Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis
Abstract
:1. Introduction
2. Oral Malignant and Potentially Malignant Disorders
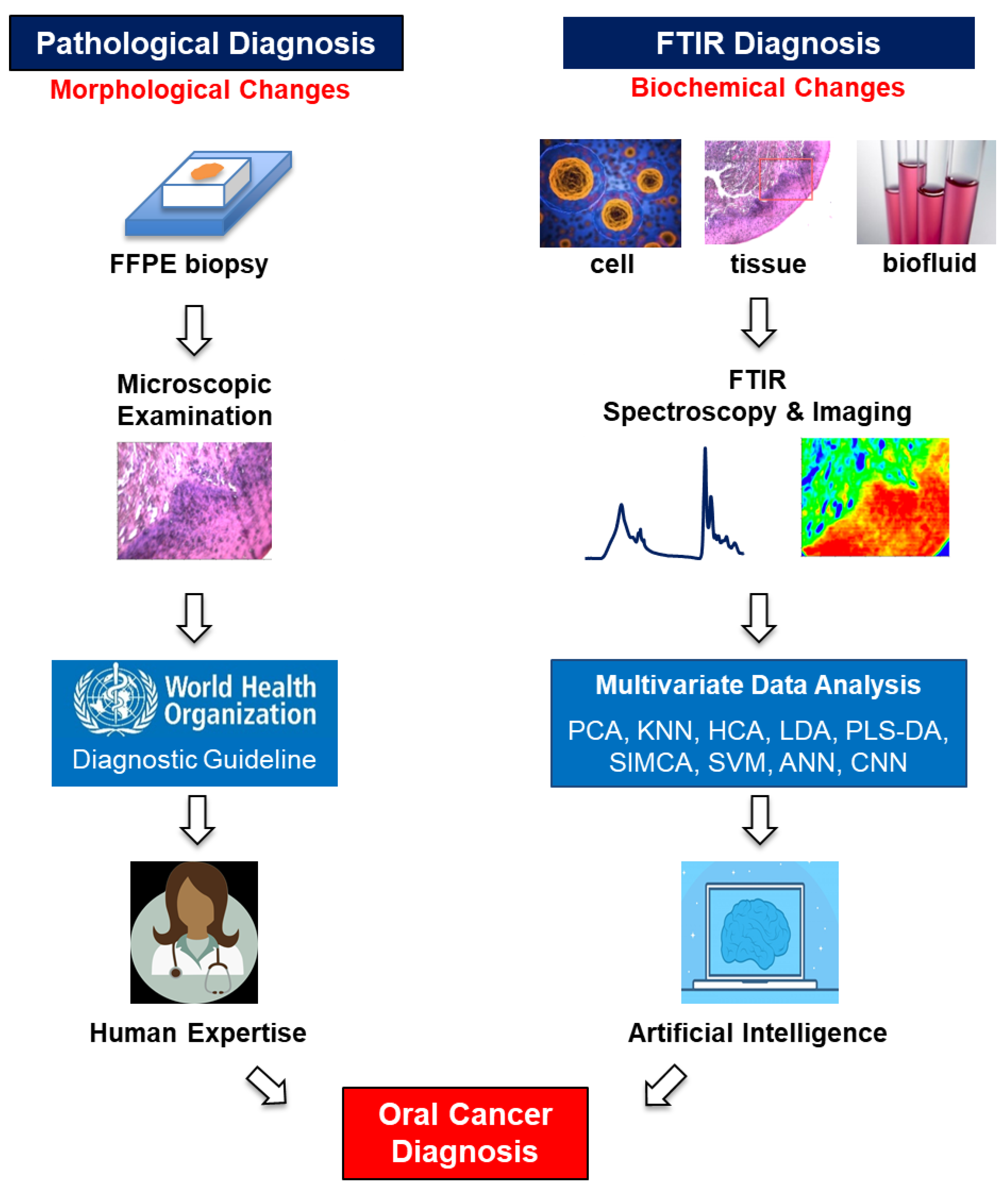
3. Current Diagnostic/Grading/Staging/Methods and Limitations
4. Fourier Transform Infrared Spectroscopy/Microspectroscopy
4.1. Fourier Transform Infrared (FTIR) Fundamentals
4.2. FTIR Sample Techniques
4.3. FTIR Microspectroscopy
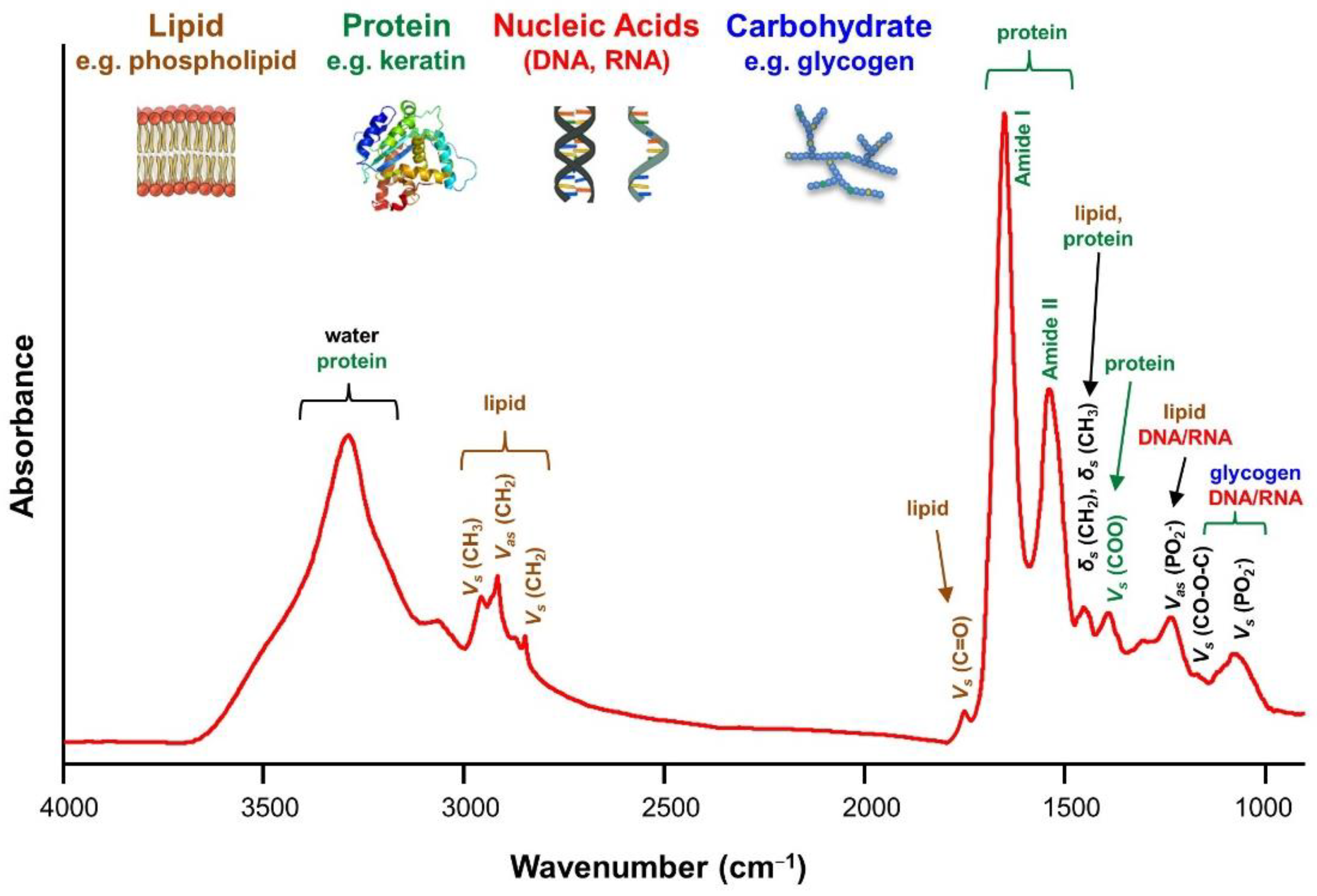
4.4. Common FTIR Bands for Biomolecules
4.5. Comparison of FTIR with Other Spectroscopic Diagnostic Techniques
5. Signal Preprocessing and Data Analysis
5.1. Signal Preprocessing
5.2. Exploratory Analysis
5.3. Classification Modeling Process
5.4. Clustering and Classification Methods
5.5. Model Performance Validation
6. FTIR for Oral Cancer Diagnosis
6.1. Oral Tissue Studies
6.2. Oral Cell Studies (FTIR Cytopathology)
6.3. Biofluid Studies
6.4. Tumor Microenvironment Study
7. Clinical Translation of FTIR
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Cancer Prevention. Available online: https://www.who.int/cancer/prevention/diagnosis-screening/oral-cancer/en/ (accessed on 20 December 2020).
- Miller, K.D.; Siegel, R.L.; Khan, R.; Jemal, A. Cancer Statistics. Cancer Rehabil. 2018, 70, 7–30. [Google Scholar] [CrossRef]
- Farah, C.S.; Woo, S.-B.; Zain, R.B.; Sklavounou, A.; McCullough, M.J.; Lingen, M. Oral Cancer and Oral Potentially Malignant Disorders. Int. J. Dent. 2014, 2014, 853479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epstein, J.B.; Güneri, P.; Boyacioglu, H.; Abt, E. The limitations of the clinical oral examination in detecting dysplastic oral lesions and oral squamous cell carcinoma. J. Am. Dent. Assoc. 2012, 143, 1332–1342. [Google Scholar] [CrossRef] [PubMed]
- Speight, P.M.; Khurram, S.A.; Kujan, O. Oral potentially malignant disorders: Risk of progression to malignancy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 612–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Ibrahim, O.; Byrne, H.J.; Mikkonen, J.W.; Koistinen, A.; Kullaa, A.M.; Lyng, F.M. Recent advances in optical diagnosis of oral cancers: Review and future perspectives. Head Neck 2015, 38, E2403–E2411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warnakulasuriya, S. Oral potentially malignant disorders: A comprehensive review on clinical aspects and management. Oral Oncol. 2020, 102, 104550. [Google Scholar] [CrossRef] [PubMed]
- Lousada-Fernandez, F.; Óscar, R.-G.; López-Cedrún, J.L.; López-López, R.; Muinelo-Romay, L.; Mercedes, S.-C.M. Liquid Biopsy in Oral Cancer. Int. J. Mol. Sci. 2018, 19, 1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasthuri, M.; Babu, N.A.; Masthan, K.M.K.; Sankari, S.L. Toludine Blue Staining in The Diagnosis of Oral Precancer and Cancer: Stains, Technique and its Uses—A Review. Biomed. Pharmacol. J. 2015, 8, 519–522. [Google Scholar] [CrossRef]
- Shashidara, R.; Sreeshyla, H.S.; Sudheendra, U.S. Chemiluminescence: A diagnostic adjunct in oral precancer and cancer: A review. J. Cancer Res. Ther. 2014, 10, 487–491. [Google Scholar]
- Cicciù, M.; Cervino, G.; Fiorillo, L.; D’Amico, C.; Oteri, G.; Troiano, G.; Zhurakivska, K.; Muzio, L.L.; Herford, A.S.; Crimi, S.; et al. Early Diagnosis on Oral and Potentially Oral Malignant Lesions: A Systematic Review on the VELscope® Fluorescence Method. Dent. J. 2019, 7, 93. [Google Scholar] [CrossRef] [Green Version]
- Nagi, R.; Reddy-Kantharaj, Y.-B.; Rakesh, N.; Janardhan-Reddy, S.; Sahu, S. Efficacy of light based detection systems for early detection of oral cancer and oral potentially malignant disorders: Systematic review. Med. Oral Patol. Oral Cir. Bucal 2016, 21, e447–e455. [Google Scholar] [CrossRef] [PubMed]
- Lingen, M.W.; Tampi, M.P.; Urquhart, O.; Abt, E.; Agrawal, N.; Chaturvedi, A.K.; Cohen, E.; D’Souza, G.; Gurenlian, J.; Kalmar, J.R.; et al. Adjuncts for the evaluation of potentially malignant disorders in the oral cavity: Diagnostic test accuracy systematic review and meta-analysis-a report of the American Dental Association. J. Am. Dent. Assoc. 2017, 148, 797–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatia, A.K.; Burtness, B. Novel Molecular Targets for Chemoprevention in Malignancies of the Head and Neck. Cancers 2017, 9, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liyanage, C.; Wathupola, A.; Muraleetharan, S.; Perera, K.; Punyadeera, C.; Udagama, P. Promoter Hypermethylation of Tumor-Suppressor Genes p16(INK4a), RASSF1A, TIMP3, and PCQAP/MED15 in Salivary DNA as a Quadruple Biomarker Panel for Early Detection of Oral and Oropharyngeal Cancers. Biomolecules 2019, 9, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yete, S.; Saranath, D. MicroRNAs in oral cancer: Biomarkers with clinical potential. Oral Oncol. 2020, 110, 105002. [Google Scholar] [CrossRef] [PubMed]
