citSATdb: Genome-Wide Simple Sequence Repeat (SSR) Marker Database of Citrus Species for Germplasm Characterization and Crop Improvement
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. In Silico Simple Sequence Repeat Mining and Primer Designing
2.3. Functional Annotation of SSR Markers
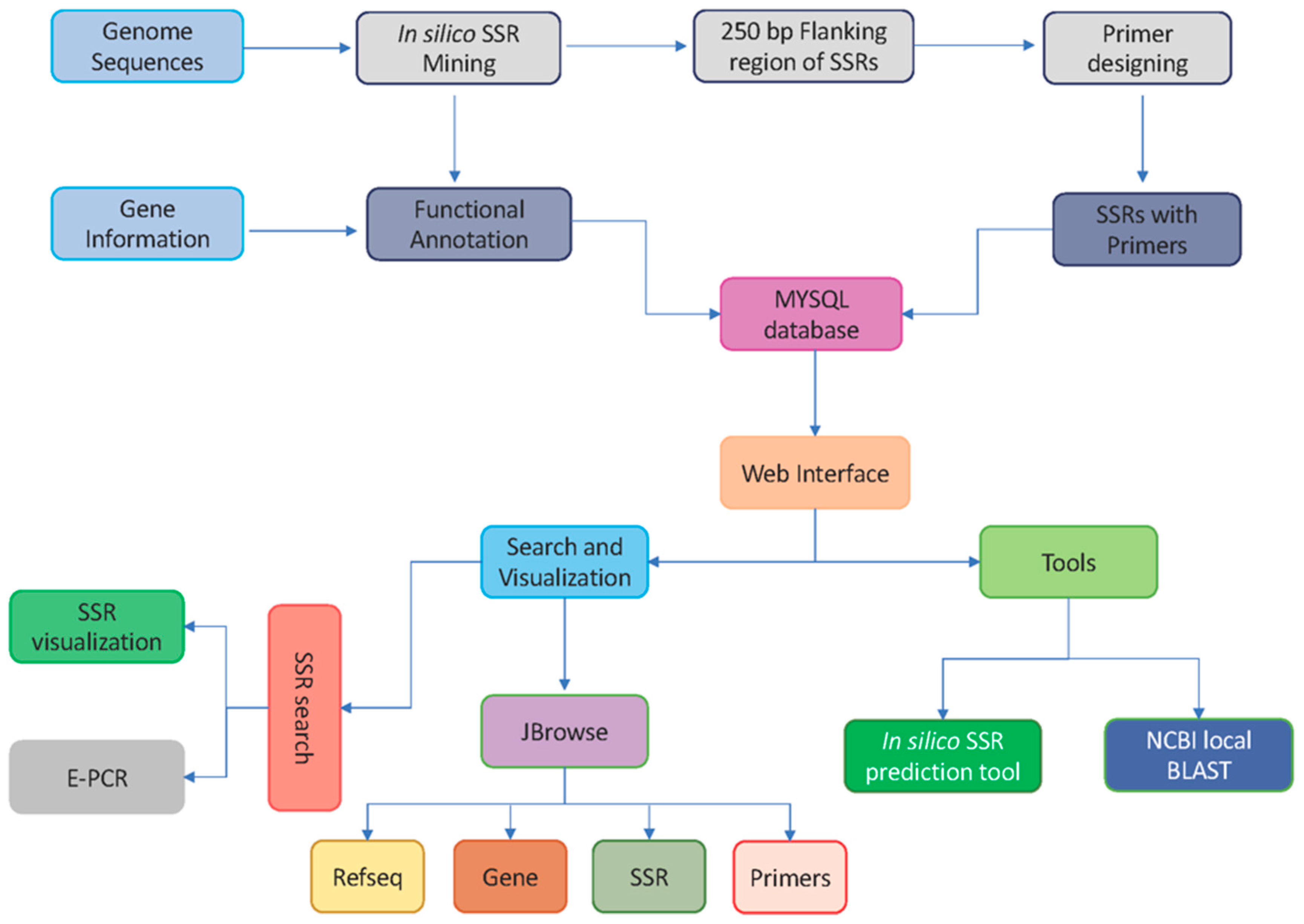
2.4. Marker and Database Development Workflow
2.5. Database Development and Web Interface
3. Results and Discussion
3.1. Cross-Species Comparison of Citrus Species SSRs
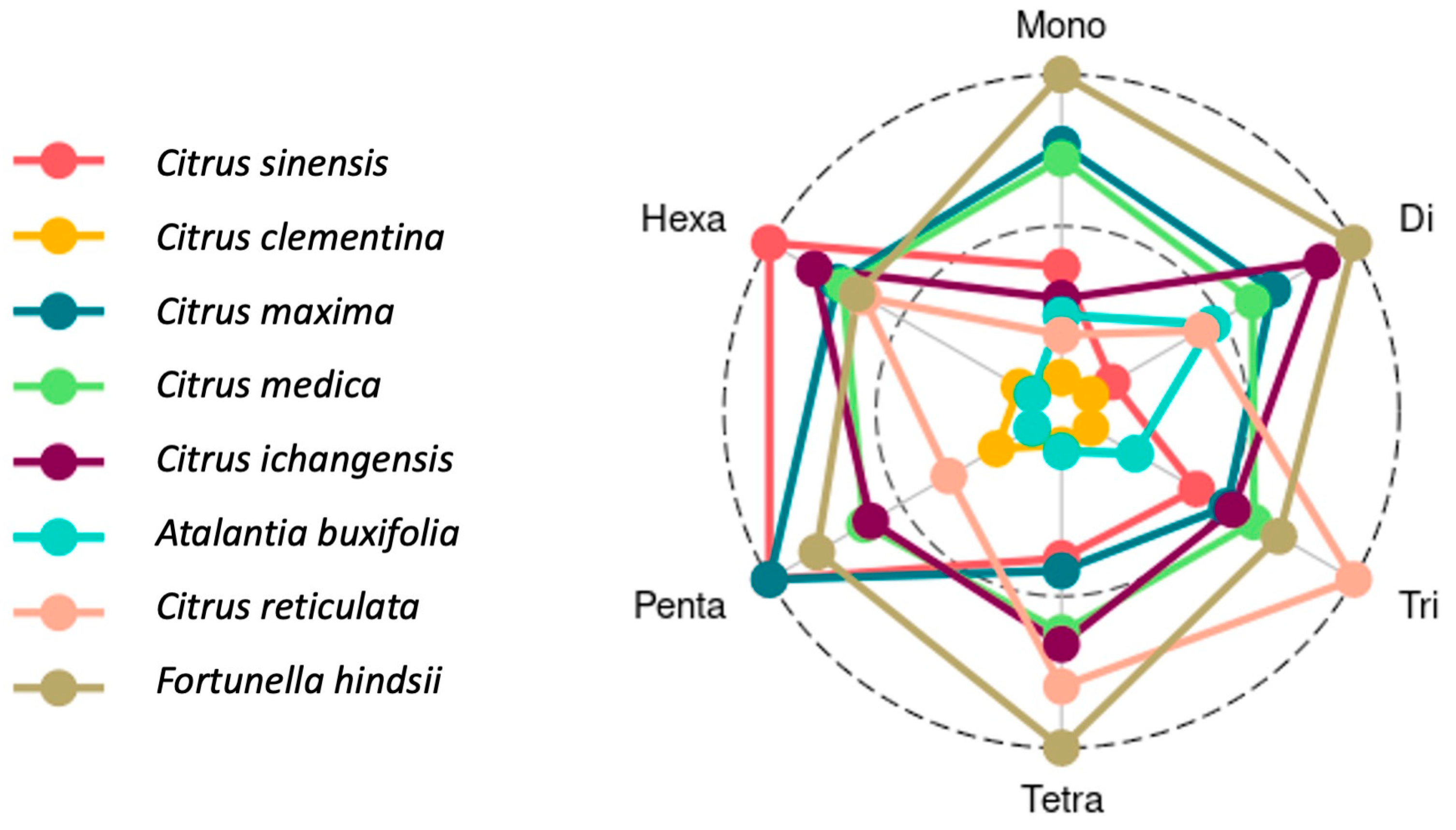
3.2. SSR Motifs Characterized by Repeat Length
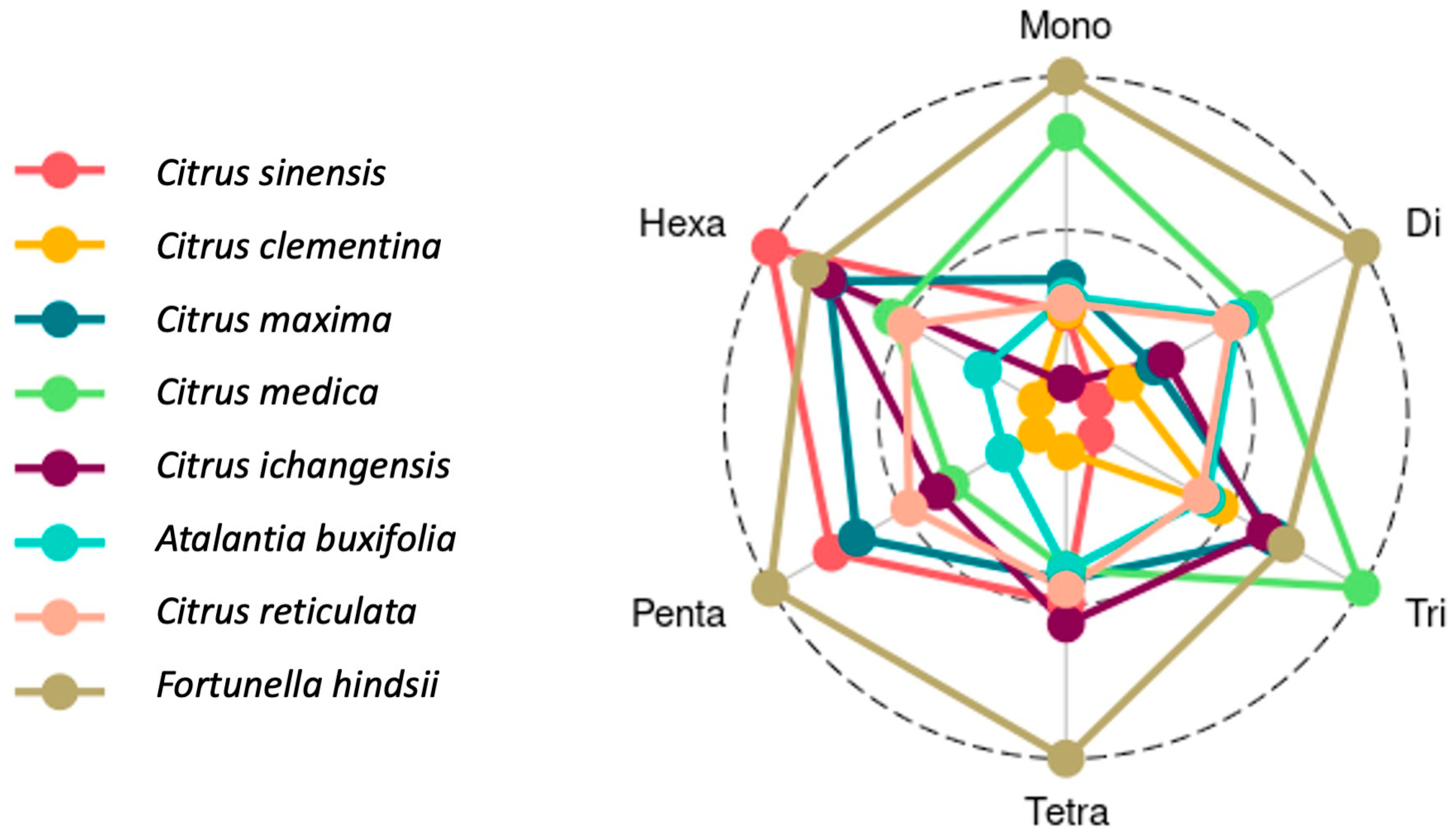
3.3. Designed SSR Primers, Motif Characterization by Repeat Length
3.4. Functional Annotation of SSRs and Markers
3.5. Comparison with Another Databases
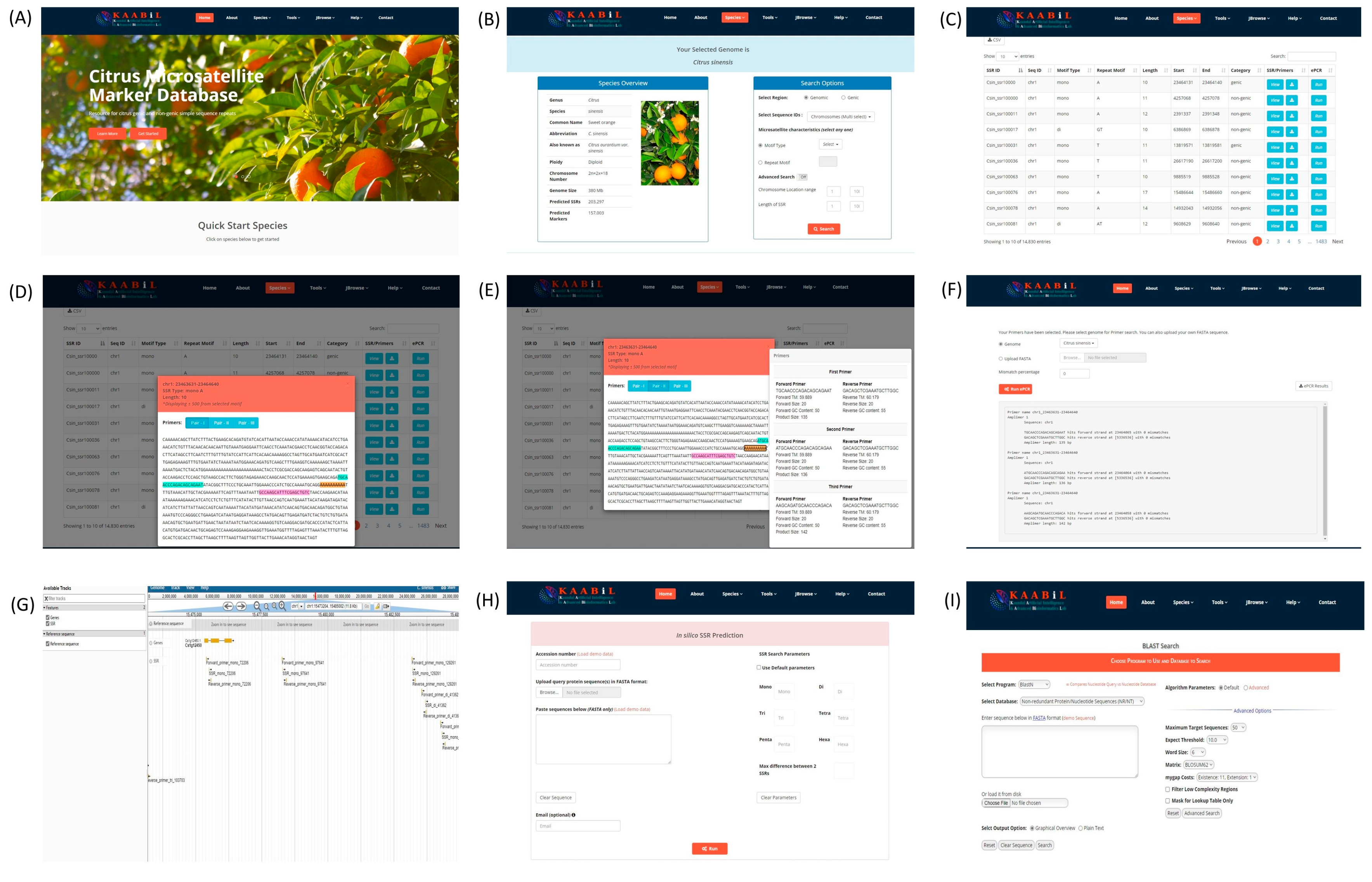
3.6. citSATdb: Citrus Microsatellite Web-Genomic Resource
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Collard, B.C.Y.; Septiningsih, E.M.; Das, S.R.; Carandang, J.J.; Pamplona, A.M.; Sanchez, D.L.; Kato, Y.; Ye, G.; Reddy, J.N.; Singh, U.S.; et al. Developing new flood-tolerant varieties at the international rice research institute (IRRI). Sabrao J. Breed. Genet. 2013, 45, 42–56. [Google Scholar]
- Xiu, X.D. Advances in worldwide citrus breeding. Acta Hortic. Sin. 2005, 32, 1140–1146. [Google Scholar]
- Johnson, R. Marker-Assisted Selection. In Plant Breeding Reviews; John Wiley & Sons Inc.: Oxford, UK, 2010; pp. 293–309. [Google Scholar]
- Cuenca, J.; Aleza, P.; Garcia-Lor, A.; Ollitrault, P.; Navarro, L. Fine Mapping for Identification of Citrus Alternaria Brown Spot Candidate Resistance Genes and Development of New SNP Markers for Marker-Assisted Selection. Front. Plant Sci. 2016, 7, 1948. [Google Scholar] [CrossRef] [PubMed]
- Gupta, H.; Agrawal, P.K.; Mahajan, V.; Bisht, G.S.; Kumar, A.; Verma, P.; Srivastava, A.; Saha, S.; Babu, R.; Pant, M.C.; et al. Quality protein maize for nutritional security: Rapid development of short duration hybrids through molecular marker assisted breeding. Curr. Sci. 2009, 96, 230–237. [Google Scholar]
- Omura, M.; Shimada, T. Citrus breeding, genetics and genomics in Japan. Breed. Sci. 2016, 66, 3–17. [Google Scholar] [CrossRef]
- Xu, Y.; Crouch, J.H. Marker-assisted selection in plant breeding: From publications to practice. Crop Sci. 2008, 48, 391–407. [Google Scholar] [CrossRef]
