; Safaralizadeh, R.; Taheri, T. Lactococcus lactis expressing sand fly ppsp15 salivary protein confers long-term protection against leishmania major in balb/c mice. PLoS Negl. Trop. Dis. 2020, 14, 1–18. [Google Scholar] [CrossRef]
- Craig, K.; Dai, X.; Li, A.; Lu, M.; Xue, M.; Rosas, L.; Gao, T.Z.; Niehaus, A.; Jennings, R.; Li, J. A lactic acid bacteria (LAB)-based vaccine candidate for Human Norovirus. Viruses 2019, 11, 213. [Google Scholar] [CrossRef]
- Diaz-Dinamarca, D.A.; Hernandez, C.; Escobar, D.F.; Soto, D.A.; Muñoz, G.A.; Badilla, J.F.; Manzo, R.A.; Carrión, F.; Kalergis, A.M.; Vasquez, A.E. Mucosal vaccination with lactococcus lactis-secreting surface immunological protein induces humoral and cellular immune protection against group b streptococcus in a murine model. Vaccines 2020, 8, 146. [Google Scholar] [CrossRef]
- Ciaćma, K.; Wiȩckiewicz, J.; Kȩdracka-Krok, S.; Kurtyka, M.; Stec, M.; Siedlar, M.; Baran, J. Secretion of tumoricidal human tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) by recombinant Lactococcus lactis: Optimization of in vitro synthesis conditions. Microb. Cell Fact. 2018, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shirdast, H.; Ebrahimzadeh, F.; Taromchi, A.H.; Mortazavi, Y.; Esmaeilzadeh, A.; Sekhavati, M.H.; Nedaei, K.; Mirabzadeh, E. Recombinant Lactococcus Lactis Displaying Omp31 Antigen of Brucella melitensis Can Induce an Immunogenic Response in BALB/c Mice. Probiotics Antimicrob. Proteins 2020. [Google Scholar] [CrossRef] [PubMed]
- Vasiee, A.; Falah, F.; Sankian, M.; Tabatabaei-Yazdi, F.; Mortazavi, S.A. Oral Immunotherapy Using Probiotic Ice Cream Containing Recombinant Food-Grade Lactococcus lactis Which Inhibited Allergic Responses in a BALB/c Mouse Model. J. Immunol. Res. 2020, 2020, 2635230. [Google Scholar] [CrossRef] [PubMed]
- Daniel, C.; Titecat, M.; Poiret, S.; Cayet, D.; Boutillier, D.; Simonet, M.; Sirard, J.-C.; Lemaitre, N.; Sebbane, F. Characterization of the protective immune response to Yersinia pseudotuberculosis infection in mice vaccinated with an LcrV-secreting strain of Lactococcus lactis. Vaccine 2016, 34, 5762–5767. [Google Scholar] [CrossRef] [PubMed]
- Daniel, C.; Dewitte, A.; Poiret, S.; Marceau, M.; Simonet, M.; Marceau, L.; Descombes, G.; Boutillier, D.; Bennaceur, N.; Bontemps-Gallo, S.; et al. Polymorphism in the yersinia LcrV antigen enables immune escape from the protection conferred by an LcrV-secreting lactococcus lactis in a pseudotuberculosis mouse model. Front. Immunol. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Bruand, C.; Le Chatelier, E.; Ehrlich, S.D.; Janniere, L. A fourth class of theta-replicating plasmids: The pAM beta 1 family from gram-positive bacteria. Proc. Natl. Acad. Sci. USA 1993, 90, 11668–11672. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Ehrlich, S.D.; Janniere, L. The pAM beta 1 CopF repressor regulates plasmid copy number by controlling transcription of the repE gene. Mol. Microbiol. 1994, 14, 463–471. [Google Scholar] [CrossRef]
