AFM and FTIR Investigation of the Effect of Water Flow on Horseradish Peroxidase
Abstract
:1. Introduction
2. Results
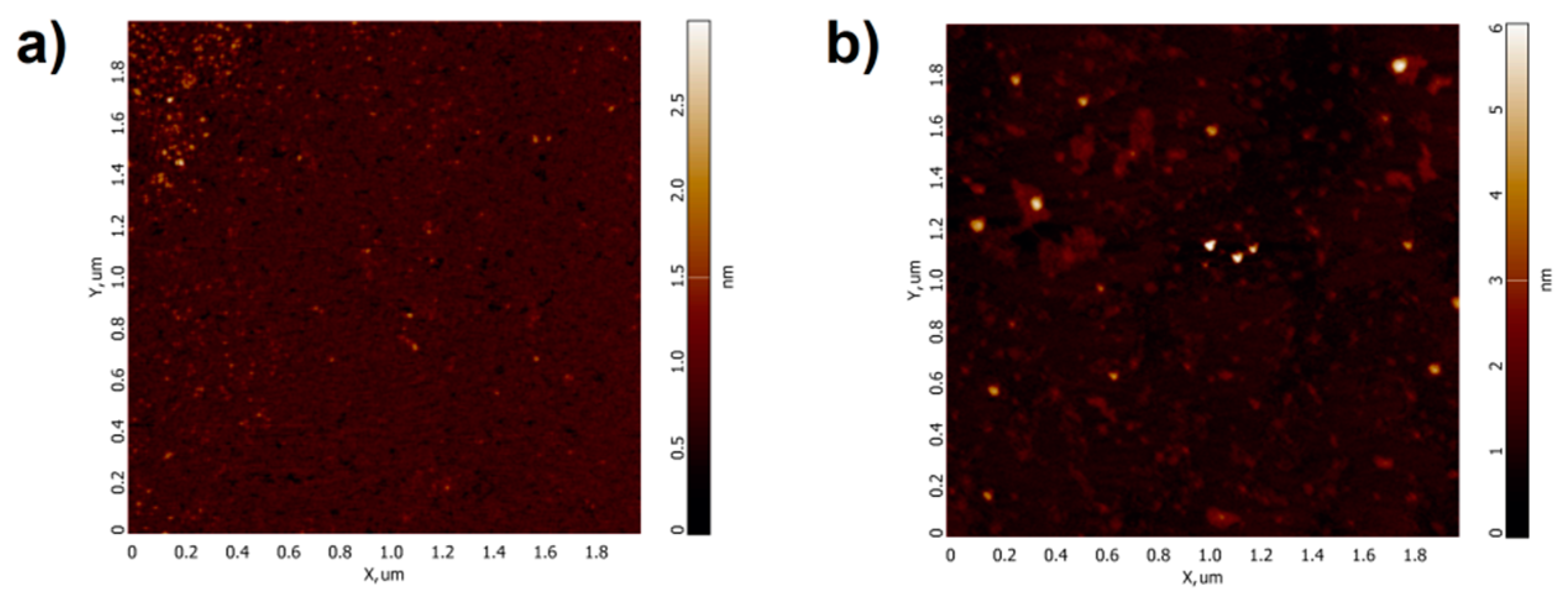
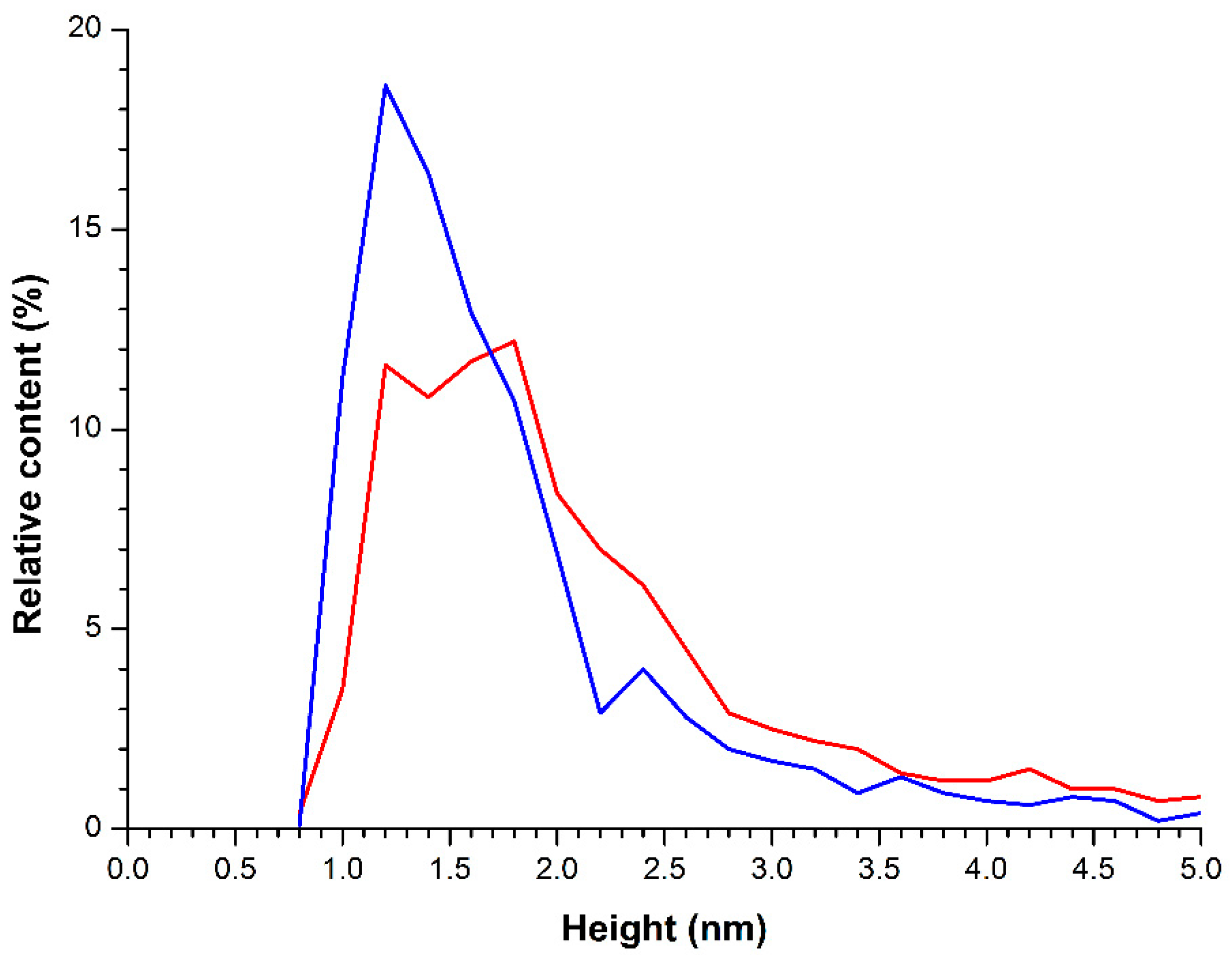
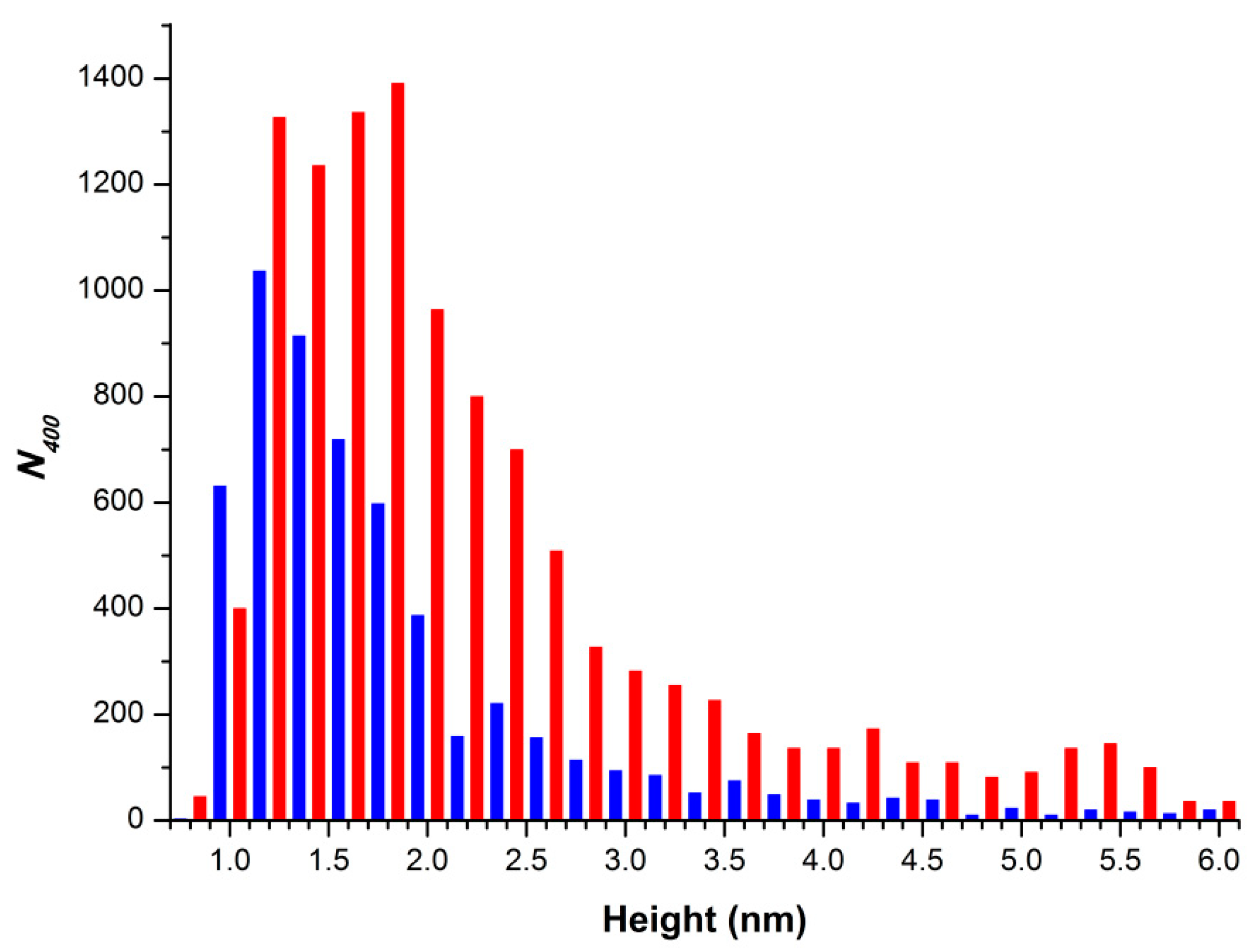
2.1. Atomic Force Microscopy Visualization of Mica-Adsorbed HRP
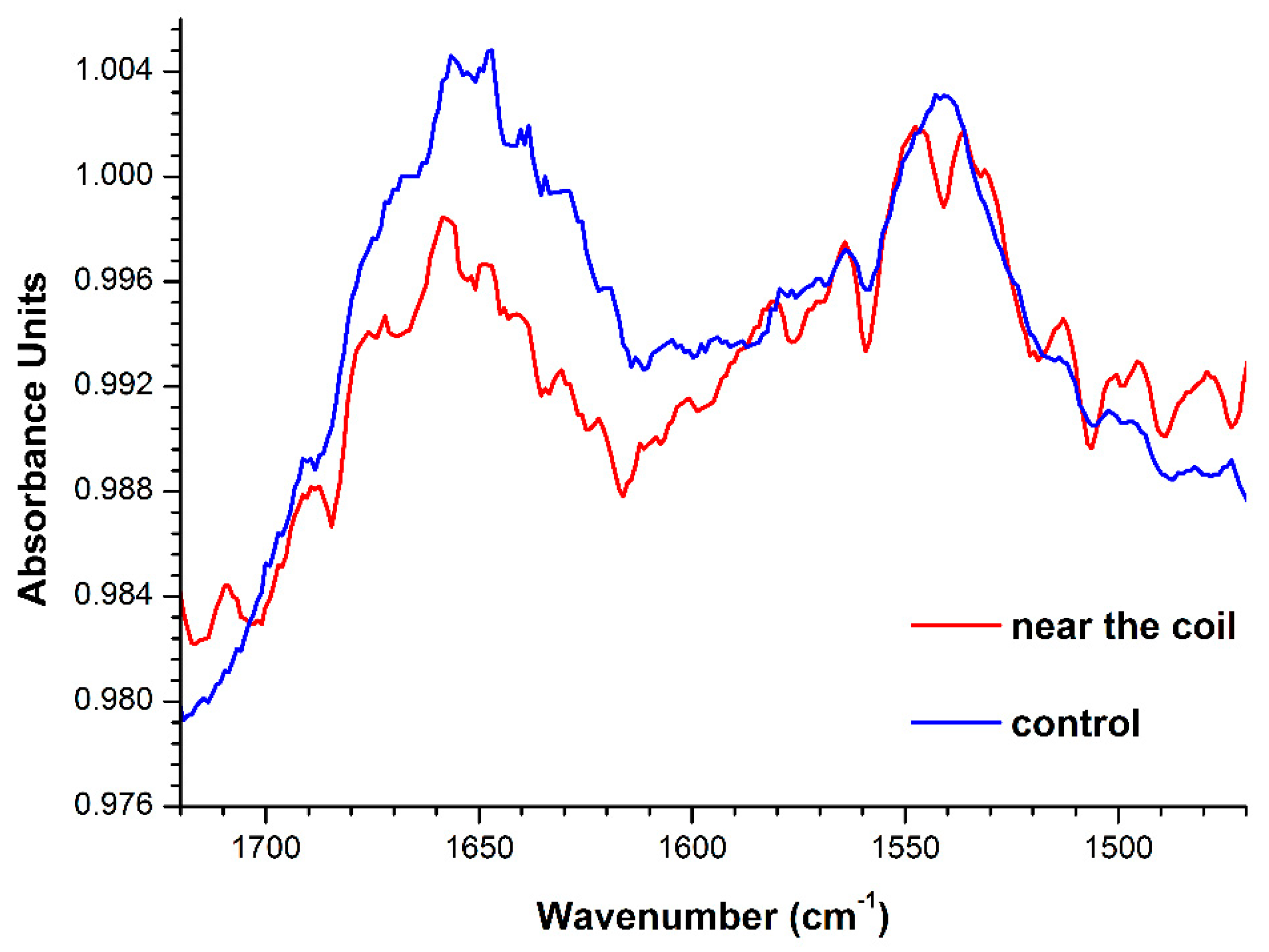
2.2. ATR-FTIR Revelation of Changes in the HRP Secondary Structure