- Yu, J.; Dossa, K.; Wang, L.; Zhang, Y.; Wei, X.; Liao, B.; Zhang, X. PMDBase: A database for studying microsatellite DNA and marker development in plants. Nucleic Acids Res. 2017, 45, D1046–D1053. [Google Scholar] [CrossRef]
- Kalia, R.K.; Rai, M.K.; Kalia, S.; Singh, R.; Dhawan, A.K. Microsatellite markers: An overview of the recent progress in plants. Euphytica 2011, 177, 309–334. [Google Scholar] [CrossRef]
- Xu, X.; Peng, M.; Fang, Z.; Xu, X. The direction of microsatellite mutations is dependent upon allele length. Nat. Genet. 2000, 24, 396–399. [Google Scholar] [CrossRef]
- Wierdl, M.; Dominska, M.; Petes, T.D. Microsatellite instability in yeast: Dependence on the length of the microsatellite. Genetics 1997, 146, 769–779. [Google Scholar]
- Akemi, A.; Pereira, J.; Macedo, P.; Alessandra, K. Microsatellites as Tools for Genetic Diversity Analysis. Genet. Divers. Microorg. 2012. [Google Scholar] [CrossRef]
- Jiao, Y.; Jia, H.; Li, X.; Chai, M.; Jia, H.; Chen, Z.; Wang, G.; Chai, C.; van de Weg, E.; Gao, Z. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genom. 2012, 13, 201. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-R.; Li, W.-Y.; Long, D.; Hu, C.-G.; Zhang, J.-Z. Development and Characterization of Genomic and Expressed SSRs in Citrus by Genome-Wide Analysis. PLoS ONE 2013, 8, e75149. [Google Scholar] [CrossRef]
- Shrestha, R.L.; Dhakal, D.D.; Gautum, D.M.; Paudyal, K.P.; Shrestha, S.; Shrestha, R.L.; Dhakal, D.D.; Gautum, D.M.; Paudyal, K.P.; Shrestha, S. Genetic Diversity Assessment of Acid Lime (Citrus aurantifolia Swingle) Landraces in Nepal, Using SSR Markers. Am. J. Plant Sci. 2012, 3, 1674–1681. [Google Scholar] [CrossRef]
- Biswas, M.K.; Xu, Q.; Mayer, C.; Deng, X. Genome Wide Characterization of Short Tandem Repeat Markers in Sweet Orange (Citrus sinensis). PLoS ONE 2014, 9, e104182. [Google Scholar] [CrossRef]
- Ollitrault, F.; Terol, J.; Pina, J.A.; Navarro, L.; Talon, M.; Ollitrault, P. Development of SSR markers from Citrus clementina (Rutaceae) BAC end sequences and interspecific transferability in Citrus. Am. J. Bot. 2010, 97, e124–e129. [Google Scholar] [CrossRef]