- Celentano, A.; Glurich, I.; Borgnakke, W.S.; Farah, C.S. World Workshop on Oral Medicine VII: Prognostic biomarkers in oral leukoplakia and proliferative verrucous leukoplakia—A systematic review of retrospective studies. Oral Dis. 2020. [Google Scholar] [CrossRef]
- Radhika, T.; Jeddy, N.; Nithya, S.; Muthumeenakshi, R. Salivary biomarkers in oral squamous cell carcinoma—An insight. J. Oral Biol. Craniofac. Res. 2016, 6, S51–S54. [Google Scholar] [CrossRef] [Green Version]
- Bogomolny, E.; Huleihel, M.; Suproun, Y.; Sahu, R.K.; Mordechai, S. Early spectral changes of cellular malignant transformation using Fourier transform infrared microspectroscopy. J. Biomed. Opt. 2007, 12, 024003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papamarkakis, K.; Bird, B.; Schubert, J.M.; Miljković, M.; Wein, R.; Bedrossian, K.; Laver, N.M.V.; Diem, M. Cytopathology by optical methods: Spectral cytopathology of the oral mucosa. Lab. Investig. 2010, 90, 589–598. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Srinivasan, A.; Nikolajeff, F. Role of Infrared Spectroscopy and Imaging in Cancer Diagnosis. Curr. Med. Chem. 2017, 25, 1055–1072. [Google Scholar] [CrossRef]
- Bellisola, G.; Sorio, C. Infrared spectroscopy and microscopy in cancer research and diagnosis. Am. J. Cancer Res. 2011, 2, 1–21. [Google Scholar] [PubMed]
- Bel’Skaya, L.V. Use of IR Spectroscopy in Cancer Diagnosis. A Review. J. Appl. Spectrosc. 2019, 86, 187–205. [Google Scholar] [CrossRef]
- Su, K.-Y.; Lee, W.-L. Fourier Transform Infrared Spectroscopy as a Cancer Screening and Diagnostic Tool: A Review and Prospects. Cancers 2020, 12, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambert, R.; Sauvaget, C.; Cancela, M.D.C.; Sankaranarayanan, R. Epidemiology of cancer from the oral cavity and oropharynx. Eur. J. Gastroenterol. Hepatol. 2011, 23, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Messadi, D.V. Diagnostic aids for detection of oral precancerous conditions. Int. J. Oral Sci. 2013, 5, 59–65. [Google Scholar] [CrossRef]
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884–11894. [Google Scholar] [PubMed]
- Mello, F.W.; Miguel, A.F.P.; Dutra, K.L.; Porporatti, A.L.; Warnakulasuriya, S.; Guerra, E.N.S.; Rivero, E.R.C. Prevalence of oral potentially malignant disorders: A systematic review and meta-analysis. J. Oral Pathol. Med. 2018, 47, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Villa, A.; Sonis, S. Oral leukoplakia remains a challenging condition. Oral Dis. 2018, 24, 179–183. [Google Scholar] [CrossRef]
- Scully, C. Challenges in predicting which oral mucosal potentially malignant disease will progress to neoplasia. Oral Dis. 2013, 20, 1–5. [Google Scholar] [CrossRef]
- Warnakulasuriya, S.; Ariyawardana, A. Malignant transformation of oral leukoplakia: A systematic review of observational studies. J. Oral Pathol. Med. 2016, 45, 155–166. [Google Scholar] [CrossRef]
- Gupta, S.; Gupta, S. Role of human papillomavirus in oral squamous cell carcinoma and oral potentially malignant disorders: A review of the literature. Indian J. Dent. 2015, 6, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Gómez, S.; Chimenos-Küstner, E.; Marí, A.; Tous, S.; Penin, R.; Clavero, O.; Quirós, B.; Pavon, M.A.; Taberna, M.; Alemany, L.; et al. Human papillomavirus in premalignant oral lesions: No evidence of association in a Spanish cohort. PLoS ONE 2019, 14, e0210070. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, Y. Human papillomavirus infection in oral potentially malignant disorders and cancer. Arch. Oral Biol. 2017, 83, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, K.; Kavitha, L. Oral epithelial dysplasia: Classifications and clinical relevance in risk assessment of oral potentially malignant disorders. J. Oral Maxillofac. Pathol. 2019, 23, 19–27. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, A.K.; Grandis, J.R.; Takata, T.; Slootweg, P.J. WHO Classification of Head and Neck Tumours; World Health Organization: Geneva, Switzerland, 2017; Volume 9. [Google Scholar]
