The Phylogenetic Structure of Reptile, Avian and Uropathogenic Escherichia coli with Particular Reference to Extraintestinal Pathotypes
Abstract
1. Introduction
2. Results
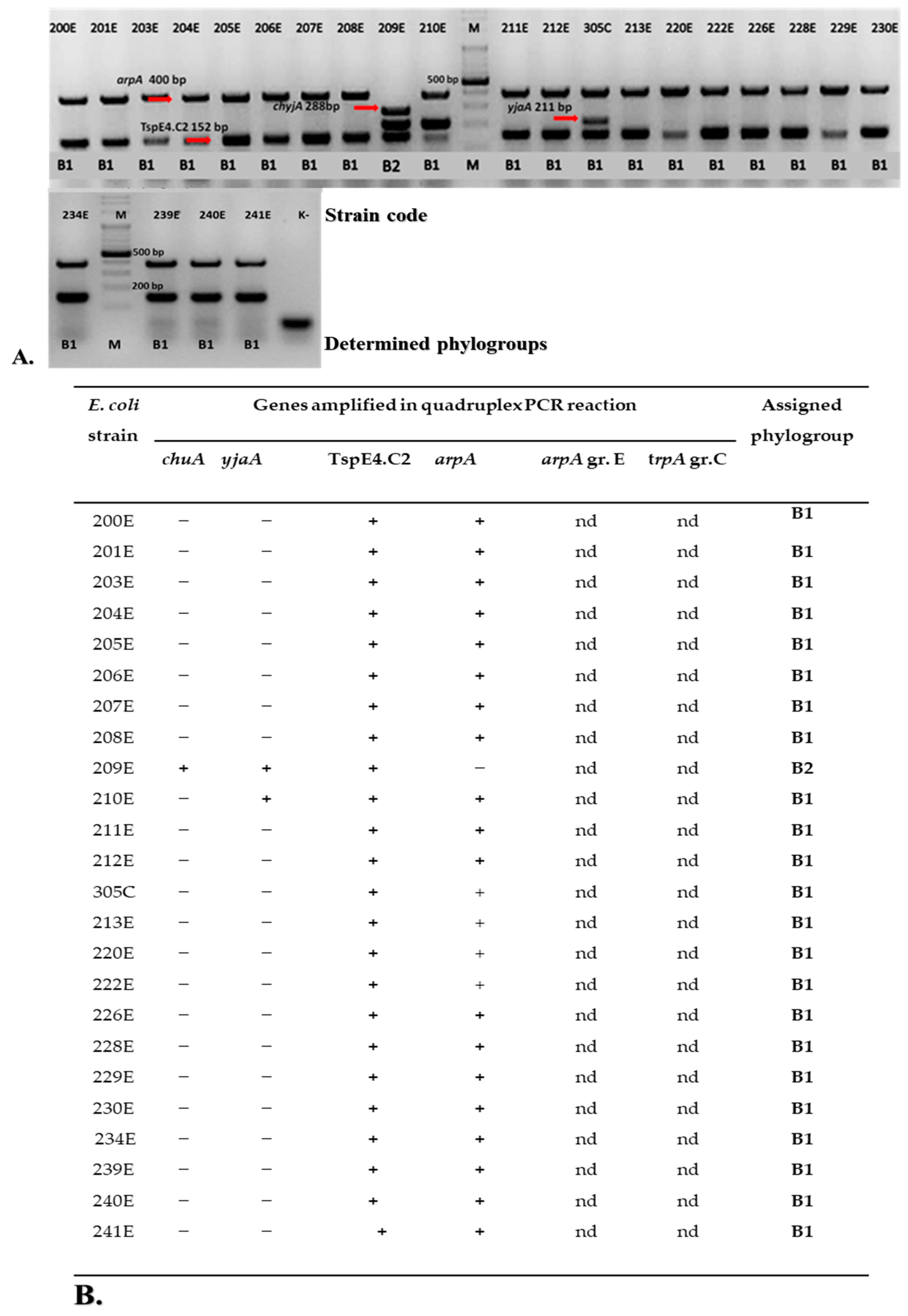
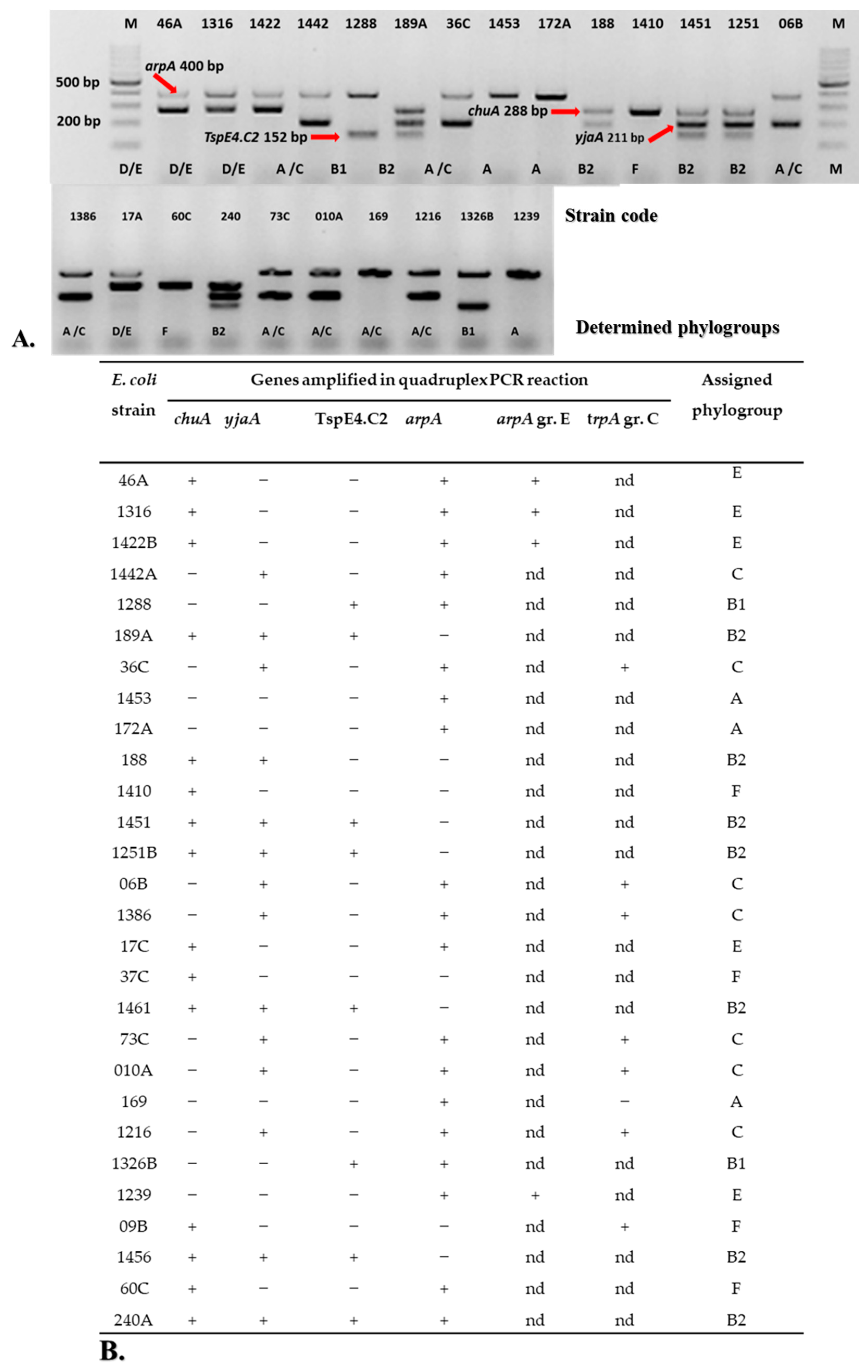
2.1. Phylogenetic Groups of E. coli
2.2. Virulence Gene Typing
2.3. Analysis of Phylogenetic Relationships between E. coli Strains from Different Hosts
2.4. Multi-Locus Sequence Typing
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains
4.2. Isolation and Identification of Bacterial Strains
4.3. DNA Extraction
4.4. Phylogenetic Groups of E. coli
4.5. Virulence Genotyping
4.6. Gel Electrophoresis of PCR Amplification Products
4.7. Restriction Analysis Combined with Pulsed-Field Gel Electrophoresis (RAE-PFGE)
4.8. Genetic Population Structure Analysis of E. coli Strains by Multi-Locus Sequence Typing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Group of Escherichia coli Strains | |||||
|---|---|---|---|---|---|
| RepEC | UPEC | APEC | |||
| Strain Code | Source of Isolated Bacteria | Strain Code | Source of Isolated Bacteria | Strain Code | Source of Isolated Bacteria |
| Escherichia coli strains isolated from feces of reptiles (snakes, lizards and turtles) | Escherichia coli isolated from urine samples of patients with symptomatic and asymptomatic urinary tract infections | Escherichia coli strains isolated from extraintestinal organs (heart, liver) of poultry with colibacillosis | |||
| 200E | Snake (Lamprophis fuliginosus) | 15641.38 | 46A | ||
| 201E | Snake (Orthriophis taeniurus) | 15514.33 | 1316 | ||
| 203E | Lizard (Pogona vitticeps) | 15165.15 | 1422B | ||
| 204E | Lizard (Chamaeleo calyptratus) | 15279.21 | 1442A | ||
| 205E | Lizard (Anolis barbatus) | 15988.54 | 1288 | ||
| 206E | Snake (Epicrates cenchria) | 15300.22 | 189A | ||
| 207E | Snake (Lamprophis fuliginosus) | 15593.39 | 36C | ||
| 208E | Snake (Lamprophis fuliginosus) | 15159.19 | 1453 | ||
| 209E* | Snake (Lamprophis fuliginosus) | 15607.35 | 172A | ||
| 210E | Snake (Lampropeltisgetula) | 15992.42 | 188 | ||
| 211E | Snake (Elaphe schrencki) | 15997.9 | 1410 | ||
| 212E | Snake (Coelognathus radiata) | 15550.25 | 1451 | ||
| 305C | Turtle (Kinixys belliana) | 15589.37 | 1251B | ||
| 213E | Turtle (Chelonoidis nigra) | 1110 | 06B | ||
| 220E | Turtle (Geochelone platynota) | 15599.34 | 1386C | ||
| 222E | Turtle (Testudo marginata) | 15067.4 | 17A | ||
| 226E | Lizard (Chamaeleo calyptratus) | 15059.6 | 60C | ||
| 228E | Turtle (Geochelone platynota) | 15320.27 | 240 | ||
| 229E | Turtle (Astrochelysradiata) | 15038.2 | 73C | ||
| 230E | Turtle (Centrochelys sulcata) | 1119 | 010A | ||
| 234E | Turtle (Testudo horsfieldii) | 15611.41 | 169 | ||
| 239E | Snake (Lampropeltis triangulum) | 15155.16 | 1216 | ||
| 240E | Lizard (Anolis baracoae) | 15035.8 | 1326B | ||
| 241E | Turtle (Chelonoidisnigra) | 15084.1 | 1239 | ||
| Gene | Sequences of Primers | PCR Product Size (bp) | Gene Bank ID |
|---|---|---|---|
| yjaA | 5′-CAAACGTGAAGTGTCAGGAG-3′ 5′-AATGCGTTCCTCAACCTGTG-3′ | 288 | 948515 |
| chuA | 5′-CAAACGTGAAGTGTCAGGAG-3′ 5′-AATGCGTTCCTCAACCTGTG-3′ | 211 | 7155958 |
| TspE4C2 | 5′-CACTATTCGTAAGGTCATCC-3′ 5′-AGTTTATCGCTGCGGGTCGC-3′ | 152 | EU240725.1 |