2.3. Spectrophotometric Estimation of the HRP Enzymatic Activity
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Chemicals and Protein
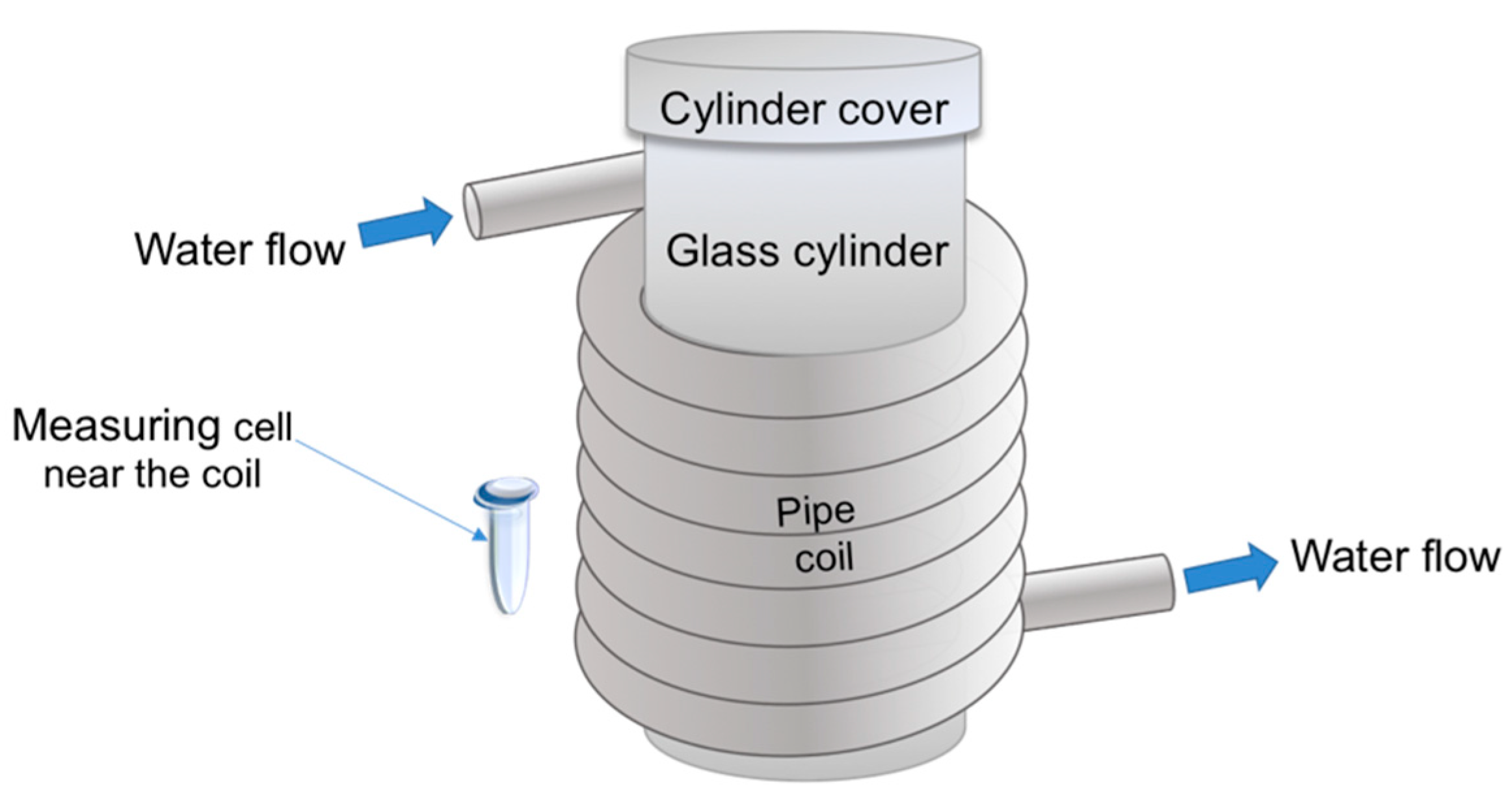
5.2. Experimental Setup
5.3. Atomic Force Microscopy
5.4. ATR-FTIR Measurements
5.5. Spectrophotometric Measurements
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J.; et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archakov, A.I.; Ivanov, Y.D.; Lisitsa, A.V.; Zgoda, V.G. AFM