- Simon, D.; Chopin, A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 1988, 70, 559–566. [Google Scholar] [CrossRef]
- Breyner, N.M.; Michon, C.; de Sousa, C.S.; Vilas Boas, P.B.; Chain, F.; Azevedo, V.A.; Langella, P.; Chatel, J.M. Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii Shows a Protective Effect on DNBS and DSS-Induced Colitis Model in Mice through Inhibition of NF-kappaB Pathway. Front. Microbiol. 2017, 8, 114. [Google Scholar] [CrossRef]
- Kasarello, K.; Szczepankowska, A.; Kwiatkowska-Patzer, B.; Lipkowski, A.W.; Gadamski, R.; Sulejczak, D.; Lachwa, M.; Bialy, M.; Bardowski, J. Effect of recombinant Lactococcus lactis producing myelin peptides on neuroimmunological changes in rats with experimental allergic encephalomyelitis. Folia Neuropathol. 2016, 54, 249–258. [Google Scholar] [CrossRef]
- Kobierecka, P.A.; Olech, B.; Książek, M.; Derlatka, K.; Adamska, I.; Majewski, P.M.; Jagusztyn-Krynicka, E.K.; Wyszyńska, A.K. Cell Wall Anchoring of the Campylobacter Antigens to Lactococcus lactis. Front. Microbiol. 2016, 7, 165. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.F.; Breyner, N.M.; Mahi, M.; Ahmed, B.; Benbouziane, B.; Boas, P.C.B.V.; Miyoshi, A.; Azevedo, V.; Langella, P.; Bermudez-Humaran, L.G.; et al. Expression of fibronectin binding protein A (FnBPA) from Staphylococcus aureus at the cell surface of Lactococcus lactis improves its immunomodulatory properties when used as protein delivery vector. Vaccine 2016, 34, 1312–1318. [Google Scholar] [CrossRef] [PubMed]
- Mancha-Agresti, P.; Drumond, M.M.; do Carmo, F.L.R.; Santos, M.M.; dos Santos, J.S.C.; Venanzi, F.; Chatel, J.-M.; Leclercq, S.Y.; Azevedo, V. A New Broad Range Plasmid for DNA Delivery in Eukaryotic Cells Using Lactic Acid Bacteria: In Vitro and In Vivo Assays. Mol. Ther. Methods Clin. Dev. 2016, 4, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Acquah, F.K.; Obboh, E.K.; Asare, K.; Boampong, J.N.; Nuvor, S.V.; Singh, S.K.; Theisen, M.; Williamson, K.C.; Amoah, L.E. Antibody responses to two new Lactococcus lactis-produced recombinant Pfs48/45 and Pfs230 proteins increase with age in malaria patients living in the Central Region of Ghana. Malar. J. 2017, 16, 306. [Google Scholar] [CrossRef]
- Mistarz, U.H.; Singh, S.K.; Nguyen, T.T.T.N.; Roeffen, W.; Yang, F.; Lissau, C.; Madsen, S.M.; Vrang, A.; Tiendrebeogo, R.W.; Kana, I.H.; et al. Expression, Purification and Characterization of GMZ2′.10C, a Complex Disulphide-Bonded Fusion Protein Vaccine Candidate against the Asexual and Sexual Life-Stages of the Malaria-Causing Plasmodium falciparum Parasite. Pharm. Res. 2017, 34, 1970–1983. [Google Scholar] [CrossRef]