| arpA | 5′-AACGCTATTCGCCAGCTTGC-3′ 5′-TCTCCCCATACCGTACGCTA-3′ | 400 | 7155679 |
| arpA | 5′-GATTCCATCTTGTCAAAATATGCC-3′ 5′-GAAAAGAAAAAGAATTCCCAAGAG-3′ | 301 | 944933 |
| trpA | 5′-AGTTTTATGCCCAGTGCGAG-3′ 5′-TCTGCGCCGGTCACGCCC-3′ | 219 | 912862 |
| trpA | 5′-CGGCGATAAAGACATCTTCAC-3′ 5′-GCAACGCGGCCTGGCGGAAG-3′ | 489 | 13702525 |
| Gene | Sequences of Primers | PCR Product Size (bp) | Gene Bank ID | Biological (Enzymatic) Function |
|---|---|---|---|---|
| astA | 5′-CCATCAACACA-3′ 5′-TCAGGTCGCGAGTG-3′ | 116 | AF143819 | Gene encoding heat-stable toxin 1 (ST1) |
| Iss | 5′-ATCACATAGGATTCTGCCG-3′ 5′-CAGCGGAGTATAGATGCCA-3′ | 309 | X52665 | Gene encoding outer membrane protein of increased serum survival |
| irp2 | 5′-GGATTCGCTGTTACCGGAC3′ 5′-AACTCCTGATACAGGTGGC-3′ | 413 | L18881 | Gene encoding iron binding protein |
| iucD | 5′-ACAAAAAGTTCTATCGCTTCC-3′ 5′-CCTGATCCAGATGATGCTC-3′ | 714 | M18968 | Gene encoding siderophore’s complex |
| tsh | 5′-ACTATTCTCTGCAGGAAGTC-3′ 5′-TTCCGATGTTCTGAA-3′ | 824 | AF218073 | Gene encoding thermolabile haemagglutinin |
| vat | 5′-TCCTGGGACATAATGGTCAG-3′ 5′- GTGTCAGAACGGAATTGT-3′ | 981 | AY151282 | Gene encoding vacuolating autotransporter toxin |
| fyuA | 5′-TGATTAACCCCGCGACGGGAA-3′ 5′-CGCAGTAGGCACGATGTTGTA-3′ | 880 | Z38064 | Gene belonging to the operon encoding aerobactin siderophore complex at the plasmid ColV-K30 |
| stx2f | 5′-ATCCTATTCCCGGGAGTTTACG-3′ 5′-GCGTCATCGTATACACAGGAGC-3′ | 338 | 912579 | Gene encoding Shiga like toxin |
| hlyE | 5′-GGTGCAGCAGAAAAAGTTGTAG-3′ 5′-TCTCGCCTGATAGTGTTTGGTA-3′ | 569 | 13701006 | Gene encoding haemolysin |
| fimC | 5′-GTTGATCAAACCGTTCAG-3′ 5′-AATAACGCGCCTGGAACG-3′ | 424 | 948843 | Gene encoding fimbriae type I |
| eae | 5′-TCAATGCAGTTCCGTTATCAGTT-3′ 5′-GTAAAGTCCGTTACCCCAACCTG-3′ | 482 | AF022236 | Gene encoding intimin |
| bfp | 5′-GGAAGTCAAATTCATGGGGGTAT-3′ 5′-GGAATCAGACGCAGACTGGTAGT 3′ | 246 | KJ020697 | Gene encoding bundle forming pilli |
| cvi/cva | 5′-TGGTAGAATGTGCCAGAGCAAG-3′ 5′-GAGCTGTTTGTAGCGAAGCC-3′ | 1181 | AJ223631 | Gene encoding colicin at the ColV plasmid |
| papC | 5′-GTGGCAGTATGAGTAATGACCGTTA-3′ 5′-ATATCCTTTCTGCAGGGATGCAATA-3′ | 200 | X61239 | Gene encoding P fimbriae subunit |
| kpsMTII | 5′-GCGCATTTGCTGATACTGTTG-3′ 5′-CATCCAGACGATAAGCATGAGCA-3′ | 272 | X53819 | Gene encoding extracellular capsules type II biosynthesis proteins |
| kpsMT (K1) | 5′-TAGCAAACGTTCTATTGGTGC-3′ 5′-CATCCAGACGATAAGCATGAGCA-3′ | 153 | M57382 | Gene encoding capsules antigen K1 |
| kpsMTIII | 5′-TCCTCTTGCTACTATTCCCCCT -3′ 5′-AGGCGTATCCATCCCTCCTAAC-3′ | 392 | AF007777 | Gene encoding extracellular capsules type III biosynthesis proteins |
| rfc | 5′-ATCCATCAGGAGGGGACTGGA-3′ 5′-AACCATACCAACCAATGCGAG -3′ | 788 | U39042 | Gene encoding O-side-chain antigen of LPS |
| traT | 5′- GGTGTGGTGCGATGAGCACAG-3′ 5′-CACGGTTCAGCCATCCCTGAG-3′ | 290 | J01769 | Gene encoding serum resistance and pathogenicity-related protein |
| Gene | Primer’s Sequences | PCR Product Size (bp) | Biological (Enzymatic) Function |
|---|---|---|---|
| adk | 5’-ATTCTGCTTGGCGCTCCGGG-3’ 5’-CCGTCAACTTTCGCGTATTT-3’ | 583 | adenylate kinase |
| fumC | 5’-TCACAGGTCGCCAGCGCTTC-3’ 5’-GTACGCAGCGAAAAAGATTC-3’ | 806 | fumarate hydratase |
| gyrB | 5’-TCGGCGACACGGATGACGGC-3’ 5’-GTCCATGTAGGCGTTCAGGG-3’ | 911 | DNA gyrase |