- Mehanna, H.; Rattay, T.; Smith, J.; McConkey, C.C. Treatment and follow-up of oral dysplasia—A systematic review and meta-analysis. Head Neck 2009, 31, 1600–1609. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S.; Reibel, J.; Bouquot, J.; Dabelsteen, E. Oral epithelial dysplasia classification systems: Predictive value, utility, weaknesses and scope for improvement. J. Oral Pathol. Med. 2008, 37, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Awadallah, M.; Idle, M.; Patel, K.; Kademani, D. Management update of potentially premalignant oral epithelial lesions. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 628–636. [Google Scholar] [CrossRef] [Green Version]
- Tilakaratne, W.M.; Sherriff, M.; Morgan, P.R.; Odell, E.W. Grading oral epithelial dysplasia: Analysis of individual features. J. Oral Pathol. Med. 2011, 40, 533–540. [Google Scholar] [CrossRef]
- Speight, P.M. Update on Oral Epithelial Dysplasia and Progression to Cancer. Head Neck Pathol. 2007, 1, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Dost, F.; Cao, K.L.; Ford, P.; Ades, C.; Farah, C.S. Malignant transformation of oral epithelial dysplasia: A real-world evaluation of histopathologic grading. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2014, 117, 343–352. [Google Scholar] [CrossRef] [Green Version]
- Silverman, S., Jr.; Gorsky, M.; Lozada, F. Oral leukoplakia and malignant transformation. A follow-up study of 257 patients. Cancer 1984, 53, 563–568. [Google Scholar] [PubMed]
- Warnakulasuriya, S.; Johnson, N.W.; van der Waal, I. Nomenclature and classification of potentially malignant disorders of the oral mucosa. J. Oral Pathol. Med. 2007, 36, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Fischer, D.J.; Epstein, J.B.; Morton, T.H., Jr.; Schwartz, S.M. Interobserver reliability in the histopathologic diagnosis of oral pre-malignant and malignant lesions. J. Oral Pathol. Med. 2004, 33, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Karabulut, A.; Reibel, J.; Therkildsen, M.H.; Praetorius, F.; Nielsen, H.W.; Dabelsteen, E. Observer variability in the histologic assessment of oral premalignant lesions. J. Oral Pathol. Med. 1995, 24, 198–200. [Google Scholar] [CrossRef] [PubMed]
- Weaver, D.L.; Rosenberg, R.D.; Barlow, W.E.; Ichikawa, L.; Carney, P.A.; Kerlikowske, K.; Buist, D.S.; Geller, B.M.; Key, C.R.; Maygarden, S.J.; et al. Pathologic findings from the Breast Cancer Surveillance Consortium: Population-based outcomes in women undergoing biopsy after screening mammography. Cancer 2006, 106, 732–742. [Google Scholar] [CrossRef]
- Almangush, A.; Mäkitie, A.A.; Triantafyllou, A.; Strojan, P.; Strojan, P.; Rinaldo, A.; Hernandez-Prera, J.C.; Suárez, C.; Kowalski, L.P.; Ferlito, A.; et al. Staging and grading of oral squamous cell carcinoma: An update. Oral Oncol. 2020, 107, 104799. [Google Scholar] [CrossRef] [PubMed]
- Roland, N.J.; Daslin, A.W.; Nash, J.; Stell, P.M. Value of grading squamous cell carcinoma of the head and neck. Head Neck 1992, 14, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta Bioenerg. 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kourkoumelis, N.; Zhang, X.; Lin, Z.; Wang, J. Fourier Transform Infrared Spectroscopy of Bone Tissue: Bone Quality Assessment in Preclinical and Clinical Applications of Osteoporosis and Fragility Fracture. Clin. Rev. Bone Miner. Metab. 2019, 17, 24–39. [Google Scholar] [CrossRef]
- Fabian, H.; Naumann, D. Methods to study protein folding by stopped-flow FT-IR. Methods 2004, 34, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, H.; Garlapati, C.; Janssen, E.A.M.; Krishnamurti, U.; Qin, G.; Aneja, R.; Perera, A.G.U. Protein Conformational Changes in Breast Cancer Sera Using Infrared Spectroscopic Analysis. Cancers 2020, 12, 1708. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.G.; Martin-Hirsch, P.L.; Martin, F.L. Discrimination of Base Differences in Oligonucleotides Using Mid-Infrared Spectroscopy and Multivariate Analysis. Anal. Chem. 2009, 81, 5314–5319. [Google Scholar] [CrossRef] [PubMed]
- Petibois, C.; Déléris, G. Evidence that erythrocytes are highly susceptible to exercise oxidative stress: FT-IR spectrometric studies at the molecular level. Cell Biol. Int. 2005, 29, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Benseny-Cases, N.; Klementieva, O.; Cotte, M.; Ferrer, I.; Cladera, J. Microspectroscopy (μFTIR) Reveals Co-localization of Lipid Oxidation and Amyloid Plaques in Human Alzheimer Disease Brains. Anal. Chem. 2014, 86, 12047–12054. [Google Scholar] [CrossRef] [PubMed]