- Sharafi, A.A.; Abkenar, A.A.; Sharafi, A.; Masaeli, M. Genetic variation assessment of acid lime accessions collected from south of Iran using SSR and ISSR molecular markers. Physiol. Mol. Biol. Plants 2016, 22, 87–95. [Google Scholar] [CrossRef]
- Barkley, N.A.; Roose, M.L.; Krueger, R.R.; Federici, C.T. Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theor. Appl. Genet. 2006, 112, 1519–1531. [Google Scholar] [CrossRef]
- Fanciullino, A.L.; Dhuique-Mayer, C.; Luro, F.; Morillon, R.; Ollitrault, P. Carotenoid biosynthetic pathway in the Citrus genus: Number of copies and phylogenetic diversity of seven genes. J. Agric. Food Chem. 2007, 55, 7405–7417. [Google Scholar] [CrossRef]
- Luro, F.L.; Costantino, G.; Terol, J.; Argout, X.; Allario, T.; Wincker, P.; Talon, M.; Ollitrault, P.; Morillon, R. Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other citrus species and their effectiveness for genetic mapping. BMC Genom. 2008, 9, 287. [Google Scholar] [CrossRef]
- Jiang, D.; Zhong, G.Y.; Hong, Q.B. Analysis of microsatellites in citrus unigenes. Acta Genet. Sin. 2006, 33, 345–353. [Google Scholar] [CrossRef]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.C.; Grover, A.; Kahl, G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007, 25, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.S.; Lee, J.M.; Yi, G.B.; Yi, S.I.; Kim, K.M.; Soh, E.H.; Bae, K.M.; Park, E.K.; Song, I.H.; Kim, B.D. Use of SSR markers to complement tests of distinctiveness, uniformity, and stability (DUS) of pepper (Capsicum annuum L.) varieties. Mol. Cells 2005, 19, 428–435. [Google Scholar] [PubMed]
- Pourabed, E.; Jazayeri Noushabadi, M.R.; Jamali, S.H.; Moheb Alipour, N.; Zareyan, A.; Sadeghi, L. Identification and DUS testing of rice varieties through microsatellite markers. Int. J. Plant Genom. 2015, 2015, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Thirusendura Selvi, D.; Srimathi, P.; Senthil, N.; Ganesan, K.N. Distinctness, uniformity and stability (DUS) characterization on phenological traits and assessing the diversity of inbreds in maize (Zea mays L.). Afr. J. Agric. Res. 2013, 8, 6086–6092. [Google Scholar] [CrossRef]
- Vishnu, K. Characterization for DUS descriptors and environmental interaction studies for grain protein and starch content in barley (H. vulgare). Sabrao J. Breed. Genet. Pagination 2015, 260, 267. [Google Scholar]