- Aliramaei, M.R.; Khorasgani, M.R.; Rahmani, M.R.; Zarkesh Esfahani, S.H.; Emamzadeh, R. Expression of Helicobacter pylori CagL gene in Lactococcus lactis MG1363 and evaluation of its immunogenicity as an oral vaccine in mice. Microb. Pathog. 2020, 142, 103926. [Google Scholar] [CrossRef]
- Rezaei, M.; Khorasgani, M.R.; Esfahani, S.H.Z.; Emamzadeh, R.; Abtahi, H. Production of brucella melitensis Omp16 protein fused to the human interleukin 2 in lactococcus lactis MG1363 toward developing a lactococcus-based vaccine against brucellosis. Can. J. Microbiol. 2020, 66, 39–45. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, D.; Sun, B.; Yu, M.; Wang, Y.; Ru, Y.; Jiang, Y.; Qiao, X.; Cui, W.; Zhou, H.; et al. Oral vaccination with the porcine circovirus type 2 (PCV-2) capsid protein expressed by Lactococcus lactis induces a specific immune response against PCV-2 in mice 2. J. Appl. Microbiol. 2019. [Google Scholar] [CrossRef]
- Takiishi, T.; Cook, D.P.; Korf, H.; Sebastiani, G.; Mancarella, F.; Cunha, J.P.M.C.M.; Wasserfall, C.; Casares, N.; Lasarte, J.J.; Steidler, L.; et al. Reversal of Diabetes in NOD Mice by Clinical-Grade Proinsulin and IL-10-Secreting Lactococcus lactis in Combination with Low-Dose Anti-CD3 Depends on the Induction of Foxp3-Positive T Cells. Diabetes 2017, 66, 448–459. [Google Scholar] [CrossRef]
- Derakhshandeh, S.; Shahrokhi, N.; Khalaj, V.; Habibi, M.; Moazzezy, N.; Karam, M.R.A.; Bouzari, S. Surface display of uropathogenic Escherichia coli FimH in Lactococcus lactis: In vitro characterization of recombinant bacteria and its protectivity in animal model. Microb. Pathog. 2020, 141, 103974. [Google Scholar] [CrossRef]
- Durmaz, E.; Hu, Y.; Aroian, R.V.; Klaenhammer, T.R. Intracellular and extracellular expression of Bacillus thuringiensis crystal protein Cry5B in Lactococcus lactis for use as an anthelminthic. Appl. Environ. Microbiol. 2016, 82, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Maddaloni, M.; Kochetkova, I.; Hoffman, C.; Pascual, D.W. Delivery of IL-35 by lactococcus lactis ameliorates collagen-induced arthritis in mice. Front. Immunol. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lertcanawanichakul, M. Construction of plasmid vector for expression of bacteriocin N15-encoding gene and effect of engineered bacteria on Enterococcus faecalis. Curr. Microbiol. 2007, 54, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Duarte, S.O.D.; Martins, M.C.; Andrade, S.M.; Prazeres, D.M.F.; Monteiro, G.A. Plasmid Copy Number of pTRKH3 in Lactococcus lactis is Increased by Modification of the repDE Ribosome-Binding Site. Biotechnol. J. 2019, 14, e1800587. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.-J.; Shin, H.-J.; Han, I.-K.; Jung, W.-W.; Kim, Y.B.; Sul, D.; Oh, Y.-K. Induction of mucosal and systemic immune responses following oral immunization of mice with Lactococcus lactis expressing human papillomavirus type 16 L1. Vaccine 2007, 25, 8049–8057. [Google Scholar] [CrossRef]