- Siesler, H.W. Near-Infrared Spectra, Interpretation. In Encyclopedia of Spectroscopy and Spectrometry, 3rd ed.; Lindon, J.C., Tranter, G.E., Koppenaal, D.W., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 30–39. [Google Scholar]
- Tsai, S.-R.; Hamblin, M.R. Biological effects and medical applications of infrared radiation. J. Photochem. Photobiol. B Biol. 2017, 170, 197–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beć, K.B.; Grabska, J.; Huck, C.W. Biomolecular and bioanalytical applications of infrared spectroscopy—A review. Anal. Chim. Acta 2020, 1133, 150–177. [Google Scholar] [CrossRef] [PubMed]
- Kazarian, S.G.; Chan, K. Applications of ATR-FTIR spectroscopic imaging to biomedical samples. Biochim. Biophys. Acta Biomembr. 2006, 1758, 858–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pallua, J.D.; Brunner, A.; Zelger, B.; Stalder, R.; Unterberger, S.H.; Schirmer, M.; Tappert, M. Clinical infrared microscopic imaging: An overview. Pathol. Res. Pract. 2018, 214, 1532–1538. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Fringeli, V.P. Membrane Spectroscopy; Springer: New York, NY, USA, 1981. [Google Scholar]
- Casal, H.L.; Mantsch, H.H. Polymorphic phase behaviour of phospholipid membranes studied by infrared spectroscopy. Biochim. Biophys. Acta Rev. Biomembr. 1984, 779, 381–401. [Google Scholar] [CrossRef]
- Siebert, P.H.F. Vibrational Spectroscopy in Life Science; John Wiley & Sons: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2017, 185, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Kendall, C.; Stone, N.; Notingher, I. Raman spectroscopy for medical diagnostics—From in-vitro biofluid assays to in-vivo cancer detection. Adv. Drug Deliv. Rev. 2015, 89, 121–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cals, F.L.; Koljenovic, S.; Hardillo, J.A.; de Jong, R.J.B.; Schut, T.C.B.; Puppels, G.J. Development and validation of Raman spectroscopic classification models to discriminate tongue squamous cell carcinoma from non-tumorous tissue. Oral Oncol. 2016, 60, 41–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Yan, B.; Xue, L.; Li, Y.; Luo, X.; Ji, P. Surface-enhanced Raman spectroscopy of blood serum based on gold nanoparticles for the diagnosis of the oral squamous cell carcinoma. Lipids Health Dis. 2017, 16, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeng, M.-J.; Sharma, M.; Sharma, L.; Chao, T.-Y.; Huang, S.-F.; Chang, L.-B.; Wu, S.-L.; Chow, L. Raman Spectros-copy Analysis for Optical Diagnosis of Oral Cancer Detection. J. Clin. Med. 2019, 8, 1313. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Elías, M.G.; González, F.J. Raman Spectroscopy for In Vivo Medical Diagnosis; InTech: London, UK, 2018. [Google Scholar]
- Gillenwater, A.; Jacob, R.; Ganeshappa, R.; Kemp, B.; El-Naggar, A.K.; Palmer, J.L.; Clayman, G.; Mitchell, M.F.; Richards-Kortum, R. Noninvasive diagnosis of oral neoplasia based on fluorescence spectroscopy and native tis-sue autofluorescence. Arch. Otolaryngol. Head Neck Surg. 1998, 124, 1251–1258. [Google Scholar] [CrossRef] [Green Version]
- Majumder, S.; Krishna, H.; Muttagi, S.; Gupta, P.; Chaturvedi, P. Fluorescence spectroscopy for noninvasive early diagnosis of oral mucosal malignant and potentially malignant lesions. J. Cancer Res. Ther. 2010, 6, 497–502. [Google Scholar] [CrossRef]
- Luo, X.; Xu, H.; He, M.; Han, Q.; Wang, H.; Sun, C.; Li, J.; Jiang, L.; Zhou, Y.; Dan, H.; et al. Accuracy of autofluorescence in diagnosing oral squamous cell carcinoma and oral potentially malignant disor-ders: A comparative study with aero-digestive lesions. Sci. Rep. 2016, 6, 29943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macey, R.; Walsh, T.; Brocklehurst, P.; Kerr, A.R.; Liu, J.L.; Lingen, M.W.; Ogden, G.R.; Warnakulasuriya, S.; Scul-ly, C. Diagnostic tests for oral cancer and potentially malignant disorders in patients presenting with clinically evi-dent lesions. Cochrane Database Syst. Rev. 2015, 5, Cd010276. [Google Scholar]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Ra-man and infrared spectroscopy. EPJ Tech. Instrum. 2015, 2, 8. [Google Scholar] [CrossRef] [Green Version]
- Harthcock, M.A.; Atkin, S.C. Imaging with Functional Group Maps Using Infrared Microspectroscopy. Appl. Spectrosc. 1988, 42, 449–455. [Google Scholar] [CrossRef]