- He, B.; Geng, R.; Cheng, L.; Yang, X.; Ge, H.; Ren, M. Genetic diversity and fingerprinting of 33 standard flue-cured tobacco varieties for use in distinctness, uniformity, and stability testing. BMC Plant Biol. 2020, 20, 378. [Google Scholar] [CrossRef]
- Bisen, A.; Khare, D.; Nair, P.; Tripathi, N. SSR analysis of 38 genotypes of soybean (Glycine Max (L.) Merr.) genetic diversity in India. Physiol. Mol. Biol. Plants 2014, 21, 109–115. [Google Scholar] [CrossRef]
- Wang, L.X.; Qiu, J.; Chang, L.F.; Liu, L.H.; Li, H.B.; Pang, B.S.; Zhao, C.P. Assessment of wheat variety distinctness using SSR markers. J. Integr. Agric. 2015, 14, 1923–1935. [Google Scholar] [CrossRef][Green Version]
- Molla, R.; Ahmed, I.; Rohman, M.; Hossain, A.; Chowdhury, A.Z. Genetic Diversity Analysis and DNA Fingerprinting of Mungbean (Vigna radiata L.) Genotypes Using SSR Markers. J. Plant Sci. 2016, 4, 153. [Google Scholar] [CrossRef]
- Selvan, R.T.; Parthiban, K.T.; Palanikumaran, B. Distinctness, Uniformity and Stability (DUS) Characterization of Neolamarckia Cadamba Genetic Resources. Curr. Agric. Res. J. 2019, 7, 268–275. [Google Scholar] [CrossRef]
- Reid, A.; Kerr, E.M. A rapid simple sequence repeat (SSR)-based identification method for potato cultivars. Plant Genet. Resour. 2007, 5, 7–13. [Google Scholar] [CrossRef]
- Moore, S.S.; Sargeant, L.L.; King, T.J.; Mattick, J.S.; Georges, M.; Hetzel, D.J. The conservation of dinucleotide microsatellites among mammalian genomes allows the use of heterologous PCR primer pairs in closely related species. Genomics 1991, 10, 654–660. [Google Scholar] [CrossRef]
- Gao, L.Z.; Zhang, C.H.; Jia, J.Z. Cross-species Transferability of Rice Microsatellites in its Wild Relatives and the Potential for Conservation Genetic Studies. Genet. Resour. Crop Evol. 2005, 52, 931–940. [Google Scholar] [CrossRef]
- Du, L.; Liu, Q.; Zhao, K.; Tang, J.; Zhang, X.; Yue, B.; Fan, Z. PSMD: An extensive database for pan-species microsatellite investigation and marker development. Mol. Ecol. Resour. 2020, 20, 283–291. [Google Scholar] [CrossRef]
- Mokhtar, M.M.; Atia, M.A.M. SSRome: An integrated database and pipelines for exploring microsatellites in all organisms. Nucleic Acids Res. 2019, 47, D244–D252. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 2013, 45, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Zhang, S.; Cao, L.; Huang, Y.; Cheng, J.; Wu, G.; Tian, S.; Chen, C.; Liu, Y.; et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017, 49, 765–772. [Google Scholar] [CrossRef]