- Mariat, D.; Robert, V.; Langella, P.; Chatel, J.-M. Plasmid transfer efficiency using Lactoccocus lactis strains depends on invasiveness status but also on plasmid copy number. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Foley, S.; Bron, S.; Venema, G.; Daly, C.; Fitzgerald, G.F. Molecular analysis of the replication origin of the Lactococcus lactis plasmid pCJ305. Plasmid 1996, 36, 125–141. [Google Scholar] [CrossRef][Green Version]
- O’Sullivan, D.J.; Klaenhammer, T.R. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 1993, 137, 227–231. [Google Scholar] [CrossRef]
- Marreddy, R.K.R.; Pinto, J.P.C.; Wolters, J.C.; Geertsma, E.R.; Fusetti, F.; Permentier, H.P.; Kuipers, O.P.; Kok, J.; Poolman, B. The response of Lactococcus lactis to membrane protein production. PLoS ONE 2011, 6, e24060. [Google Scholar] [CrossRef]
- Emond, E.; Lavallee, R.; Drolet, G.; Moineau, S.; LaPointe, G. Molecular characterization of a theta replication plasmid and its use for development of a two-component food-grade cloning system for Lactococcus lactis. Appl. Environ. Microbiol. 2001, 67, 1700–1709. [Google Scholar] [CrossRef]
- del Rio, B.; Redruello, B.; Fernandez, M.; Martin, M.C.; Ladero, V.; Alvarez, M.A. Lactic Acid Bacteria as a Live Delivery System for the in situ Production of Nanobodies in the Human Gastrointestinal Tract. Front. Microbiol. 2019, 9, 3179. [Google Scholar] [CrossRef]
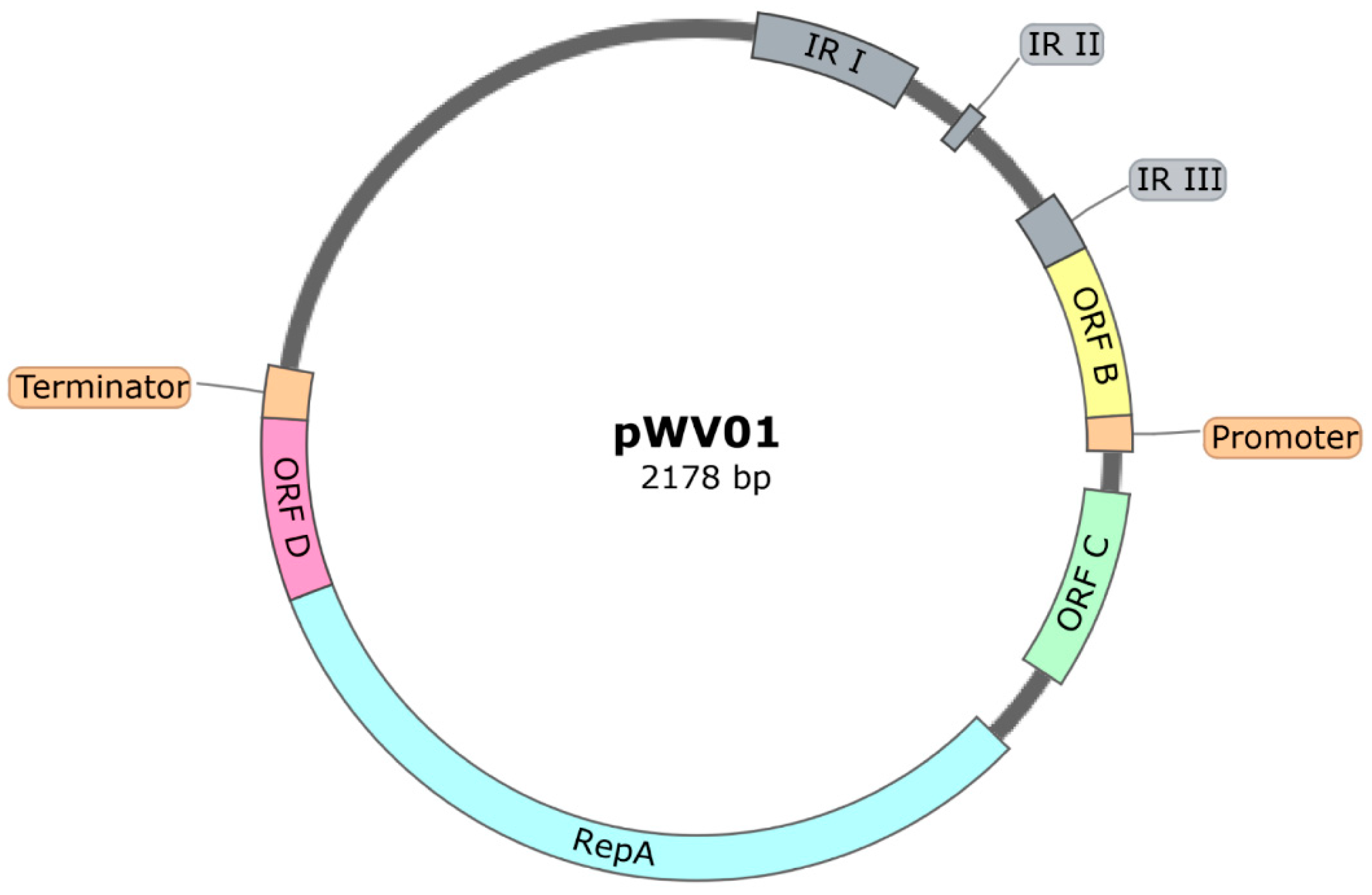
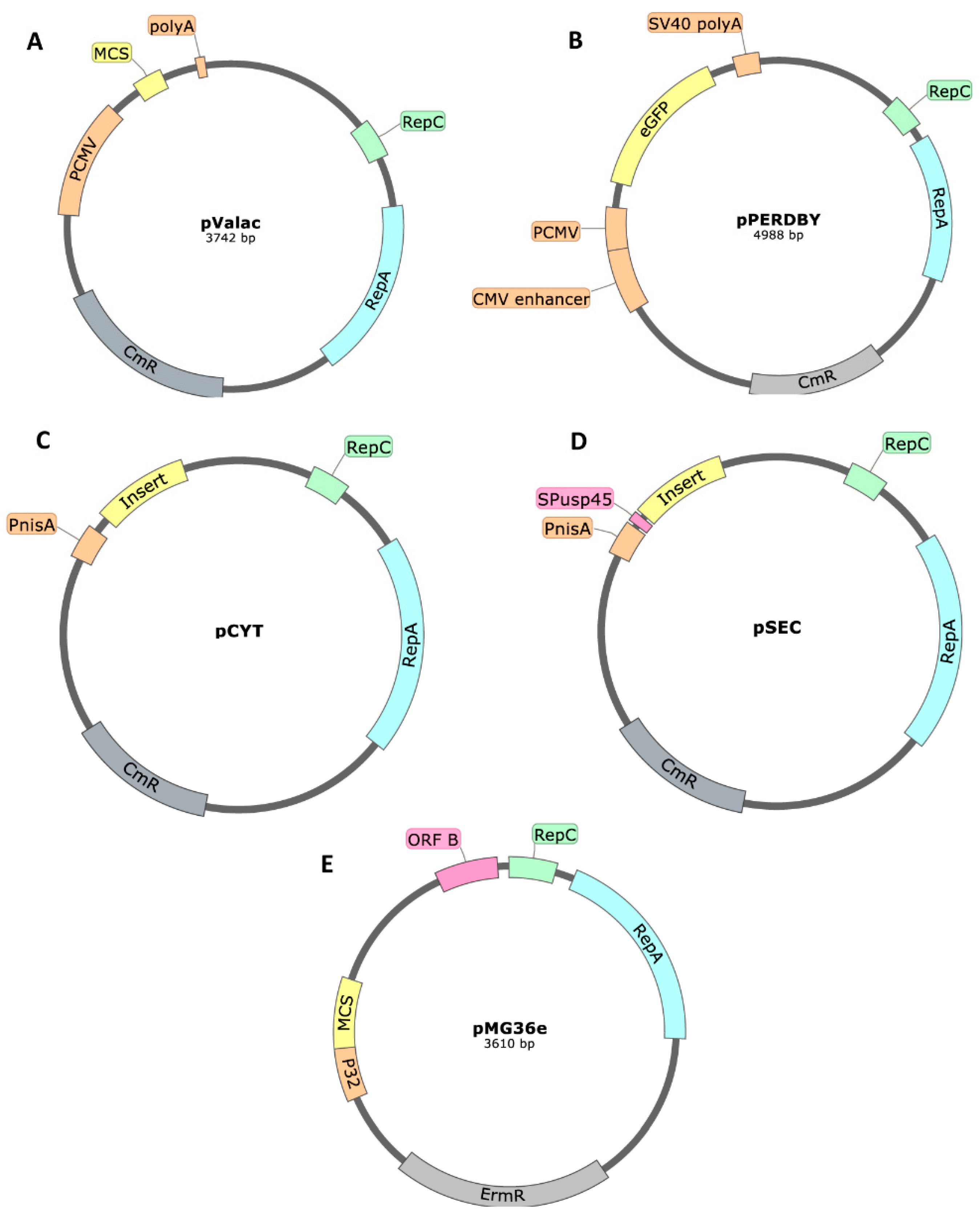
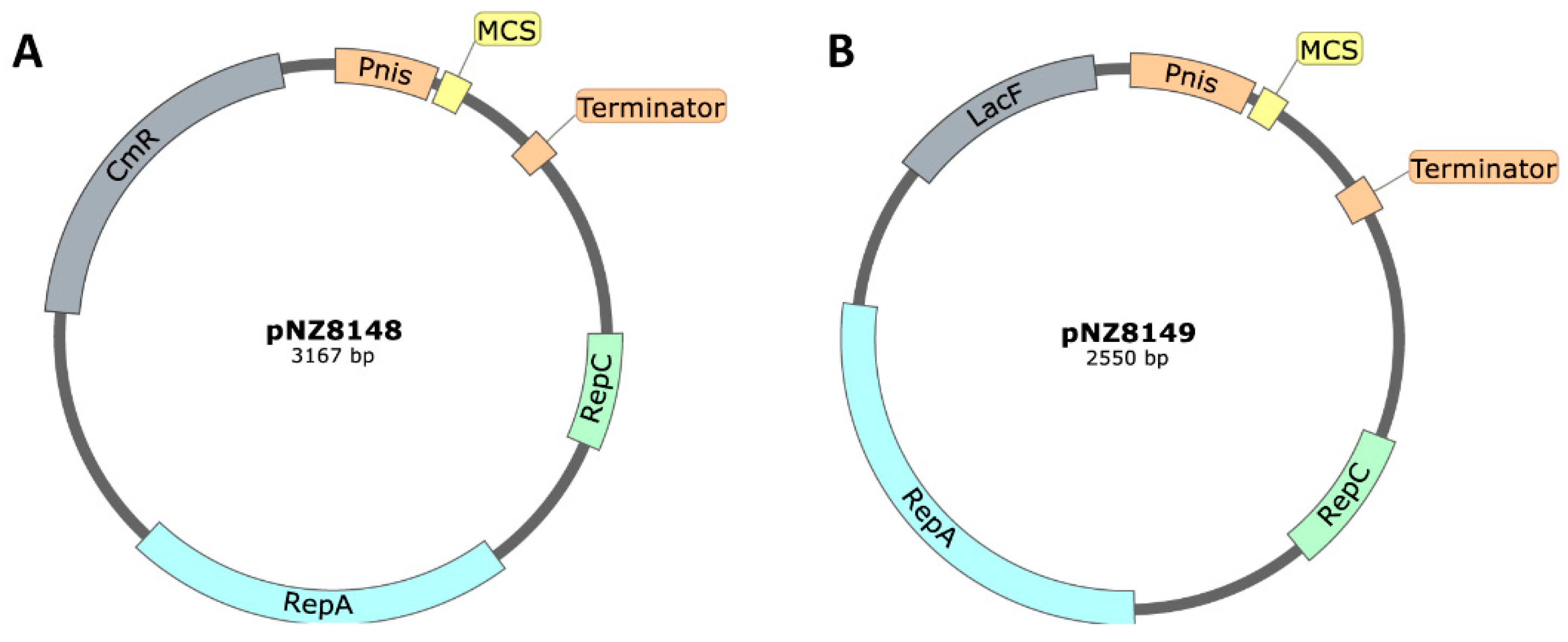
| Product | Plasmid | Replicon | Reference |
|---|---|---|---|
| L-alanine | pNZ2650 (pNZ8020 derived) | pSH71 | [16] |
| Linalool | pNZ7640 (pNZ8150 derived) | pSH71 | [17] |
| Germacrene D | pNZ8048 | pSH71 | [18] |
| β-Sesquiphellandrene | pNZ8048 | pSH71 | [19] |
| Hyaluronic acid | pSJR6 (pNZ8048 derived) | pSH71 | [20] |
| Folate (vitamin B11) | pNZ7011 (pNZ8048 derived) | pSH71 | [21,22] |
| Mabinlin II | pNZ8149 | pSH71 | [23] |
| β-Cyclodextrin glucanotransferase | pNZ8048 | pSH71 | [24] |
| Coumarate CoA ligase (4CL) | pFI2413 | pSH71 | [25] |
| Alcohol acyltransferase (SAAT) | pNZ7630 (pNZ8150 derived) | pSH71 | [17] |
| Sesquiterpene synthase | pNZ8048 | pSH71 | [18] |
| 3-Hydroxy-3-methylglutaryl CoA reductase (HMGR) | pNZ8048 | pSH71 | [19] |
| Bile salt hydrolase (BSH) | pNZ8149 | pSH71 | [26] |
| Acid urease | pNZ8148/pNZ8149 | pSH71 | [27] |
| Acetoin/diacetyl | pCIT264 | Rep264 | [28] |
| Lactic acid | - | - | [29] |
| Riboflavin (vitamin B2) | - | - | [30] |
| Ethanol | - | - | [31] |