| icd | 5’-ATGGAAAGTAAAGTAGTTGTTCCGGCACA-3’ 5’-GGACGCAGCAGGATCTGTT-3’ | 878 | isocitrate dehydrogenase |
| mdh | 5’-AGCGCGTTCTGTTCAAATGC-3’ 5’-CAGGTTCAGAACTCTCTCTGT-3’ | 932 | malate dehydrogenase |
| purA | 5’-CGCGCTGATGAAAGAGATGA-3’ 5’-CATACGGTAAGCCACGCAGA-3’ | 816 | adenylosuccinate synthetase |
| recA | 5’-ACCTTTGTAGCTGTACCACG-3’ 5’-TCGTCGAAATCTACGGACCGGA-3’ | 780 | ATP/GTP binding motif |

References
- Whittam, T.S.; Ochman, H.; Selander, R.K. Multilocus genetic structure in natural populations of Escherichia coli. Proc. Natl. Acad. Sci. USA 1983, 80, 1751–1755. [Google Scholar] [CrossRef] [PubMed]
- Milkman, R. Electrophoretic variation in Escherichia coli from natural sources. Science 1973, 182, 1024–1026. [Google Scholar] [CrossRef] [PubMed]
- Salinas, L.; Cárdenas, P.; Johnson, T.J.; Vasco, K.; Graham, J.; Trueba, G. Diverse Commensal Escherichia coli Clones and Plasmids Disseminate Antimicrobial Resistance Genes in Domestic Animals and Children in a Semirural Community in Ecuador. mSphere 2019, 4, 00316–00319. [Google Scholar] [CrossRef] [PubMed]
- Bok, E.; Kożańska, A.; Mazurek-Popczyk, J.; Wojciech, M.; Baldy-Chudzik, K. Extended Phylogeny and Extraintestinal Virulence Potential of Commensal Escherichia coli from Piglets and Sows. Int. J. Environ. Res. Public Health 2020, 17, 366. [Google Scholar] [CrossRef] [PubMed]
- Blyton, M.D.; Pi, H.; Vangchhia, B.; Abraham, S.; Trott, D.J.; Johnson, J.R.; Gordon, D.M. Genetic Structure and Antimicrobial Resistance of Escherichia coli and Cryptic Clades in Birds with Diverse Human Associations. Appl. Environ. Microbiol. 2015, 81, 5123–5133. [Google Scholar] [CrossRef]
- Nakamura, K.; Murase, K.; Sato, M.P.; Toyoda, A.; Itoh, T.; Mainil, J.G.; Piérard, D.; Yoshino, S.; Kimata, K.; Isobe, J.; et al. Differential dynamics and impacts of prophages and plasmids on the pangenome and virulence factor repertoires of Shiga toxin-producing Escherichia coli O145:H28. Microb Genom. 2020, 6, e000323. [Google Scholar] [CrossRef]
- Forde, B.M.; Roberts, L.W.; Phan, M.; Peters, K.M.; Fleming, B.A.; Russell, C.W.; Lenherr, S.M.; Myers, J.B.; Barker, A.P.; Fisher, M.A.; et al. Population dynamics of an Escherichia coli ST131 lineage during recurrent urinary tract infection. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Petty, N.K.; Zakour, N.L.B.; Stanton-Cook, M.; Skippington, E.; Totsika, M.; Forde, B.M.; Phan, M.D.; Moriel, D.G.; Peters, K.M.; Davies, M.; et al. Global dissemination of E. coli ST131. Proc. Natl. Acad. Sci. USA 2014, 111, 5694–5699. [Google Scholar] [CrossRef]
- Baldy-Chudzik, K.; Bok, E.; Mazurek, J. Well-known and new variants of pathogenic Escherichia coli as a consequence of the plastic genome. Postep. Hig. Med. Dosw. 2015, 17, 345–361. [Google Scholar] [CrossRef]
- Donnenberg, M.S.; Whittam, T.S. Pathogenesis and evolution of virulence in enteropathogenic and enterohemorrhagic Escherichia coli. J. Clin. Invest. 2001, 107, 539–548. [Google Scholar] [CrossRef]
- Johnson, J.R.; Owens, K.L.; Clabots, C.R.; Weissman, S.J.; Cannon, S.B. Phylogenetic relationships among clonal groups of extraintestinal pathogenic Escherichia coli as assessed by multi-locus sequence analysis. Microbes Infect. 2006, 8, 1702–1713. [Google Scholar] [CrossRef] [PubMed]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef] [PubMed]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef] [PubMed]