fishing nanotechnology is the way to reverse the Avogadro number in proteomics. Proteomics 2007, 7, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Pleshakova, T.O.; Shumov, I.D.; Ivanov, Y.D.; Malsagova, K.A.; Kaysheva, A.L.; Archakov, A.I. AFM-Based technologies as the way towards the reverse Avogadro number. Biochem. Suppl. Ser. B Biomed. Chem. 2015, 9, 244–257. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Danichev, V.V.; Pleshakova, T.O.; Shumov, I.D.; Ziborov, V.S.; Krokhin, N.V.; Zagumenniy, M.N.; Ustinov, V.S.; Smirnov, L.P.; Shironin, A.V.; et al. Irreversible chemical AFM-based fishing for detection of low-copied proteins. Biochem. Suppl. Ser. B Biomed. Chem. 2013, 7, 46–61. [Google Scholar] [CrossRef]
- Archakov, A.I.; Ivanov, Y.D.; Lisitsa, A.V.; Zgoda, V.G. Biospecific irreversible fishing coupled with atomic force microscopy for detection of extremely low-abundant proteins. Proteomics 2009, 9, 1326–1343. [Google Scholar] [CrossRef]
- Haas, P.; Then, P.; Wild, A.; Grange, W.; Zorman, S.; Hegner, M.; Calame, M.; Aebi, U.; Flammer, J.; Hecht, B. Fast quantitative single-molecule detection at ultralow concentrations. Anal. Chem. 2010, 82, 6299–6302. [Google Scholar] [CrossRef]
- IAsys Cuvette System User’s Guide, 1st ed.; Fisons plc.: Loughborough, UK, 1993.
