Karyotype Evolution in 10 Pinniped Species: Variability of Heterochromatin versus High Conservatism of Euchromatin as Revealed by Comparative Molecular Cytogenetics
Abstract
1. Introduction
2. Materials and Methods
2.1. Species Sampled and an Ethics Statement
2.2. Cell Culture and Chromosome Preparation
2.3. Differential Staining
2.4. Preparation and Characterization of Chromosome-Specific Painting Probes
2.5. Detection of Nucleolus Organizer Regions (NORs) and Telomeric Repeats
2.6. Image Acquisition and Data Processing
3. Results
3.1. Hybridization of Dog Probes onto Chromosomes of Four Pinniped Species
3.2. GTG Banding of Spotted Seal and Harbor Seal Chromosomes
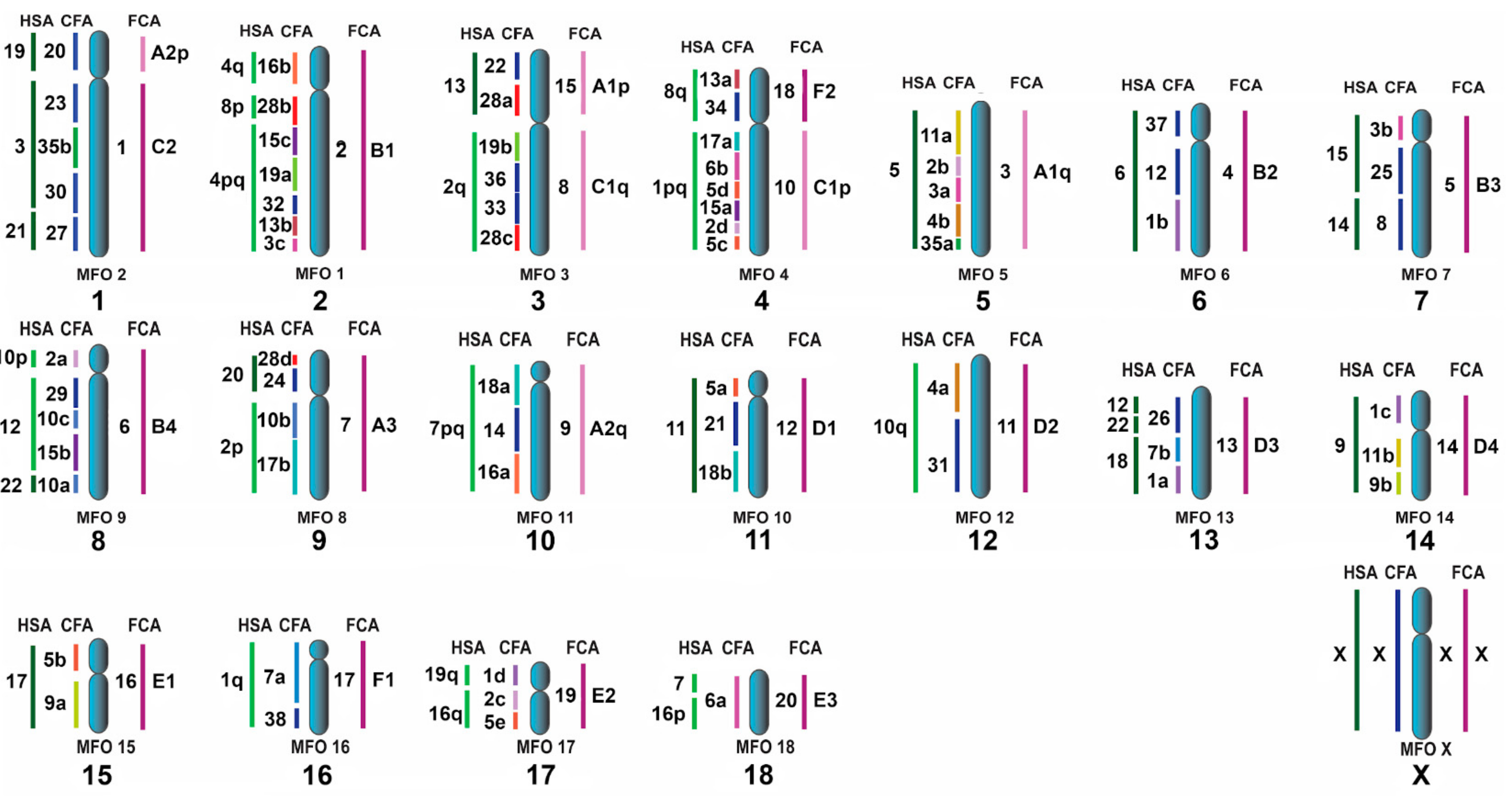
3.3. Homologous Segments in Pinniped Genomes as Determined by FISH
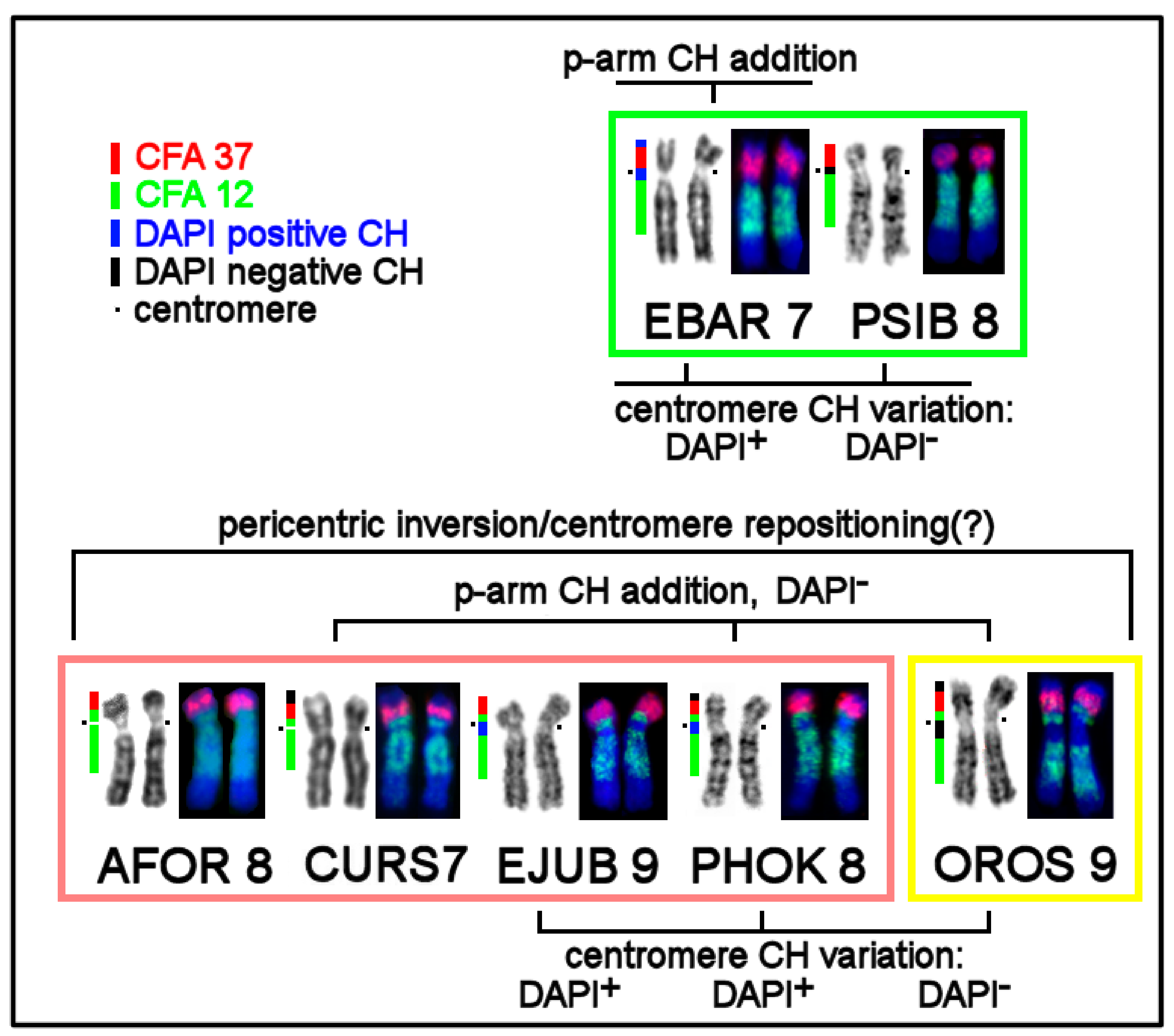
3.4. CH in Pinniped Genomes
3.5. The First Intrachromosomal Rearrangement Detected in Pinnipeds
3.6. The Y-Chromosome Features of Pinnipeds
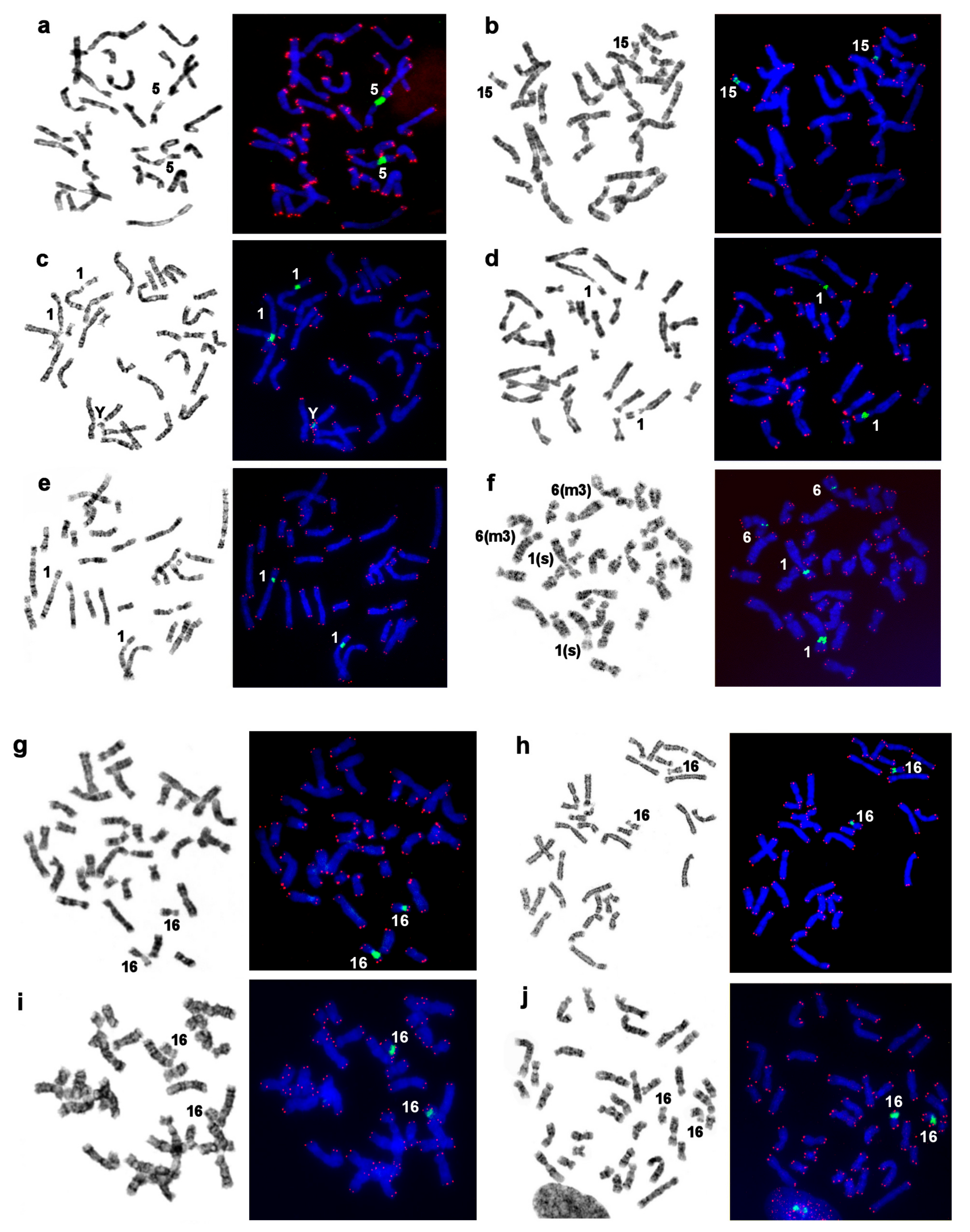
3.7. Localization of NORs and Telomeric Repeats
4. Discussion
4.1. Heterochromatin Diversity among Pinniped Karyotypes