- Stoppe, N.C.; Silva, J.S.; Carlos, C.; Sato, M.I.Z.; Saraiva, A.M.; Ottoboni, L.M.M.; Torres, T.T. Worldwide Phylogenetic Group Patterns of Escherichia coli from Commensal Human and Wastewater Treatment Plant Isolates. Front. Microbiol. 2017, 8, 2512. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Olier, M.; Hoede, C.; Diancourt, L.; Brisse, S.; Keroudean, M.; Glodt, J.; Picard, B.; Oswald, E.; Denamur, E. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 2011, 11, 654–662. [Google Scholar] [CrossRef]
- Walk, S.T. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 2009, 75, 6534–6544. [Google Scholar] [CrossRef]
- Clermont, O.; Dixit, O.V.A.; Vangchhia, B.; Condamine, B.; Dion, S.; Bridier-Nahmias, A.; Denamur, E.; Gordon, D. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ. Microbiol. 2019, 21, 3107–3117. [Google Scholar] [CrossRef]
- Rodriguez-Siek, K.E.; Giddings, C.W.; Doetkott, C.; Johnson, T.J.; Fakhr, M.K.; Nolan, L.K. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 2005, 151, 2097–2110. [Google Scholar] [CrossRef]
- Johnson, T.J.; Kariyawasam, S.; Wannemuehler, Y.; Mangiamele, P.; Johnson, S.J.; Doetkott, C.; Skyberg, J.A.; Lynne, A.M.; Johnson, J.R.; Nolan, L.K. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J. Bacteriol. 2007, 189, 3228–3236. [Google Scholar] [CrossRef]
- Brzuszkiewicz, E.; Gottschalk, G.; Ron, E.; Hacker, J.; Dobrindt, U. Adaptation of Pathogenic E. coli to Various Niches: Genome Flexibility is the Key. Genome Dyn. 2009, 6, 110–125. [Google Scholar] [PubMed]
- Coombes, B.K.; Gilmour, M.W.; Goodman, C.D. The evolution of virulence in non-o157 shiga toxin-producing Escherichia coli. Front. Microbiol. 2011, 2, 90. [Google Scholar] [CrossRef]
- Russo, T.A.; Johnson, J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes Infect. 2003, 5, 449–456. [Google Scholar] [CrossRef]
- Lutful Kabir, S.M. Avian Colibacillosis and Salmonellosis: A Closer Look at Epidemiology, Pathogenesis, Diagnosis, Control and Public Health Concerns. Int. J. Environ. Res. Public Health 2010, 7, 89–114. [Google Scholar] [CrossRef] [PubMed]
- Martínez, R.; Sánchez, S.; Alonso, J.M.; Herrera-León, S.; Rey, J.; Echeita, M.A.; Morán, J.M.; García-Sánchez, A. Salmonella spp. and Shiga toxin-producing Escherichia coli prevalence in an ocellated lizard (Timon lepidus) research center in Spain. Foodborne Pathog. Dis. 2011, 8, 1309–1311. [Google Scholar] [CrossRef]
- Gopee, N.V.; Adesiyun, A.A.; Caesar, K. A longitudinal study of Escherichia coli strains isolated from captive mammals, birds, and reptiles in Trinidad. J. Zoo Wildl. Med. Off. Publ. Am. Assoc. Zoo Vet. 2000, 31, 353–360. [Google Scholar]
- Russo, T.A.; Johnson, J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 2000, 181, 1753–1754. [Google Scholar] [CrossRef]
- Bélanger, L.; Garenaux, A.; Harel, J.; Boulianne, M.; Nadeau, E.; Dozois, C.M. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Microbiol. 2011, 62, 1–10. [Google Scholar] [CrossRef]
- Enterobase, enterobase-web, wiki, ecoli MLST Legacy Info RST—Bitbucket. Available online: http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search (accessed on 22 August 2020).