- Citrus Genome Database. Available online: https://www.citrusgenomedb.org/ (accessed on 13 November 2020).
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Katti, M.V.; Ranjekar, P.K.; Gupta, V.S. Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol. Biol. Evol. 2001, 18, 1161–1167. [Google Scholar] [CrossRef] [PubMed]
- Tóth, G.; Gáspári, Z.; Jurka, J. Microsatellites in different eukaryotic genomes: Survey and analysis. Genome Res. 2000, 10, 967–981. [Google Scholar] [CrossRef]
- Srivastava, S.; Avvaru, A.K.; Sowpati, D.T.; Mishra, R.K. Patterns of microsatellite distribution across eukaryotic genomes. BMC Genom. 2019, 20, 153. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Tian, Y.; Yang, R.; Feng, H.; Ouyang, Q.; Tian, Y.; Tan, Z.; Li, M.; Niu, Y.; Jiang, J.; et al. Coevolution between simple sequence repeats (SSRs) and virus genome size. BMC Genom. 2012, 13, 435. [Google Scholar] [CrossRef]
- Portis, E.; Portis, F.; Valente, L.; Moglia, A.; Barchi, L.; Lanteri, S.; Acquadro, A. A genome-wide survey of the microsatellite content of the globe artichoke genome and the development of a web-based database. PLoS ONE 2016, 11, e0162841. [Google Scholar] [CrossRef]
- Portis, E.; Lanteri, S.; Barchi, L.; Portis, F.; Valente, L.; Toppino, L.; Rotino, G.L.; Acquadro, A. Comprehensive Characterization of Simple Sequence Repeats in Eggplant (Solanum melongena L.) Genome and Construction of a Web Resource. Front. Plant Sci. 2018, 9, 401. [Google Scholar] [CrossRef]
- Haseneyer, G.; Schmutzer, T.; Seidel, M.; Zhou, R.; Mascher, M.; Schön, C.C.; Taudien, S.; Scholz, U.; Stein, N.; Mayer, K.F.X.; et al. From RNA-seq to large-scale genotyping—Genomics resources for rye (Secale cereale L.). BMC Plant Biol. 2011, 11, 131. [Google Scholar] [CrossRef]
- Kariin, S.; Burge, C. Dinucleotide relative abundance extremes: A genomic signature. Trends Genet. 1995, 11, 283–290. [Google Scholar] [CrossRef]
- Shioiri, C.; Takahata, N. Skew of mononucleotide frequencies, relative abundance of dinucleotides, and DNA strand asymmetry. J. Mol. Evol. 2001, 53, 364–376. [Google Scholar] [CrossRef] [PubMed]