4.2. The Intrachromosomal Rearrangement Detected in Pinnipeds
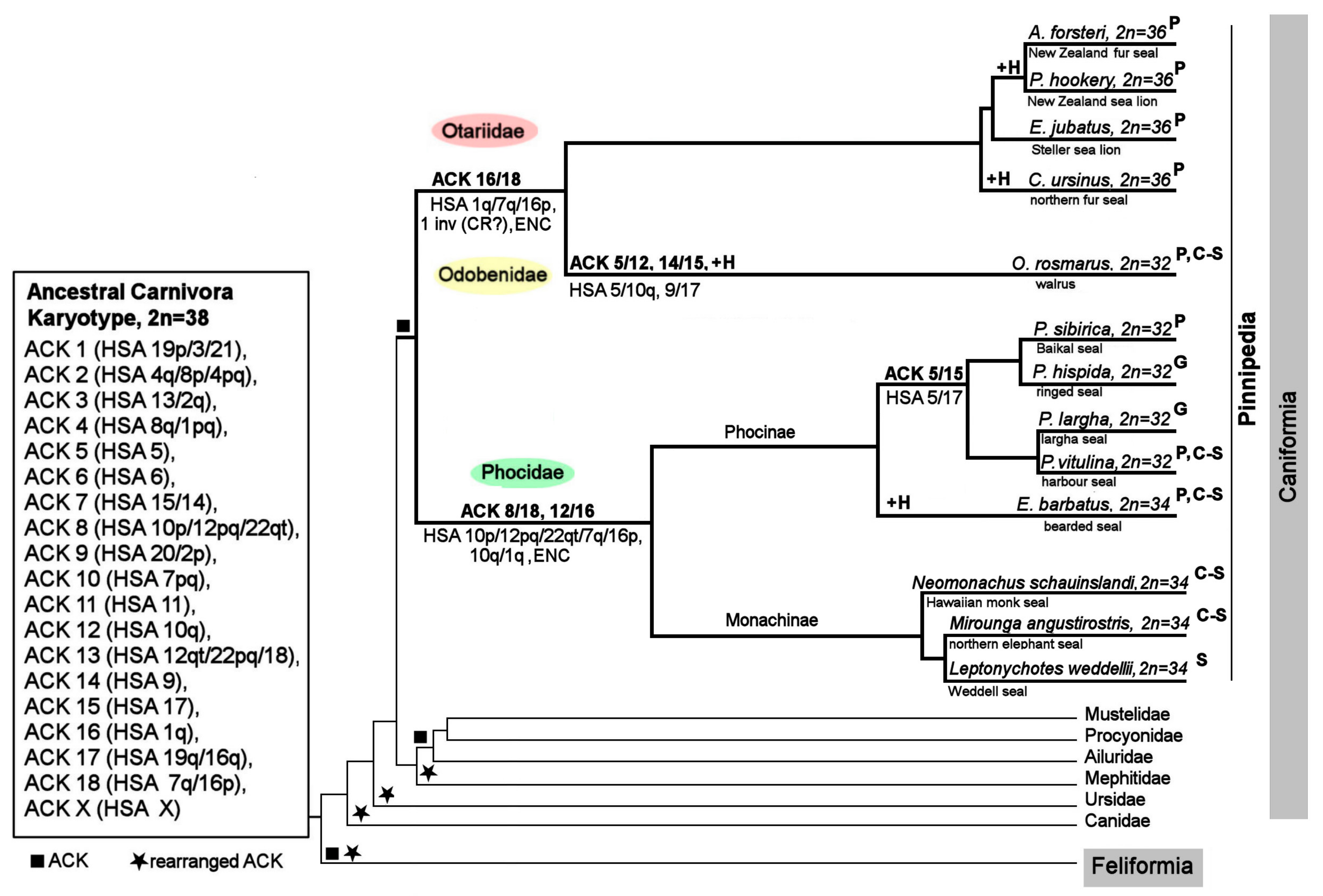
4.3. The ACK Validated by the New Pinniped Painting Data
4.4. Chromosome Evolution in Pinnipedia
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Janssen, A.; Colmenares, S.U.; Karpen, G.H. Heterochromatin: Guardian of the genome. Annu. Rev. Cell Dev. Biol. 2018, 34, 265–288. [Google Scholar] [CrossRef] [PubMed]
- Okita, A.K.; Zafar, F.; Su, J.; Weerasekara, D.; Kajitani, T.; Takahashi, T.S.; Kimura, H.; Murakami, Y.; Masukata, H.; Nakagawa, T. Heterochromatin suppresses gross chromosomal rearrangements at centromeres by repressing Tfs1/TFIIS-dependent transcription. Commun. Biol. 2019, 2, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Smalec, B.M.; Heider, T.N.; Flynn, B.L.; O’Neill, R.J. A centromere satellite concomitant with extensive karyotypic diversity across the Peromyscus genus defies predictions of molecular drive. Chromosome Res. 2019, 27, 237–252. [Google Scholar] [CrossRef]
- Graphodatsky, A.; Perelman, P.; OBrien, S.J. Atlas of Mammalian Chromosomes; John Wiley & Sons, Incorporated: Hoboken, NJ, USA, 2020. [Google Scholar]