- Roer, L.; Overballe-Petersen, S.; Hansen, F. Escherichia coli Sequence Type 410 Is Causing New International High-Risk Clones. mSphere 2018, 3, 1–14. [Google Scholar] [CrossRef]
- Riley, L.W. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 2014, 20, 380–390. [Google Scholar] [CrossRef]
- Day, M.J.; Rodríguez, I.; van Essen-Zandbergen, A. Diversity of STs, plasmids and ESBL genes among Escherichia coli from humans, animals and food in Germany, the Netherlands and the UK. J Antimicrob. Chemother. 2016, 71, 1178–1182. [Google Scholar] [CrossRef] [PubMed]
- Ha Lau, S.; Reddy, S.; Cheesbrough, J.; Bolton, F.J.; Willshaw, G.; Cheasty, T.; Fox, A.J.; Upton, M. Major Uropathogenic Escherichia coli Strain Isolated in the Northwest of England Identified by Multilocus Sequence Typing. J. Clin. Microbiol. 2008, 46, 1076–1080. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alghoribi, M.F.; Doumith, M.; Upton, M. Complete Genome Sequence of a Colistin-Resistant Uropathogenic Escherichia coli Sequence Type 131 fimH22 Strain Harboring mcr-1 on an IncHI2 Plasmid, Isolated in Riyadh, Saudi Arabia. Microbiol. Resour. Announc. 2019, 8, e00104–e00119. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Karns, J.S.; Van Kessel, J.A.S.; Haley, B.J. Genome Sequences of Five Multidrug-Resistant Escherichia coli Sequence Type 117 Isolates Recovered from Dairy Calves. Genome Announc. 2017, 5, e00732-17. [Google Scholar] [CrossRef]
- Long, H.; Feng, Y.; Ma, K.; Liu, L.; McNally, A.; Zong, Z. The co-transfer of plasmid-borne colistin-resistant genes mcr-1 and mcr-3.5, the carbapenemase gene blaNDM-5 and the 16S methylase gene rmtB from Escherichia coli. Sci. Rep. 2019, 9, 1–6. [Google Scholar] [CrossRef]
- Bonnedahl, J.; Drobni, M.; Gauthier-Clerc, M.; Hernandez, J.; Granholm, S. Dissemination of Escherichia coli with CTX-M Type ESBL between Humans and Yellow-Legged Gulls in the South of France. PLoS ONE 2009, 4, e5958. [Google Scholar] [CrossRef]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Johnson, J.R.; Russo, T.A. Extraintestinal pathogenic Escherichia coli: ‘the other bad E coli’. J. Lab. Clin. Med. 2002, 139, 155–162. [Google Scholar] [CrossRef]
- Croxen, M.A.; Finlay, B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef]
- Pawlak, A. Reptile-associated salmonellosis as an important epidemiological problem. Postępy Hig. Med. Dośw. 2014, 17, 1335–1342. [Google Scholar] [CrossRef]
- Schmidt, V.; Mock, R.; Burgkhardt, E.; Junghanns, A.; Ortlieb, F.; Szabo, I. Cloacal aerobic bacterial flora and absence of viruses in free-living slow worms (Anguis fragilis), grass snakes (Natrix natrix) and European Adders (Vipera berus) from Germany. EcoHealth 2014, 11, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Unger, F.; Eisenberg, T.; Prenger-Berninghoff, E.; Leidner, U.; Ludwig, M.L.; Rothe, M. Imported reptiles as a risk factor for the global distribution of Escherichia coli harbouring the colistin resistance gene mcr-1. Int. J. Antimicrob. Agents 2017, 49, 122–123. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, A.M.; Júnior, A.K.P.; Andrade, M.A.; de Jayme, V.S. Prevalence of Enterobacteriaceae in Tupinambis merianae (Squamata: Teiidae) from a captive facility in Central Brazil, with a profile of antimicrobial drug resistance in Salmonella enterica. Phyllomedusa J. Herpetol. 2013, 12, 57–67. [Google Scholar] [CrossRef][Green Version]
- Gordon, D.M.; Cowling, A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: Host and geographic effects. Microbiol Read. Engl. 2003, 149, 3575–3586. [Google Scholar] [CrossRef] [PubMed]
- Książczyk, M.; Kuczkowski, M.; Dudek, B.; Korzekwa, K.; Tobiasz, A.; Korzeniowska-Kowal, A.; Paluch, E.; Wieliczko, A.; Bugla-Płoskońska, G. Application of Routine Diagnostic Procedure, VITEK 2 Compact, MALDI-TOF MS, and PCR Assays in Identification Procedure of Bacterial Strain with Ambiguous Phenotype. Curr. Microbiol. 2016, 72, 570–582. [Google Scholar] [CrossRef]
- Vizcarra, I.A.; Hosseini, V.; Kollmannsberger, P.; Meier, S.; Weber, S.S.; Arnoldini, M. How type 1 fimbriae help Escherichia coli to evade extracellular antibiotics. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef]
- Anderson, G.G.; Palermo, J.J.; Schilling, J.D.; Roth, R.; Heuser, J.; Hultgren, S.J. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 2003, 301, 105–107. [Google Scholar] [CrossRef]
- Brooks, D.E.; Cavanagh, J.; Jayroe, D.; Janzen, J.; Snoek, R.; Trust, T.J. Involvement of the MN blood group antigen in shear-enhanced hemagglutination induced by the Escherichia coli F41 adhesin. Infect. Immun. 1989, 57, 377–383. [Google Scholar] [CrossRef]
- Hultgren, S.J.; Porter, T.N.; Schaeffer, A.J.; Duncan, J.L. Role of type 1 pili and effects of phase variation on lower urinary tract infections produced by Escherichia coli. Infect. Immun. 1985, 50, 370–377. [Google Scholar] [CrossRef]
- Baorto, D.M.; Gao, Z.; Malaviya, R.; Dustin, M.L.; van der Merwe, A.; Lublin, D.M. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature 1997, 389, 636–639. [Google Scholar] [CrossRef]
- Croxen, M.A.; Law, R.J.; Scholz, R.; Keeney, K.M.; Wlodarska, M.; Finlay, B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013, 26, 822–880. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Dobrindt, U.; Brüggemann, H.; Nagy, G.; Janke, B.; Blum-Oehler, G.; Buchrieser, C.; Gottschalk, G.; Emödy, L.; Hacker, J. The Pathogenicity Island-Associated K15 Capsule Determinant Exhibits a Novel Genetic Structure and Correlates with Virulence in Uropathogenic Escherichia coli Strain 536. Infect. Immun. 2004, 72, 5993–6001. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.D.; Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011, 301, 642–647. [Google Scholar]
- Castellanos, L.R.; Donado-Godoy, P.; León, M. High Heterogeneity of Escherichia coli Sequence Types Harbouring ESBL/AmpC Genes on IncI1 Plasmids in the Colombian Poultry Chain. PLoS ONE 2017, 12, e0170777. [Google Scholar] [CrossRef] [PubMed]
- Maluta, R.P.; Logue, C.M.; Casas, M.R.T.; Meng, T.; Guastalli, E.A.L.; Rojas, T.C.G. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE 2014, 9, e105016. [Google Scholar] [CrossRef] [PubMed]
- Manges, A.R.; Harel, J.; Masson, L.; Edens, T.J.; Portt, A.; Reid-Smith, R.J. Multilocus sequence typing and virulence gene profiles associated with Escherichia coli from human and animal sources. Foodborne Pathog. Dis. 2015, 12, 302–310. [Google Scholar] [CrossRef]
- Collins, C.H.; Lyne, P.M. Microbiological Methods, 5th ed.; Butterworths: London, UK, 1985. [Google Scholar]
- Miller, J.M. Quality Control in Microbiology; Centers for Disease Control and Prevention: Atlanta, GA, USA, 1987.