- Iquebal, M.A.; Arora, V.; Verma, N.; Rai, A.; Kumar, D. First whole genome based microsatellite DNA marker database of tomato for mapping and variety identification. BMC Plant Biol. 2013, 13, 197. [Google Scholar] [CrossRef]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Karakousis, A.; Barr, A.R.; Chalmers, K.J.; Ablett, G.A.; Holton, T.A.; Henry, R.J.; Lim, P.; Langridge, P. Potential of SSR markers for plant breeding and variety identification in Australian barley germplasm. Aust. J. Agric. Res. 2003, 54, 1197–1210. [Google Scholar] [CrossRef]
- Ali, A.; Jin-Da, W.; Yong-Bao, P.; Zu-Hu, D.; Zhi-Wei, C.; Ru-Kai, C.; San-Ji, G. Molecular Identification and Genetic Diversity Analysis of Chinese Sugarcane (Saccharum spp. Hybrids) Varieties using SSR Markers. Trop. Plant Biol. 2017, 10, 194–203. [Google Scholar] [CrossRef]
- Stàgel, A.; Portis, E.; Toppino, L.; Rotino, G.L.; Lanteri, S. Gene-based microsatellite development for mapping and phylogeny studies in eggplant. BMC Genom. 2008, 9, 357. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Ishii, K.; Kim, C.; Ban, T.; Suzuki, M.; Ito, T.; Muranaka, T.; Kobayashi, M.; Nagata, N.; Isobe, S.; et al. Development of Capsicum EST-SSR markers for species identification and in silico mapping onto the tomato genome sequence. Mol. Breed. 2013, 31, 101–110. [Google Scholar] [CrossRef]
- Bhattacharyya, U.; Pandey, S.K.; Dasgupta, T. Identification of EST-SSRs and FDM in sesame (Sesamum indicum L.) through data mining. Sch. J. Agric. Sci. 2014, 4, 60–69. [Google Scholar]
- Goto, S.; Yoshioka, T.; Ohta, S.; Kita, M.; Hamada, H.; Shimizu, T. QTL mapping of male sterility and transmission pattern in progeny of Satsuma mandarin. PLoS ONE 2018, 13, e0200844. [Google Scholar] [CrossRef]
- Hong, Q.B.; Ma, X.J.; Gong, G.Z.; Peng, Z.C.; He, Y.R. QTL mapping of citrus freeze tolerance. Acta Hortic. 2015, 1065, 467–474. [Google Scholar] [CrossRef]
- Biswas, M.K.; Liu, Y.; Li, C.; Sheng, O.; Mayer, C.; Yi, G. Genome-Wide Computational Analysis of Musa Microsatellites: Classification, Cross-Taxon Transferability, Functional Annotation, Association with Transposons & miRNAs, and Genetic Marker Potential. PLoS ONE 2015, 10, e0131312. [Google Scholar] [CrossRef]
| Species | Assembly Version | Assembly Level | Genome Size (Mb) | GC% |