- Ivanov, Y.D.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Romanova, T.S.; Valueva, A.A.; Tatur, V.Y.; Stepanov, I.N.; Ziborov, V.S. Investigation of the influence of liquid motion in a flow-based system on an enzyme aggregation state with an atomic force microscopy sensor: The effect of water flow. Appl. Sci. 2020, 10, 4560. [Google Scholar] [CrossRef]
- Davies, P.F.; Rennke, H.G.; Cotran, R.S. Influence of molecular charge upon the endocytosis and intracellular fate of peroxidase activity in cultured arterial endothelium. J. Cell Sci. 1981, 49, 69–86. [Google Scholar]
- Welinder, K.G. Amino acid sequence studies of horseradish peroxidase: Amino and carboxyl termini, cyanogen bromide and tryptic fragments, the complete sequence, and some structural characteristics of horseradish peroxidase C. Eur. J. Biochem. 1979, 96, 483–502. [Google Scholar] [CrossRef]
- Rogozhin, V.V.; Kutuzova, G.D.; Ugarova, N.N. Inhibition of horseradish peroxidase by N-ethylamide of o-sulfobenzoylacetic acid. Bioorganic Chem. 2000, 26, 156–160. [Google Scholar]
- Veitch, N.C.; Smith, A.T. Horseradish peroxidase. Adv. Inorg. Chem. 2001, 51, 107–162. [Google Scholar] [CrossRef]
- Shannon, L.M.; Kay, E.; Lew, J.Y. Peroxidase isozymes from horseradish roots I. Isolation and physical properties. J. Biol. Chem. 1966, 241, 2166–2172. [Google Scholar] [CrossRef]
- Tams, J.W.; Welinder, K.G. Mild chemical deglycosylation of horseradish peroxidase yields a fully active, homogeneous enzyme. Anal. Biochem. 1995, 228, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Kiselyova, O.I.; Yaminsky, I.V.; Ivanov, Y.D.; Kanaeva, I.P.; Kuznetsov, V.Y.; Archakov, A.I. AFM study of membrane proteins, cytochrome P450 2B4, and NADPH–Cytochrome P450 reductase and their complex formation. Arch. Biochem. Biophys. 1999, 371, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Braga, P.C.; Ricci, D. Methods in molecular biology. In Atomic Force Microscopy. Biomedical Methods and Applications; Humana Press: Totowa, NJ, USA, 2003; Volume 242. [Google Scholar]
- Ivanov, Y.D.; Bukharina, N.S.; Frantsuzov, P.A.; Pleshakova, T.O.; Kanashenko, S.L.; Medvedeva, N.V.; Argentova, V.V.; Zgoda, V.G.; Munro, A.W.; Archakov, A.I. AFM study of cytochrome CYP102A1 oligomeric state. Soft Matter 2012, 8, 4602–4608. [Google Scholar] [CrossRef]
- Usoltsev, D.; Sitnikova, V.; Kajava, A.; Uspenskaya, M. Systematic FTIR spectroscopy study of the secondary structure changes in human serum albumin under various denaturation conditions. Biomolecules 2019, 9, 359. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Yang, S.; Kong, J.; Dong, A.; Yu, S. Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 2015, 10, 382–396. [Google Scholar] [CrossRef]
- Arunkumar, R.; Drummond, C.J.; Greaves, T.L. FTIR spectroscopic study of the secondary structure of globular proteins in aqueous protic ionic liquids. Front. Chem. 2019, 7, 14. [Google Scholar] [CrossRef]
- Kariakin, A.; Davydov, D.; Peterson, J.A.; Jung, C. A new approach to the study of protein–protein interaction by FTIR: Complex formation between cytochrome P450BM-3 heme domain and FMN reductase domain. Biochemistry 2002, 41, 13514–13525. [Google Scholar] [CrossRef]
- Haris, P.I. Probing protein–protein interaction in biomembranes using Fourier transform infrared spectroscopy. Biochim. Biophys. Acta 2013, 1828, 2265–2271. [Google Scholar] [CrossRef]