- August, M.J. Quality Control and Quality Assurance Practices in Clinical Microbiology. Am. Soc. for Microbiol. 1990, 3A, 1–14. [Google Scholar]
- Basics of Quality Assurance for Intermediate and Peripheral Laboratories, 2nd ed.; WHO Regional Office for the Eastern Mediterranean: Alexandria, Egypt, 2000.
- Koppes, L.H.; Woldringh, C.L.; Nanninga, N. Size variations and correlation of different cell cycle events in slow-growing Escherichia coli. J. Bacteriol. 1978, 134, 423–433. [Google Scholar] [CrossRef]
- Stenutz, R.; Weintraub, A.; Widmalm, G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 2006, 30, 382–403. [Google Scholar] [CrossRef]
- Pulsed-field Gel Electrophoresis (PFGE). PulseNet Methods, PulseNet, CDC. 2017. Available online: https://www.cdc.gov/pulsenet/pathogens/pfge.html (accessed on 22 August 2020).

 = presence of the gene and
= presence of the gene and  = absence of the gene.
= absence of the gene.
 = presence of the gene and
= presence of the gene and  = absence of the gene.
= absence of the gene.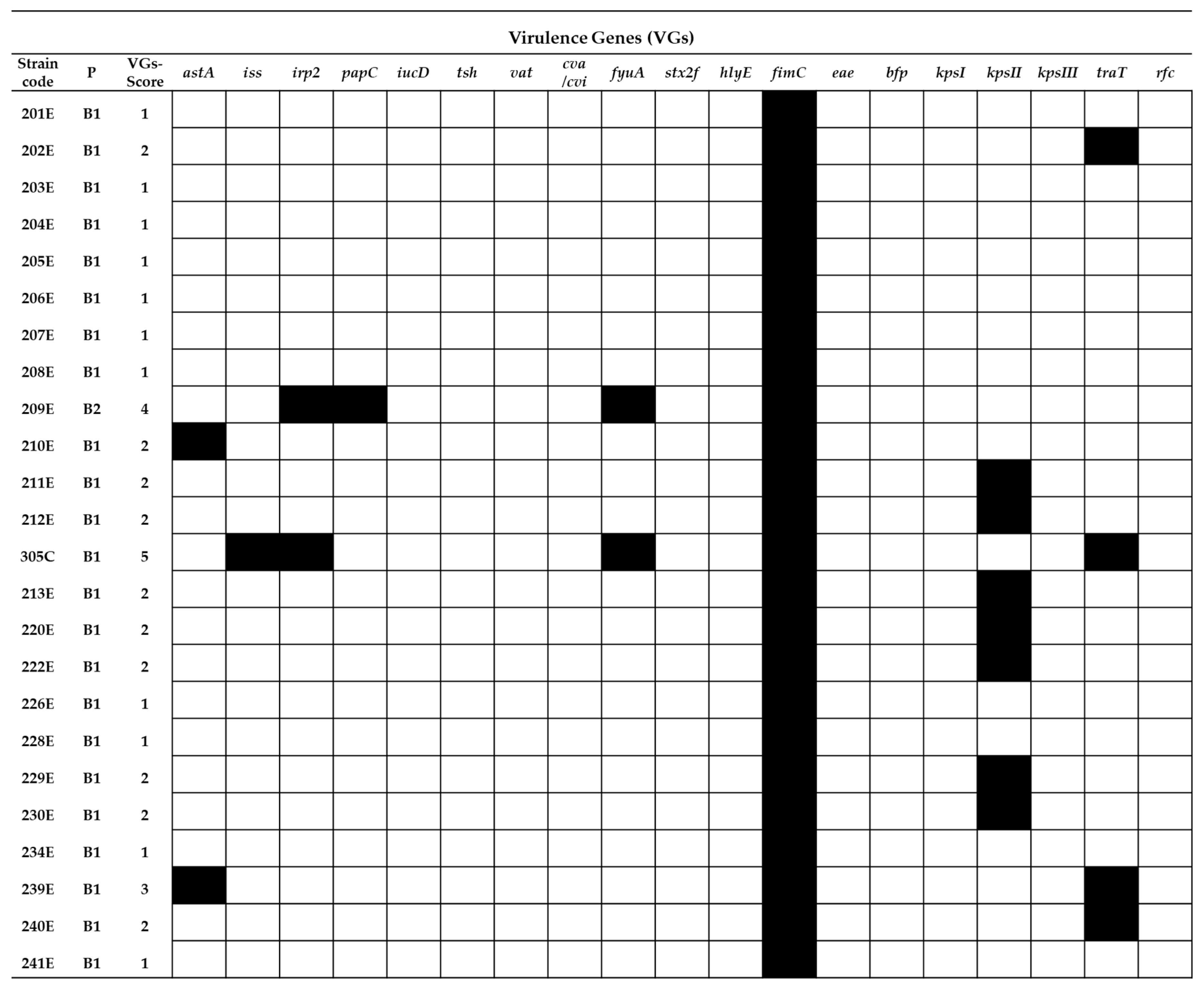
 = presence of the gene and
= presence of the gene and  = absence of the gene.