|---|---|---|---|---|
| Citrus sinensis | v2.0 (HZAU) | Chromosome | 327.945 | 34.06% |
| Citrus clementina | v1.0 (JGI) | Scaffold | 301.387 | 34.96% |
| Citrus maxima | v1.0 (HZAU) | Chromosome | 345.78 | 34.99% |
| Citrus medica | v1.1 (HZAU) | Scaffold | 406.058 | 35.16% |
| Citrus ichangensis | v1.0 (HZAU) | Scaffold | 357.621 | 34.21% |
| Atalantia buxifolia | v1.1 (HZAU) | Scaffold | 315.821 | 33.55% |
| Citrus reticulata | v1.1(HZAU) | Pseudomolecule | 334.2 | - |
| Fortunella hindsii | V1.1(HZAU) | Contig | 373.6 | 34.49% |
| Species | Predicted SSRs | Designed Markers | SSRs/MB | Genome Size (MB) |
|---|---|---|---|---|
| Citrus sinensis | 203,297 | 157,003 | 619.91 | 380 |
| Citrus clementina | 187,778 | 150,593 | 638.15 | 370 |
| Citrus maxima | 224,961 | 184,840 | 650.59 | 328 |
| Citrus medica | 210,590 | 143,826 | 552.12 | 380 |
| Citrus ichangensis | 226,950 | 157,810 | 648.06 | 407 |
| Atalantia buxifolia | 204,687 | 147,140 | 675.26 | 391 |
| Citrus reticulata | 201,408 | 158,143 | 602.66 | 370 |
| Fortunella hindsii | 240,182 | 197,145 | 642.89 | 370 |
| Species | Mono | Di | Tri | Tetra | Penta | Hexa |
|---|---|---|---|---|---|---|
| Citrus sinensis | 121,051 | 54,874 | 23,568 | 3050 | 473 | 281 |
| Citrus clementina | 115,888 | 50,108 | 18,553 | 2620 | 436 | 173 |
| Citrus maxima | 142,446 | 57,059 | 21,668 | 2954 | 539 | 295 |
| Citrus medica | 125,467 | 60,040 | 21,259 | 2975 | 534 | 315 |
| Citrus ichangensis | 144,115 | 57,930 | 21,149 | 2844 | 613 | 299 |
| Atalantia buxifolia | 129,304 | 51,030 | 20,572 | 2822 | 613 | 346 |
| Citrus reticulata | 123,469 | 55,336 | 19,400 | 2631 | 408 | 164 |
| Fortunella hindsii | 152,611 | 61,408 | 22,143 | 3159 | 576 | 285 |
| Species | Mono | Di | Tri | Tetra | Penta | Hexa |
|---|---|---|---|---|---|---|
| Citrus sinensis | 97,149 | 44,198 | 13,069 | 2022 | 385 | 180 |
| Citrus clementina | 96,191 | 38,870 | 13,418 | 1722 | 285 | 107 |
| Citrus maxima | 120,885 | 45,399 | 16,037 | 1979 | 352 | 188 |
| Citrus medica | 85,972 | 40,922 | 14,248 | 2097 | 363 | 224 |
| Citrus ichangensis | 100,422 | 40,333 | 14,404 | 2002 | 426 | 223 |
| Atalantia buxifolia | 95,905 | 37,363 | 11,120 | 2050 | 446 | 256 |
| Citrus reticulata | 98,016 | 44,528 | 13,171 | 1981 | 310 | 137 |