- Ami, D.; Natalello, A.; Doglia, S.M. Fourier Transform Infrared Microspectroscopy of Complex Biological Systems: From Intact Cells to Whole Organisms. In Stem Cell Niche; Springer Science and Business Media LLC: Berlin/Hedelberg, Germany, 2012; Volume 895, pp. 85–100. [Google Scholar]
- Deming, S.N. Chemometrics: An overview. Clin. Chem. 1986, 32, 1702–1706. [Google Scholar] [CrossRef] [PubMed]
- Morais, C.L.M.; Lima, K.M.G.; Singh, M.; Martin, F.L. Tutorial: Multivariate classification for vibrational spec-troscopy in biological samples. Nat. Protoc. 2020, 15, 2143–2162. [Google Scholar] [CrossRef]
- Berisha, S.; Lotfollahi, M.; Jahanipour, J.; Gurcan, I.; Walsh, M.; Bhargava, R.; van Nguyen, H.; Mayerich, D. Deep learning for FTIR histology: Leveraging spatial and spectral features with convolutional neural networks. Analyst 2019, 144, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Šimundić, A.-M. Measures of Diagnostic Accuracy: Basic Definitions. EJIFCC 2009, 19, 203–211. [Google Scholar] [PubMed]
- Schultz, C.P.; Mantsch, H.H. Biochemical imaging and 2D classification of keratin pearl structures in oral squa-mous cell carcinoma. Cell. Mol. Biol. 1998, 44, 203–210. [Google Scholar]
- Schultz, C.P.; Liu, K.Z.; Kerr, P.D.; Mantsch, H.H. In situ infrared histopathology of keratinization in human oral/oropharyngeal squamous cell carcinoma. Oncol. Res. Feature Preclin. Clin. Cancer Ther. 1998, 10, 277–286. [Google Scholar]
- Fukuyama, Y.; Yoshida, S.; Yanagisawa, S.; Shimizu, M. A study on the differences between oral squamous cell carcinomas and normal oral mucosas measured by Fourier transform infrared spectroscopy. Biospectroscopy 1999, 5, 117–126. [Google Scholar] [CrossRef]
- Wu, J.-G.; Xu, Y.-Z.; Sun, C.-W.; Soloway, R.D.; Xu, D.-F.; Wu, Q.-G.; Sun, K.-H.; Weng, S.-F.; Xu, G.-X. Distinguishing malignant from normal oral tissues using FTIR fiber-optic techniques. Biopolymers 2001, 62, 185–192. [Google Scholar] [CrossRef]
- Bruni, P.; Conti, C.; Giorgini, E.; Pisani, M.; Rubini, C.; Tosi, G. Histological and microscopy FT-IR imaging study on the proliferative activity and angiogenesis in head and neck tumours. Faraday Discuss. 2003, 126, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Conti, C.; Giorgini, E.; Pieramici, T.; Rubini, C.; Tosi, G. FT-IR microscopy imaging on oral cavity tumours, II. J. Mol. Struct. 2005, 187–193. [Google Scholar] [CrossRef]
- Conti, C.; Ferraris, P.; Giorgini, E.; Pieramici, T.; Possati, L.; Rocchetti, R.; Rubini, C.; Sabbatini, S.; Tosi, G.; Morabito, C.; et al. Microimaging FT-IR of oral cavity tumours. Part III: Cells, inoculated tissues and human tissues. J. Mol. Struct. 2007, 86–94. [Google Scholar] [CrossRef]
- Sabbatini, S.; Conti, C.; Rubini, C.; Librando, V.; Tosi, G.; Giorgini, E. Infrared microspectroscopy of Oral Squamous Cell Carcinoma: Spectral signatures of cancer grading. Vib. Spectrosc. 2013, 68, 196–203. [Google Scholar] [CrossRef]
- Pallua, J.D.; Pezzei, C.; Zelger, B.; Schaefer, G.; Bittner, L.K.; Huck-Pezzei, V.A.; Schoenbichler, S.A.; Hahn, H.; Kloss-Brandstaetter, A.; Kloss, F.; et al. Fourier transform infrared imaging analysis in discrimi-nation studies of squamous cell carcinoma. Analyst 2012, 137, 3965–3974. [Google Scholar] [CrossRef]
- Banerjee, S.; Pal, M.; Chakrabarty, J.; Petibois, C.; Paul, R.R.; Giri, A.; Chatterjee, J. Fouri-er-transform-infrared-spectroscopy based spectral-biomarker selection towards optimum diagnostic differentia-tion of oral leukoplakia and cancer. Anal. Bioanal. Chem. 2015, 407, 7935–7943. [Google Scholar] [CrossRef]
- Naurecka, M.L.; Sierakowski, B.M.; Kasprzycka, W.; Dojs, A.; Dojs, M.; Suszyński, Z.; Kwaśny, M. FTIR-ATR and FT-Raman Spectroscopy for Biochemical Changes in Oral Tissue. Am. J. Anal. Chem. 2017, 8, 180–188. [Google Scholar] [CrossRef] [Green Version]
- Miljkovic, M.; Bird, B.; Lenau, K.; Mazur, A.I.; Diem, M. Spectral cytopathology: New aspects of data collection, manipulation and confounding effects. Analyst 2013, 138, 3975–3982. [Google Scholar] [CrossRef] [PubMed]
- Townsend, D.; Lenau, K.; Old, O.; Almond, M.; Shepherd, N.; Stone, N.; Diem, M.; Miljkovic, M.; Bird, B.; Kendall, C.; et al. Infrared micro-spectroscopy for cyto-pathological classification of esophageal cells. Analyst 2014, 140, 2215–2223. [Google Scholar] [CrossRef]
- Diem, M.; Mazur, A.I.; Schubert, J.M.; Townsend, D.; Laver, N.; Almond, M.; Old, O.; Miljkovic, M.; Bird, B. Cancer screening via infrared spectral cytopathology (SCP): Results for the upper respiratory and digestive tracts. Analyst 2015, 141, 416–428. [Google Scholar] [CrossRef]