= absence of the gene.
 = presence of the gene and
= presence of the gene and  = absence of the gene.
= absence of the gene.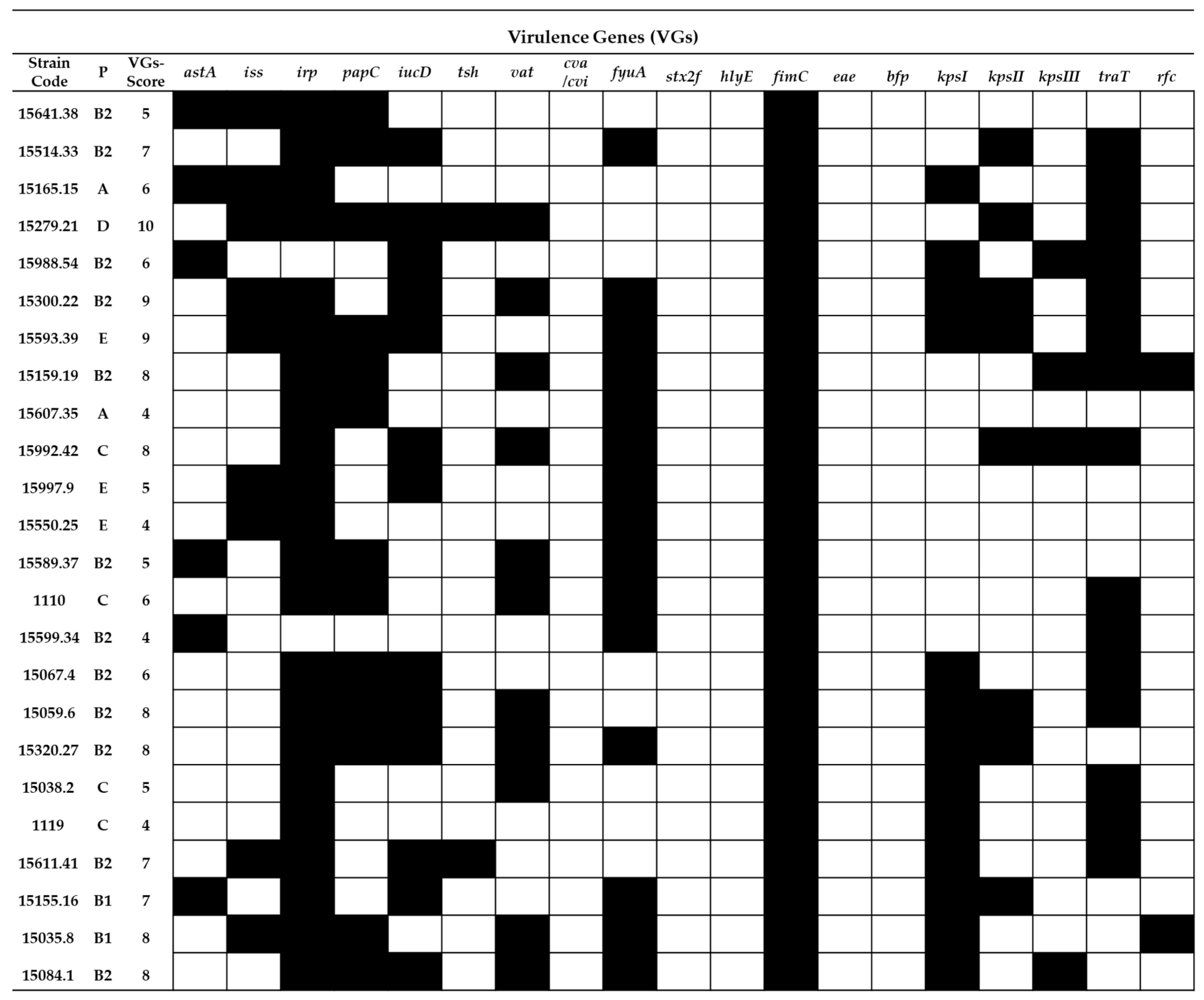
 = presence of the gene and
= presence of the gene and  = absence of the gene.
= absence of the gene.
 = presence of the gene and
= presence of the gene and  = absence of the gene.