| Disease | Antigen/Cytokine | pWV01 Based-Plasmid | Reference |
|---|---|---|---|
| Tuberculosis | Mycobacterium tuberculosis ESAT-6 | pValac | [10,68] |
| Tuberculosis | Mycobacterium tuberculosis Ag85A | pValac | [69] |
| Inflammatory bowel disease | IL-4 | pValac | [70] |
| Inflammatory bowel disease | Anti-Tumor Necrosis Factor-alpha | pValac | [71] |
| Inflammatory bowel disease | IL-10 of Mus musculus | pValac | [72] |
| Colorectal cancer | Catalase, superoxide dismutase or IL-10 | pValac/ pGroESL (pSEC derived) | [73] |
| Shigella dysenteriae diarrhea | Outer membrane protein A of S. dysenteriae | pSEC | [74] |
| Oral vaccines against cancer or infectious diseases | Carcinoembryonic antigen | pSEC | [75] |
| Inflammatory bowel disease | Mycobacterium heat shock protein 65 (Hsp65) | pSEC | [76] |
| Bovine tuberculosis | Mycobacterium leprae Hsp65 | pSEC | [77] |
| Hemorrhagic shock | Heme oxygenase-1 | pSEC | [78] |
| Intestinal mucositis | Human pancreatitis-associated protein I | pSEC | [79,80] |
| Oral vaccines against infectious diseases | Staphylococcus aureus nuclease fused with OmpH β1α1 domain of Yersinia enterocolitica | pSEC | [81] |
| Arthritis | Mycobacterium leprae Hsp65 | pXylT:SEC (pSEC derived) | [82] |
| Allergic asthma | Mycobacterium Hsp65 | pXylT:SEC (pSEC derived) | [83] |
| Shigellosis | Outer membrane protein A | pPERDBY (LacVax®, pSEC derived) | [84] |
| DNA vaccination | Enhanced Green Fluorescence Protein (EGFP) | pPERDBY (pSEC derived) | [53,54] |
| Streptococcus iniae infection | S. iniae M-like protein antigen | pCYT | [85] |
| Type 1 diabetes mellitus | Hsp65 and tandemly repeated IA2P2 | pCYT | [86] |
| Type 1 diabetes mellitus | Staphylococcal nuclease (SNase) | pCYT | [87] |
| Fish edwardsiellosis | OmpA and flagellar hook protein D antigens from Edwardsiella tarda | pCYT | [88] |
| Inflammatory bowel diseases | Pancreatitis-associated protein | pSEC and pCYT | [89] |
| Gastrointestinal lead poisoning | Human metallothionein-I fusion protein | pMG36e | [90] |
| Cysticercosis | Taenia solium TSOL18 antigen | pMG36e | [91] |
| Hand, foot, and mouth disease | Viral protein 1 of enterovirus 71 | pMG36e | [92] |
| Alzheimer’s and Parkinson’s diseases | Glucagon-like peptide-1 | pMG36e | [93] |
| Disease | Antigen/Cytokine | pSH71 Based-Plasmid | Reference |
|---|---|---|---|
| Helicobacter pylory | H. pylori adhesin A (HpaA) | pNZ8110 | [100] |
| H. pylory, allergic diseases and carcinomas | H. pylori neutrophil-activating protein A subunit | pNZ8110 | [101] |
| H. pylory vaccine adjuvant | Heat-labile enterotoxin B subunit | pNZ8149 | [102] |
| H. pylori infection | H. pylori Lpp20 antigen | pNZ8149 | [103] |
| Infectious bursal disease virus (chickens) | OmpH of an M cell-targeting ligand and IBDV-VP2 protein | pNZ8149 | [104] |