- Murphy, W.J.; Stanyon, R.; O’Brien, S.J. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. 2001, 2. [Google Scholar] [CrossRef] [PubMed]
- Ferguson-Smith, M.A.; Trifonov, V. Mammalian karyotype evolution. Nat. Rev. Genet. 2007, 8, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, A.; Farré, M.; Robinson, T.J. Molecular cytogenetic and genomic insights into chromosomal evolution. Heredity 2012, 108, 28–36. [Google Scholar] [CrossRef]
- Frönicke, L.; Müller-Navia, J.; Romanakis, K.; Scherthan, H. Chromosomal homeologies between human, harbor seal (Phoca vitulina) and the putative ancestral carnivore karyotype revealed by Zoo-FISH. Chromosoma 1997, 106, 108–113. [Google Scholar] [PubMed]
- Beklemisheva, V.R.; Perelman, P.L.; Lemskaya, N.A.; Kulemzina, A.I.; Proskuryakova, A.A.; Burkanov, V.N.; Graphodatsky, A.S. The ancestral carnivore karyotype as substantiated by comparative chromosome painting of three pinnipeds, the walrus, the steller sea lion and the Baikal seal (Pinnipedia, Carnivora). PLoS ONE 2016, 11, e0147647. [Google Scholar] [CrossRef]