= absence of the gene.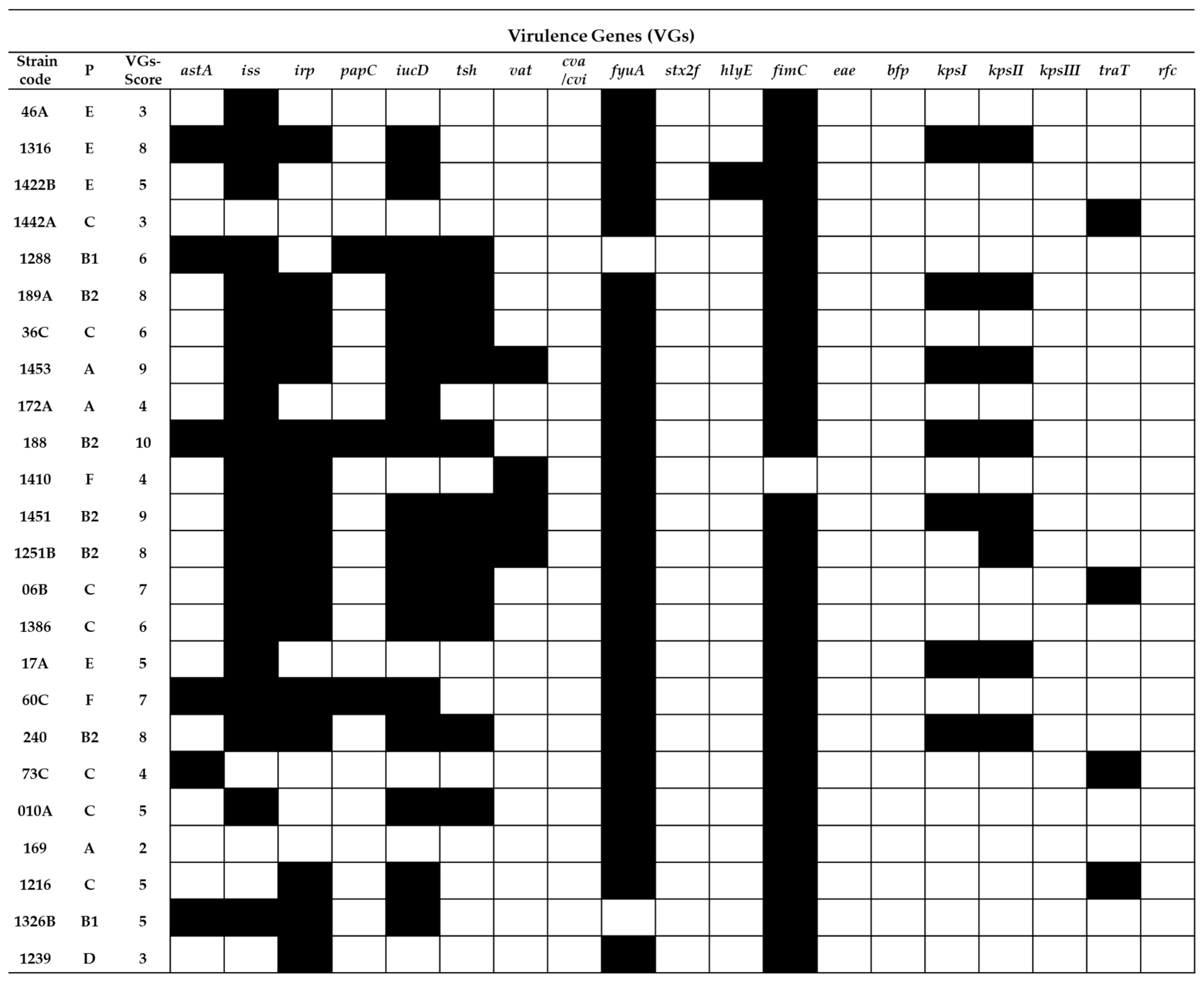


| Group of E. coli | E. coli Strain Code | P | Asigned Sequence Type | Description of STs | Citations |
|---|---|---|---|---|---|
| UPEC (n = 3) | 15279.21 | D | ST321 | ST321 is characteristic of nonpathogenic E. coli strains isolated from wild animals. | [29] |
| 1119 | C | ST410 | which includes pathogenic E. coli strains (human UPEC strains, predominantly). | [29,30] | |
| 15035.8 | B1 | ST12 | ST12 is typical of extraintestinal pathogenic E. coli strains causing systemic infections in warm-blooded animals and also typical of E. coli causing urinary tract infections (UTIs) among humans. | [29,31] | |
| APEC (n = 4) | 1288 | B1 | ST1582 | ST1582 is characteristic for UPEC strains causing UTIs in horses. | [29] |
| 1239 | D | ST665 | ST665 is typical of nonpathogenic E. coli strains isolated from poultry. | [29,32] | |
| 189A | B2 | ST131 | ST131 is a globally disseminated clone that poses a substantial health risk to humans, causing acute extraintestinal infections, such as serious UTI, urosepsis and sepsis. | [8,29,33,34] | |
| 60C | ST117 | ST117, which is a heterogenic group of extraintestinal pathogenic E. coli strains. | [29,35] | ||
| REPEC (n = 4) | 200E | B1 | ST446 | ST446 is typical of APEC strains and also of other human pathogenic extraintestinal E. coli. | [29,36] |
| 209E | B2 | ST681 | ST 681 includes E. coli strains pathogenic for humans and other animals. | [29,37] | |
| 212E | B1 | ST212 | This heterogenous ST mainly includes UPEC strains as well as other pathogenic and nonpathogenic E. coli strains. | [29,38] | |
| 305C | B1 | New ST! | Strain 305C could not be assigned to any known ST, although it could be a member of the ST88 clonal complex (with differences in two alleles adk and mdh; unconfirmed data). | - |
| Percentage Prevalence of VGs | Frequency of VGs |
|---|---|
| 0–10% | Very low |
| 11–20% | Low |
| 21–45% | Medium |
| 46–80% | High |
| 81–100% | Very high |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Książczyk, M.; Dudek, B.; Kuczkowski, M.; O’Hara, R.; Korzekwa, K.; Wzorek, A.; Korzeniowska-Kowal, A.; Upton, M.; Junka, A.; Wieliczko, A.; et al. The Phylogenetic Structure of Reptile, Avian and Uropathogenic Escherichia coli with Particular Reference to Extraintestinal Pathotypes. Int. J. Mol. Sci. 2021, 22, 1192. https://doi.org/10.3390/ijms22031192
Książczyk M, Dudek B, Kuczkowski M, O’Hara R, Korzekwa K, Wzorek A, Korzeniowska-Kowal A, Upton M, Junka A, Wieliczko A, et al. The Phylogenetic Structure of Reptile, Avian and Uropathogenic Escherichia coli with Particular Reference to Extraintestinal Pathotypes. International Journal of Molecular Sciences. 2021; 22(3):1192. https://doi.org/10.3390/ijms22031192
Chicago/Turabian StyleKsiążczyk, Marta, Bartłomiej Dudek, Maciej Kuczkowski, Robert O’Hara, Kamila Korzekwa, Anna Wzorek, Agnieszka Korzeniowska-Kowal, Mathew Upton, Adam Junka, Alina Wieliczko, and et al. 2021. "The Phylogenetic Structure of Reptile, Avian and Uropathogenic Escherichia coli with Particular Reference to Extraintestinal Pathotypes" International Journal of Molecular Sciences 22, no. 3: 1192. https://doi.org/10.3390/ijms22031192
APA StyleKsiążczyk, M., Dudek, B., Kuczkowski, M., O’Hara, R., Korzekwa, K., Wzorek, A., Korzeniowska-Kowal, A., Upton, M., Junka, A., Wieliczko, A., Ratajszczak, R., & Bugla-Płoskońska, G. (2021). The Phylogenetic Structure of Reptile, Avian and Uropathogenic Escherichia coli with Particular Reference to Extraintestinal Pathotypes. International Journal of Molecular Sciences, 22(3), 1192. https://doi.org/10.3390/ijms22031192