| Infectious bursal disease virus (IBDV) (chickens) | Antigen VP2 of IBDV and RCK protein of Salmonella enterica | pNZ8149 | [105] |
| Proteus mirabilis | OmpA of P. mirabilis | pNZ8149 | [106] |
| Brucellosis | Brucella melitensis bp26 gene | pNZ8149 | [107] |
| Streptococcus pyogenes infections | M-protein antigens derived from Group A Streptococcus | pNZ8149 | [108] |
| Bordetella pertussis infection | F1S1 fusion protein (N-terminal region of S1 subunit from pertussis toxin and filamentous hemagglutinin type 1 immunodominant domain) | pNZ8149 | [109,110] |
| Porcine epidemic diarrhea | Porcine Epidemic Diarrhea Virus S1Gene | pNZ8149 | [111] |
| Duck hepatitis A virus (DHAV) | VP1 protein of DHAV type 3 | pNZ8149 | [112] |
| Cancer | Azurin | pNZ8149 | [113] |
| Campylobacter jejuni infection | C. jejuni cjaA gene | pNZ8149 | [114] |
| Obesity and diabetes | Fibroblast growth factor 21 | pNZ8149 | [115] |
| Atherosclerosis and nonalcoholic fatty liver disease | Ling Zhi 8 protein | pNZ8149 | [116] |
| Enterotoxigenic Escherichia coli | Trivalent enterotoxin protein STa-LTB-STb and the F5 fimbrial antigen with OmpH of Yersinia enterocolitica | pNZ8149 | [117] |
| Bird flu H5N1 | Hemagglutinin from an Avian H5N1 isolate | pNZ8150 | [118] |
| H1N1 | H1N1 2009 haemagglutinin1 and nisP anchor fusion protein | pNZ8048 | [119] |
| Type 2 diabetes | Exendin-4 (Exd4) | pNZ8048 | [120] |
| Pathogenic avian influenzavirus infection | Ectodomain of influenza matrix protein 2 and neuraminidase | pNZ8048 | [121] |
| Bovine tuberculosis | Mycobacterium bovis antigens MPB70 and MPB83 | pNZ8048 | [122] |
| C. jejuni infection | C. jejuni surface lipoprotein A | pNZ8048 | [123] |
| Avian coccidiosis (Eimeria tenella infection) | E. tenella 3-1E protein | pTX8048 (pNZ8048 derived) | [59,124] |
| Avian coccidiosis (E. tenella infection) | Apical membrane antigen 1 of E. tenella | pTX8048 (pNZ8048 derived) | [125] |
| Hepatitis-splenomegaly syndrome in chickens | Avian hepatitis E virus truncated ORF2 protein | pTX8048 (pNZ8048 derived) | [126] |
| Thrombosis | Subtilisin QK-2 | pRF01 (pNZ8149 derived) and pRF03 (pNZ8048 derived) | [57] |
| Diabetes | Single-chain insulin (SCI-59) analog | pMRF5018 (pNZ8149 derived) and pMRF5019 (pNZ8048 derived) | [58] |
| Human colon carcinoma | Bioactive kisspeptin (KiSS1) | pNZ401 (pNZ8048 derived) | [127] |
| Shiga Toxin 1 B subunit, from Shiga toxin-producing bacteria, like enterohemorrhagic E. coli and Shigella dysenteriae | Albumin-binding domain variants | pNZ8148 | [128] |
| Inflammatory diseases, autoimmune diseases and cancer | Single-chain variable fragment antibody against mouse interleukin 6 | pNZ8148 | [129] |
| Inflammatory bowel diseases | Porcine insulin-like growth factor I | pNZ8148 | [130] |