- Ghosh, A.; Raha, S.; Dey, S.; Chatterjee, K.; Chowdhury, A.R.; Barui, A. Chemometric analysis of integrated FTIR and Raman spectra obtained by non-invasive exfoliative cytology for the screening of oral cancer. Analyst 2019, 144, 1309–1325. [Google Scholar] [CrossRef] [PubMed]
- Giorgini, E.; Sabbatini, S.; Rocchetti, R.; Notarstefano, V.; Rubini, C.; Conti, C.; Orilisi, G.; Mitri, E.; Bedolla, D.E.; Vaccari, L. In vitro FTIR microspectroscopy analysis of primary oral squamous carcinoma cells treated with cispla-tin and 5-fluorouracil: A new spectroscopic approach for studying the drug-cell interaction. Analyst 2018, 143, 3317–3326. [Google Scholar] [CrossRef] [PubMed]
- Chiu, L.-F.; Huang, P.-Y.; Chiang, W.-F.; Wong, T.-Y.; Lin, S.-H.; Lee, Y.-C.; Shieh, D.-B. Oral cancer diagnostics based on infrared spectral markers and wax physisorption kinetics. Anal. Bioanal. Chem. 2013, 405, 1995–2007. [Google Scholar] [CrossRef] [PubMed]
- Gajjar, K.; Trevisan, J.; Owens, G.; Keating, P.J.; Wood, N.J.; Stringfellow, H.; Martin-Hirsch, P.L.; Martin, F.L. Fourier-transform infrared spectroscopy coupled with a classification machine for the analysis of blood plasma or serum: A novel diagnostic approach for ovarian cancer. Analyst 2013, 138, 3917–3926. [Google Scholar] [CrossRef]
- Dong, L.; Sun, X.; Chao, Z.; Zhang, S.; Zheng, J.; Gurung, R.; Du, J.; Shi, J.; Xu, Y.; Zhang, Y.; et al. Evaluation of FTIR spectroscopy as diagnostic tool for colorectal cancer using spectral analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 122, 288–294. [Google Scholar] [CrossRef]
- Lewis, K.; Lewis, K.; Ghosal, R.; Bayliss, S.C.; Lloyd, A.J.; Wills, J.; Godfrey, R.; Kloer, P.; Mur, L.A.J. Evaluation of FTIR Spectroscopy as a diagnostic tool for lung cancer using sputum. BMC Cancer 2010, 10, 640. [Google Scholar] [CrossRef] [Green Version]
- Hands, J.R.; Clemens, G.; Stables, R.; Ashton, K.; Brodbelt, A.; Davis, C.; Dawson, T.P.; Jenkinson, M.D.; Lea, R.W.; Walker, C.; et al. Brain tumour differentiation: Rapid stratified serum diagnostics via attenuated total reflection Fourier-transform infrared spectroscopy. J. Neuro Oncol. 2016, 127, 463–472. [Google Scholar] [CrossRef] [Green Version]
- Sitnikova, V.E.; Kotkova, M.A.; Nosenko, T.N.; Kotkova, T.N.; Martynova, D.M.; Uspenskaya, M.V. Breast can-cer detection by ATR-FTIR spectroscopy of blood serum and multivariate data-analysis. Talanta 2020, 214, 120857. [Google Scholar] [CrossRef]
- Menzies, G.E.; Fox, H.R.; Marnane, C.; Pope, L.; Prabhu, V.; Winter, S.; Derrick, A.V.; Lewis, P.D. Fourier trans-form infrared for noninvasive optical diagnosis of oral, oropharyngeal, and laryngeal cancer. Transl. Res. 2014, 163, 19–26. [Google Scholar] [CrossRef]
- Rai, V.; Mukherjee, R.; Routray, A.; Ghosh, A.K.; Roy, S.; Ghosh, B.P.; Mandal, P.B.; Bose, S.; Chakraborty, C. Se-rum-based diagnostic prediction of oral submucous fibrosis using FTIR spectrometry. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 189, 322–329. [Google Scholar] [CrossRef]
- Adeeba-Siddiqui, A.J.; Sherazi, S.T.H.; Ahmed, S.; Iqbal-Choudhary, M.; Atta-Ur, R.; Musharraf, S.G. A compara-tive profiling of oral cancer patients and high risk niswar users using FT-IR and chemometric analysis. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2018, 203, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Möller, A.; Lobb, R.J. The evolving translational potential of small extracellular vesicles in cancer. Nat. Rev. Cancer 2020, 20, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Li, J.W.; Chan, J.; Meehan, K. Extracellular vesicles in head and neck cancer: A potential new trend in di-agnosis, prognosis, and treatment. Int. J. Mol. Sci. 2020, 21, 8260. [Google Scholar] [CrossRef] [PubMed]
- Zlotogorski-Hurvitz, A.; Dekel, B.Z.; Malonek, D.; Yahalom, R.; Vered, M. FTIR-based spectrum of salivary exo-somes coupled with computational-aided discriminating analysis in the diagnosis of oral cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 685–694. [Google Scholar] [CrossRef]
- Kumar, S.; Desmedt, C.; Larsimont, D.; Sotiriou, C.; Goormaghtigh, E. Change in the microenvironment of breast cancer studied by FTIR imaging. Analyst 2013, 138, 4058–4065. [Google Scholar] [CrossRef] [PubMed]