- Nash, W.G.; Menninger, J.C.; Padilla-Nash, H.M.; Stone, G.; Perelman, P.L.; O’Brien, S.J. The ancestral carnivore karyotype (2n = 38) lives today in ringtails. J. Hered. 2008, 99, 241–253. [Google Scholar] [CrossRef]
- Perelman, P.L.; Beklemisheva, V.R.; Yudkin, D.V.; Petrina, T.N.; Rozhnov, V.V.; Nie, W.; Graphodatsky, A.S. Comparative chromosome painting in Carnivora and Pholidota. Cytogenet. Genome Res. 2012, 137, 174–193. [Google Scholar]
- Arnason U The role of chromosomal rearrangement in mammalian speciation with special reference to Cetacea and Pinnipedia. Hereditas 1972, 70, 113–118.
- Lushnikova, T.P.; Grafodatskiĭ, A.S.; Ivanov, S.V.; Romashchenko, A.G.; Ternovskiĭ, L.V.; Ternovskaia, I.G.; Radzhabli, S.I. [EcoRI- and BamHI-families of repeated sequences in mustelids]. Genetika 1989, 25, 1449–1461. [Google Scholar] [PubMed]
- Fanning, T.G.; Modi, W.S.; Wayne, R.K.; O’Brien, S.J. Evolution of heterochromatin-associated satellite DNA loci in felids and canids (Carnivora). Cytogenet. Cell Genet. 1988, 48, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Modi, W.S.; Fanning, T.G.; Wayne, R.K.; O’Brien, S.J. Chromosomal localization of satellite DNA sequences among 22 species of felids and canids (Carnivora). Cytogenet. Genome Res. 1988, 48, 208–213. [Google Scholar] [CrossRef]