| House dust mite allergy | House dust mite allergen | pNZ8148 | [131] |
| Liver detoxification | CYP3A4 and two isoforms of MGST1 | pNZ8148 | [56] |
| Japanese cedar pollinosis | Gene encoding fused T cell epitopes from the Cry j 1 and Cry j 2 antigens | pNZ8148 | [132] |
| Enteropathogenic E. coli K.1.1 and human cervical carcinoma (HeLa) cells | Signal peptide gene (plnA) and bacteriocin encoding gene, plantaricin E (plnE) | pNZ8148 | [133] |
| Gastrointestinal inflammation | Pentadecapeptide BPC-157 | pNZ8148 | [134] |
| Ulcerative colitis | Interleukin (IL)-35 | pNZ8148 | [135] |
| Inflammatory bowel disease | IL-23 Receptor-Targeted REX Protein Blockers | pNZ8148 | [136] |
| Shiga toxins of E. coli O157:H7 and S. dysenteriae | Shiga toxin Stx2a1 | pNZ8148 | [137] |
| E. coli O157 and Shigella flexneri | Intimin and IpaB | pNZ8148 | [138] |
| Cancer | Anti‑human cytotoxic T lymphocyte-associated antigen 4 Single‑Chain Fragment Variable | pNZ8148 | [139] |
| H9N2 AvianInfluenza Virus (chickens) | M1 and HA2 proteins derived from an antigenically conserved endemic H9N2 virus strain | pNZ8148 | [140] |
| Ulcerative colitis | Human defensin-5 | pNZ8148 | [141] |
| Foot-and-mouth disease | Foot-and-mouth disease virus VP1 gene | pNZ8148 | [142] |
| Viral haemorrhagic septicaemia (fish) | G gene of viral hemorrhagic septicaemia virus | pNZ8148 | [143] |
| H5N1 | H5N1 infuenza virus haemagglutinin | pNZ8148 | [144] |
| Spring viremia of carp virus (fish) | Spring viremia of carp virus glycoprotein | pNZ8148 | [145] |
| Allergies | Bioactive anti‑interleukin‑4 single‑chain fragment variable | pNZ8148#2 | [146] |
| Sepsis | Recombinant bovine lactoferrin | pNZ8148:CYT | [147] |
| Emphysema | Heme Oxygenase-1 | pNZ8148#2:SEC | [148] |
| Inflammatory bowel disease | Interleukin 1 receptor antagonist | pNZ8148#2:SEC | [149] |
| Hirame novirhabdovirus | Hirame novirhabdovirus (HIRRV) G protein | pSLC (pNZ8148-derived) | [150] |
| Group A Streptococcus (GAS) | Fibronectin-collagen-T-antigen (FCT)-FCT-3 and FCT-4 | pLZ12km2 | [151] |
| Peptide vaccine | GAS serotype M1 pilus (PilM1) encoded in the FCT-2 region and Ova324–339 | pilVax (pLZ12km2 derived) | [152] |
| Staphylococcus aureus infection | Linear B-cell epitope, D3(22–33), from the fibronectin-binding protein A of Staphylococcus aureus | pilVax (pLZ12km2 derived) | [153] |
| Human papillomavirus type 16-induced tumors | E6 oncogene | pNZ8123 | [154,155] |
| HPV-16 | Codon optimized E7 oncogenes isolated from 30 Iranian HPV-16 | pNZ8148/pNZ8123 | [156,157,158,159,160] |
| Cutaneous leishmaniasisis | PpSP15-EGFP | pNZ8121 | [161] |
| Non-bacterial acute gastroenteritis | VP1 gene of a Human Norovirus GII.4 | pNZ8150 | [162] |
| Group B Streptococcus infections | Surface immune protein from Group B Streptococcus | pNZ8124 | [163] |