- Moyo, M. Horseradish peroxidase biosensor to detect zinc ions in aqueous solutions. Open J. Appl. Biosens. 2014, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Shivu, B.; Seshadri, S.; Li, J.; Oberg, K.A.; Uversky, V.N.; Fink, A.L. Distinct β-sheet structure in protein aggregates determined by ATR–FTIR spectroscopy. Biochemistry 2013, 52, 5176–5183. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, V.Y.; Ivanov, Y.D.; Bykov, V.A.; Saunin, S.A.; Fedorov, I.A.; Lemeshko, S.V.; Hui Bon Hoa, G.; Archakov, A.I. Atomic force microscopy detection of molecular complexes in multiprotein P450cam-containing monooxygenase system. Proteomics 2002, 2, 1699–1705. [Google Scholar] [CrossRef]
- Unger, B.P.; Gunsalus, I.C.; Sligar, S.G. Nucleotide sequence of the Pseudomonas putida cytochrome P-450cam gene and its expression in Escherichia coli. J. Biol. Chem. 1986, 261, 1158–1163. [Google Scholar] [PubMed]
- Ivanov, Y.D.; Frantsuzov, P.A.; Zöllner, A.; Medvedeva, N.V.; Archakov, A.I.; Reinle, W.; Bernhardt, R. Atomic force microscopy study of protein-protein interactions in the cytochrome CYP11A1 (P450scc)- containing steroid hydroxylase system. Nanoscale Res. Lett. 2011, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Chu, J.-W.; Kimura, T. Studies on adrenal steroid hydroxylases. Molecular and catalytic properties of adrenodoxin reductase (a flavoprotein). J. Biol. Chem. 1973, 248, 2089–2094. [Google Scholar] [CrossRef]
- Choi, D.; Lee, H.; Im, D.J.; Kang, I.S.; Lim, G.; Kim, D.S.; Kang, K.H. Spontaneous electrical charging of droplets by conventional pipetting. Sci. Rep. 2013, 3, 1–7. [Google Scholar] [CrossRef]
- Ivanova, Y.D.; Kozlov, A.F.; Galiullin, R.A.; Kanashenko, S.L.; Usanov, S.A.; Ivanova, N.D.; Ziborov, V.S.; Pleshakova, T.O. Spontaneous generation of charge in the flow-based AFM fishing system. J. Electrost. 2018, 91, 16–20. [Google Scholar] [CrossRef]
- Stetten, A.Z.; Golovko, D.S.; Weber, S.A.L.; Butt, H.-J. Slide electrification: Charging of surfaces by moving water drops. Soft Matter 2019, 15, 8667–8679. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, Y.D.; Kozlov, A.F.; Galiullin, R.A.; Valueva, A.A.; Pleshakova, T.O. The dependence of spontaneous charge generation in water on its flow rate in a flow-based analytical system. Appl. Sci. 2020, 10, 2444. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Liu, L.; Yang, X.; Hong, H.; Yang, Q.; Wang, J.; Tang, Q. Cumulative charging behavior of water droplets driven freestanding triboelectric nanogenerator toward hydrodynamic energy harvesting. J. Mater. Chem. A 2020, 8, 7880–7888. [Google Scholar] [CrossRef]
- Haque, R.I.; Arafat, A.; Briand, D. Triboelectric effect to harness fluid flow energy. J. Phys. Conf. Ser. 2019, 1407, 012084. [Google Scholar] [CrossRef]
- Ivanov, Y.D.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Ivanova, I.A.; Valueva, A.V.; Tatur, V.Y.; Smelov, M.V.; Ivanova, N.D.; Ziborov, V.S. AFM imaging of protein aggregation in studying the impact of knotted electromagnetic field on a peroxidase. Sci. Rep. 2020, 10, 9022. [Google Scholar] [CrossRef] [PubMed]
- Ziborov, V.S.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Ivanova, I.A.; Valueva, A.A.; Tatur, V.Y.; Negodailov, A.N.; Lukyanitsa, A.A.; Ivanov, Y.D. Investigation of the influence of liquid motion in a flow-based system on an enzyme aggregation state with an atomic force microscopy sensor: The effect of glycerol flow. Appl. Sci. 2020, 10, 4825. [Google Scholar] [CrossRef]
- Sun, J.; Sun, F.; Xu, B.; Gu, N. The quasi-one-dimensional assembly of horseradish peroxidase molecules in presence of the alternating magnetic field. Coll. Surf. A Physicochem. Eng. Asp. 2010, 360, 94–98. [Google Scholar] [CrossRef]
- Sun, J.; Zhou, H.; Jin, Y.; Wang, M.; Gu, N. Magnetically enhanced dielectrophoretic assembly of horseradish peroxidase molecules: Chaining and molecular monolayers. Chem. Phys. Chem. 2008, 9, 1847–1850. [Google Scholar] [CrossRef]