- Chrabaszcz, K.; Kaminska, K.; Augustyniak, K.; Kujdowicz, M.; Smeda, M.; Jasztal, A.; Stojak, M.; Marzec, K.M.; Malek, K. Tracking Extracellular Matrix Remodeling in Lungs Induced by Breast Cancer Metastasis. Fourier Transform Infrared Spectroscopic Studies. Molecules 2020, 25, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiwari, S.; Triulzi, T.; Holton, S.; Regondi, V.; Paolini, B.; Tagliabue, E.; Bhargava, R. Infrared Spectroscopic Imag-ing Visualizes a Prognostic Extracellular Matrix-Related Signature in Breast Cancer. Sci. Rep. 2020, 10, 5442. [Google Scholar] [CrossRef]
- Ukkonen, H.; Kumar, S.; Mikkonen, J.J.; Salo, T.; Singh, S.P.; Koistinen, A.; Goormaghtigh, E.; Kullaa, A. Changes in the microenvironment of invading melanoma and carcinoma cells identified by FTIR imaging. Vib. Spectrosc. 2015, 79, 24–30. [Google Scholar] [CrossRef]
- Yunker, P.J.; Still, T.; Lohr, M.A.; Yodh, A.G. Suppression of the coffee-ring effect by shape-dependent capillary interactions. Nat. Cell Biol. 2011, 476, 308–311. [Google Scholar] [CrossRef]
- Bassan, P.; Byrne, H.J.; Bonnier, F.; Lee, J.; Dumas, P.; Gardner, P. Resonant Mie scattering in infrared spectros-copy of biological materials-understanding the ‘dispersion artefact’. Analyst 2009, 134, 1586–1593. [Google Scholar] [CrossRef]
- Mayerhöfer, T.G.; Popp, J. The electric field standing wave effect in infrared transflection spectroscopy. Spectrochim. Acta Part. A Mol. Biomol. Spectrosc. 2018, 191, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Lovergne, L.; Bouzy, P.; Untereiner, V.; Garnotel, R.; Baker, M.J.; Thiéfin, G.; Sockalingum, G.D. Biofluid infrared spectro-diagnostics: Pre-analytical considerations for clinical applications. Faraday Discuss. 2015, 187, 521–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Afseth, N.K.; Kohler, A. Extended multiplicative signal correction in vibrational spectroscopy, a tutorial. Chemom. Intell. Lab. Syst. 2012, 117, 92–99. [Google Scholar] [CrossRef]
- Tiwari, S.; Bhargava, R. Extracting Knowledge from Chemical Imaging Data Using Computational Algorithms for Digital Cancer Diagnosis. Yale J. Boil. Med. 2015, 88, 131–143. [Google Scholar]
- Yao, Y.; Hoffman, A.J.; Gmachl, C.F. Mid-infrared quantum cascade lasers. Nat. Photon. 2012, 6, 432–439. [Google Scholar] [CrossRef]
- Pilling, M.J.; Henderson, A.; Gardner, P. Quantum Cascade Laser Spectral Histopathology: Breast Cancer Diagnos-tics Using High Throughput Chemical Imaging. Anal. Chem. 2017, 89, 7348–7355. [Google Scholar] [CrossRef] [Green Version]
- Mittal, S.; Yeh, K.; Leslie, L.S.; Kenkel, S.; Kajdacsy-Balla, A.; Bhargava, R. Simultaneous cancer and tumor microenvironment subtyping using confocal infrared microscopy for all-digital molecular histopathology. Proc. Natl. Acad. Sci. USA 2018, 115, E5651–E5660. [Google Scholar] [CrossRef] [Green Version]
- Butler, H.; Brennan, P.M.; Cameron, J.M.; Finlayson, D.; Hegarty, M.G.; Jenkinson, M.D.; Palmer, D.S.; Smith, B.A.H.; Baker, M.J. Development of high-throughput ATR-FTIR technology for rapid triage of brain cancer. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wiens, R.; Findlay, C.R.-J.; Baldwin, S.G.; Kreplak, L.; Lee, J.M.; Veres, S.P.; Gough, K. High spatial resolution (1.1 μm and 20 nm) FTIR polarization contrast imaging reveals pre-rupture disorder in damaged tendon. Faraday Discuss. 2016, 187, 555–573. [Google Scholar] [CrossRef] [Green Version]
- Atefi, N.; Vakil, T.; Abyat, Z.; Ramlochun, S.K.; Bakir, G.; Dixon, I.M.C.; Albensi, B.C.; Dahms, T.E.S.; Gough, K.M. Infrared spectroscopy: New frontiers both near and far. Spectroscopy 2018, 33, 34–38. [Google Scholar]
- Bassan, P.; Mellor, J.; Shapiro, J.; Williams, K.J.; Lisanti, M.P.; Gardner, P. Transmission FT-IR Chemical Imaging on Glass Substrates: Applications in Infrared Spectral Histopathology. Anal. Chem. 2014, 86, 1648–1653. [Google Scholar] [CrossRef] [PubMed]
- Pilling, M.J.; Henderson, A.; Shanks, J.H.; Brown, M.D.; Clarke, N.W.; Gardner, P. Infrared spectral histopathol-ogy using haematoxylin and eosin (H&E) stained glass slides: A major step forward towards clinical translation. Analyst 2017, 142, 1258–1268. [Google Scholar] [PubMed] [Green Version]
- Ogunleke, A.; Recur, B.; Balacey, H.; Chen, H.H.; Delugin, M.; Hwu, Y.; Javerzat, S.; Petibois, C. 3D chemical im-aging of the brain using quantitative IR spectro-microscopy. Chem. Sci. 2018, 9, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelp, G.; Arju, N.; Lee, A.; Esquivel, E.; Delgado, R.; Yu, Y.; Dutta-Gupta, S.; Sokolov, K.V.; Shvets, G. Application of metasurface-enhanced infra-red spectroscopy to distinguish between normal and cancerous cell types. Analyst 2019, 144, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Sieger, M.; Balluff, F.; Wang, X.; Kim, S.-S.; Leidner, L.; Gauglitz, G.; Mizaikoff, B. On-Chip Integrated Mid-Infrared GaAs/AlGaAs Mach–Zehnder Interferometer. Anal. Chem. 2012, 85, 3050–3052. [Google Scholar] [CrossRef] [PubMed]