| Colorectal cancer | Tumoricidal human tumor necrosis factor-related apoptosis-inducing ligand | pNZ8124 | [164] |
| Brucellosis | Omp31 Antigen of Brucella melitensis | pNZ7021 | [165] |
| Allergy to Amaranthus retroflexus pollens | Ama r 2 gene | pNZ7025 | [166] |
| Yersinia pseudotuberculosis infection | LcrV from Y. pseudotuberculosis | pMEC237 (pNZYR derived) | [167,168] |
| Disease | Antigen/Cytokine | pAMβ1 Based-Plasmid | Reference |
|---|---|---|---|
| Campylobacter jejuni infection | C. jejuni antigens | pIL253 | [174] |
| Multiple sclerosis | Myelin peptides | pIL253 | [173] |
| Colitis | cDNA of Microbial Anti-inflammatory Molecule (MAM) from Faecalibacterium prausnitzii | pILMAM (pIL253 derived) | [172] |
| HPV-16-induced tumors | HPV-16-E7 antigen | pOri23 (pIL253 derived) | [175] |
| DNA vaccine | eGFP | pExu (pOri253 derived) | [176] |
| Malaria | Pfs230 and Pfs48/45 | pLEA2 (pAMJ328 derivative) | [177] |
| Malaria | Epitopes of key malaria parasite antigens: glutamate-rich protein (GLURP), merozoite surface protein 3 (MSP3), and the highly disulphide bonded Pfs48/45 (10C) | pSS3 (pAMJ328 derivative) | [178] |
| H. pylory infection | H. pylori cagL gene | pAMJ2008 | [179] |
| Brucellosis | Omp16-IL2 fusion protein antigen | pAMJ2008 | [180] |
| Porcine circovirus type 2 (PCV2) | Cap protein of PCV2 | pAMJ399 | [181] |
| Diabetes | Human proinsulin and IL-10 | pT1NX (pTREX derivative) | [182] |
| Urinary tract infections | Uropathogenic E. coli FimH | pT1NX (pTREX derivative) | [183] |
| Parasitic worms | Bacillus thuringiensis crystal protein Cry5B | pTRK593 (pTRKH2 derived) | [184] |
| Rheumatoid arthritis | Murine IL-35 | pMSP3535H3 | [185] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duarte, S.O.D.; Monteiro, G.A. Plasmid Replicons for the Production of Pharmaceutical-Grade pDNA, Proteins and Antigens by Lactococcus lactis Cell Factories. Int. J. Mol. Sci. 2021, 22, 1379. https://doi.org/10.3390/ijms22031379
Duarte SOD, Monteiro GA. Plasmid Replicons for the Production of Pharmaceutical-Grade pDNA, Proteins and Antigens by Lactococcus lactis Cell Factories. International Journal of Molecular Sciences. 2021; 22(3):1379. https://doi.org/10.3390/ijms22031379
Chicago/Turabian StyleDuarte, Sofia O.D., and Gabriel A. Monteiro. 2021. "Plasmid Replicons for the Production of Pharmaceutical-Grade pDNA, Proteins and Antigens by Lactococcus lactis Cell Factories" International Journal of Molecular Sciences 22, no. 3: 1379. https://doi.org/10.3390/ijms22031379
APA StyleDuarte, S. O. D., & Monteiro, G. A. (2021). Plasmid Replicons for the Production of Pharmaceutical-Grade pDNA, Proteins and Antigens by Lactococcus lactis Cell Factories. International Journal of Molecular Sciences, 22(3), 1379. https://doi.org/10.3390/ijms22031379