- Narhi, L.O.; Fulco, A.J. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P-450 monooxygenase induced by barbiturates in Bacillus megaterium. J. Biol. Chem. 1986, 261, 7160–7169. [Google Scholar] [CrossRef]
- Sevrioukova, I.; Li, H.; Zhang, H.; Peterson, J.; Poulos, T. Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. USA 1999, 96, 1863–1868. [Google Scholar] [CrossRef] [Green Version]
- Black, S.; Martin, S. Evidence for conformational dynamics and molecular aggregation in cytochrome P450 102 (BM-3). Biochemistry 1994, 33, 12056–12062. [Google Scholar] [CrossRef]
- Neeli, R.; Girvan, H.M.; Lawrence, A.; Warren, M.J.; Leys, D.; Scrutton, N.S.; Munro, A.W. The dimeric form of flavocytochrome P450 CYP102A1 is catalytically functional as a fatty acid hydroxylase. FEBS Lett. 2005, 579, 5582–5588. [Google Scholar] [CrossRef]
- Gavrilenko, T.I.; Ryzhkova, N.A.; Parkhomenko, A.N. Myeloperoxidase and its role in the development of coronary heart disease. Ukr. J. Cardiol. 2014, 4, 119–126. [Google Scholar]
- Cook, D.; Nuro, E.; Jones, E.V.; Altimimi, H.F.; Farmer, T.; Gandin, V.; Hanna, E.; Zong, R.; Barbon, A.; Nelson, D.L.; et al. FXR1P limits long-term memory, long-lasting synaptic potentiation, and de novo GluA2 translation. Cell Rep. 2014, 9, 1402–1416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del’Guidice, T.; Latapy, C.; Rampino, A.; Khlghatyan, J.; Lemasson, M.; Gelao, B.; Quarto, T.; Rizzo, G.; Barbeau, A.; Lamarre, C.; et al. FXR1P is a GSK3β substrate regulating mood and emotion processing. Proc. Natl. Acad. Sci. USA 2015, 112, E4610–E4619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sopova, J.V.; Koshel, E.I.; Belashova, T.A.; Zadorsky, S.P.; Sergeeva, A.V.; Siniukova, V.A.; Shenfeld, A.A.; Velizhanina, M.E.; Volkov, K.V.; Nizhnikov, A.A.; et al. RNA-binding protein FXR1 is presented in rat brain in amyloid form. Sci. Rep. 2019, 9, 18983. [Google Scholar] [CrossRef] [Green Version]
- Keller, D. Reconstruction of STM and AFM images distorted by finite-size tips. Surf. Sci. 1991, 253, 353–364. [Google Scholar] [CrossRef]
- Sanders, S.A.; Bray, R.C.; Smith, A.T. pH-dependent properties of a mutant horseradish peroxidase isoenzyme C in which Arg38 has been replaced with lysine. Eur. J. Biochem. 1994, 224, 1029–1037. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanov, Y.D.; Pleshakova, T.O.; Shumov, I.D.; Kozlov, A.F.; Valueva, A.A.; Ivanova, I.A.; Ershova, M.O.; Larionov, D.I.; Repnikov, V.V.; Ivanova, N.D.; et al. AFM and FTIR Investigation of the Effect of Water Flow on Horseradish Peroxidase. Molecules 2021, 26, 306. https://doi.org/10.3390/molecules26020306
Ivanov YD, Pleshakova TO, Shumov ID, Kozlov AF, Valueva AA, Ivanova IA, Ershova MO, Larionov DI, Repnikov VV, Ivanova ND, et al. AFM and FTIR Investigation of the Effect of Water Flow on Horseradish Peroxidase. Molecules. 2021; 26(2):306. https://doi.org/10.3390/molecules26020306
Chicago/Turabian StyleIvanov, Yuri D., Tatyana O. Pleshakova, Ivan D. Shumov, Andrey F. Kozlov, Anastasia A. Valueva, Irina A. Ivanova, Maria O. Ershova, Dmitry I. Larionov, Victor V. Repnikov, Nina D. Ivanova, and et al. 2021. "AFM and FTIR Investigation of the Effect of Water Flow on Horseradish Peroxidase" Molecules 26, no. 2: 306. https://doi.org/10.3390/molecules26020306
APA StyleIvanov, Y. D., Pleshakova, T. O., Shumov, I. D., Kozlov, A. F., Valueva, A. A., Ivanova, I. A., Ershova, M. O., Larionov, D. I., Repnikov, V. V., Ivanova, N. D., Tatur, V. Y., Stepanov, I. N., & Ziborov, V. S. (2021). AFM and FTIR Investigation of the Effect of Water Flow on Horseradish Peroxidase. Molecules, 26(2), 306. https://doi.org/10.3390/molecules26020306