- Mittal, V.; Mashanovich, G.Z.; Wilkinson, J.S. Perspective on Thin Film Waveguides for on-Chip Mid-Infrared Spectroscopy of Liquid Biochemical Analytes. Anal. Chem. 2020, 92, 10891–10901. [Google Scholar] [CrossRef]
- Rai, V.; Mukherjee, R.; Ghosh, A.K.; Routray, A.; Chakraborty, C. “Omics” in oral cancer: New approaches for bi-omarker discovery. Arch. Oral Biol. 2018, 87, 15–34. [Google Scholar] [CrossRef]
| Type of Study | Title of Study | References |
|---|---|---|
| Biochemical imaging and 2D classification of keratin pearl structures in oral squamous cell carcinoma | [84] | |
| In situ infrared histopathology of keratinization in human oral/oropharyngeal squamous cell carcinoma | [85] | |
| A study on the differences between oral squamous cell carcinomas and normal oral mucosas measured by Fourier transform infrared spectroscopy | [86] | |
| Distinguishing malignant from normal oral tissues using FTIR fiber-optic techniques | [87] | |
| Histological and microscopy FT-IR imaging study on the proliferative activity and angiogenesis in head and neck tumors | [88] | |
| Oral tissue studies | FT-IR microscopy imaging on oral cavity tumors, II. | [89] |
| Microimaging FT-IR of oral cavity tumors. Part III: Cells, inoculated tissues and human tissues. | [90] | |
| Infrared microspectroscopy of Oral Squamous Cell Carcinoma: Spectral signatures of cancer grading | [91] | |
| Fourier transform infrared imaging analysis in discrimination studies of squamous cell carcinoma | [92] | |
| Fourier-transform-infrared-spectroscopy based spectral-biomarker selection towards optimum diagnostic differentiation of oral leukoplakia and cancer | [93] | |
| FTIR-ATR and FT-Raman Spectroscopy for Biochemical Changes in Oral Tissue | [94] | |
| Spectral cytopathology: new aspects of data collection, manipulation and confounding effects | [95] | |
| Infrared micro-spectroscopy for cyto-pathological classification of esophageal cells | [96] | |
| Oral cell studies | Cancer Screening via Infrared Spectral Cytopathology (SCP): Results for the Upper Respiratory and Digestive Tracts | [97] |
| Chemometric analysis of integrated FTIR and Raman spectra obtained by non-invasive exfoliative cytology for the screening of oral cancer | [98] | |
| In vitro FTIR microspectroscopy analysis of primary oral squamous carcinoma cells treated with cisplatin and 5-fluorouracil: a new spectroscopic approach for studying the drug-cell interaction | [99] | |
| Oral cancer diagnostics based on infrared spectral markers and wax physisorption kinetics | [100] | |
| Fourier transform infrared for noninvasive optical diagnosis of oral, oropharyngeal, and laryngeal cancer | [106] | |
| Serum-based diagnostic prediction of oral submucous fibrosis using FTIR spectrometry | [107] | |
| Biofluid studies | A comparative profiling of oral cancer patients and high risk niswar users using FT-IR and chemometric analysis | [108] |
| Extracellular vesicles in head and neck cancer: A potential new trend in diagnosis, prognosis, and treatment | [110] | |
| FTIR-based spectrum of salivary exosomes coupled with computational-aided discriminating analysis in the diagnosis of oral cancer | [111] | |
| Tumor microenvironment study | Changes in the microenvironment of invading melanoma and carcinoma cells identified by FTIR imaging | [112] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Wang, Y. Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1206. https://doi.org/10.3390/ijms22031206
Wang R, Wang Y. Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis. International Journal of Molecular Sciences. 2021; 22(3):1206. https://doi.org/10.3390/ijms22031206
Chicago/Turabian StyleWang, Rong, and Yong Wang. 2021. "Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis" International Journal of Molecular Sciences 22, no. 3: 1206. https://doi.org/10.3390/ijms22031206
APA StyleWang, R., & Wang, Y. (2021). Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis. International Journal of Molecular Sciences, 22(3), 1206. https://doi.org/10.3390/ijms22031206