| Fortunella hindsii | 128,597 | 50,791 | 14,639 | 2390 | 494 | 234 |
| Species | Predicted Markers | Designed Markers | ||
|---|---|---|---|---|
| Genic | Non-Genic | Genic | Non-Genic | |
| Citrus sinensis | 62,563 | 140,734 | 48,804 | 108,199 |
| Citrus clementina | 45,975 | 141,803 | 42,476 | 108,117 |
| Citrus maxima | 60,654 | 164,307 | 60,267 | 124,573 |
| Citrus medica | 49,403 | 161,187 | 50,690 | 93,136 |
| Citrus ichangensis | 52,336 | 174,314 | 54,065 | 103,745 |
| Atalantia buxifolia | 53,937 | 150,750 | 46,373 | 100,767 |
| Citrus reticulata | 53,832 | 147,576 | 48,203 | 109,940 |
| Fortunella hindsii | 73,901 | 166,281 | 67,288 | 129,857 |
| Features | citSATdb | PMDBase | PSMD | SSRome |
|---|---|---|---|---|
| Citrus species | 8 | 2 | 8 | 2 |
| Search option with ID | √ | × | √ | × |
| Search by motif type | √ | × | √ | √ |
| Search by repeat type | √ | × | √ | √ |
| Genic and genomic search | √ | × | √ | √ |
| Search on user-defined location | √ | × | √ | × |
| Length of SSR | √ | × | √ | × |
| ePCR option | √ | × | × | × |
| Non-nuclear (mitochondrion and chloroplast) | × | √ | × | × |
| JBrowse visualization of SSRs | √ | × | × | × |
| BLAST | √ | × | × | × |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duhan, N.; Meshram, M.; Loaiza, C.D.; Kaundal, R. citSATdb: Genome-Wide Simple Sequence Repeat (SSR) Marker Database of Citrus Species for Germplasm Characterization and Crop Improvement. Genes 2020, 11, 1486. https://doi.org/10.3390/genes11121486
Duhan N, Meshram M, Loaiza CD, Kaundal R. citSATdb: Genome-Wide Simple Sequence Repeat (SSR) Marker Database of Citrus Species for Germplasm Characterization and Crop Improvement. Genes. 2020; 11(12):1486. https://doi.org/10.3390/genes11121486
Chicago/Turabian StyleDuhan, Naveen, Manish Meshram, Cristian D. Loaiza, and Rakesh Kaundal. 2020. "citSATdb: Genome-Wide Simple Sequence Repeat (SSR) Marker Database of Citrus Species for Germplasm Characterization and Crop Improvement" Genes 11, no. 12: 1486. https://doi.org/10.3390/genes11121486
APA StyleDuhan, N., Meshram, M., Loaiza, C. D., & Kaundal, R. (2020). citSATdb: Genome-Wide Simple Sequence Repeat (SSR) Marker Database of Citrus Species for Germplasm Characterization and Crop Improvement. Genes, 11(12), 1486. https://doi.org/10.3390/genes11121486