- Potapov, V.A.; Solov’ev, V.V.; Romashchenko, A.G.; Sosnovtsev, S.V.; Ivanov, S.V. Features of the structure and evolution of complex, tandemly organized Bsp-repeats in the fox genome. I. Structure and internal organization of the BamHI-dimer. Mol. Biol. 1990, 24, 1649–1665. [Google Scholar]
- Potapov, V.A.; Sosnovtsev, S.V.; Solov’ev, V.V.; Ivanov, S.V.; Romashchenko, A.G. The structure of complex repeats in the fox pericentromere heterochromatin: Regulatory elements of replication, recombination and gene expression control. Dokl. Akad. Nauk SSSR 1988, 299, 1250–1255. [Google Scholar]
- Graphodatsky, A.S.; Potapov, V.A.; Lushnikova, T.P. Comparative cytogenetics of 3 canids species (Carnivora, Canidae). 4. Distribution of repetitive DNA-sequences in the chromosomes. Genetika 1985, 21, 420–423. [Google Scholar]
- DNA Zoo. Available online: https://www.dnazoo.org (accessed on 14 October 2020).
- Árnason, Ú. Comparative chromosome studies in Pinnipedia. Hereditas 1974, 76, 179–225. [Google Scholar] [CrossRef]
- Seabright, M. A rapid banding technique for human chromosomes. Lancet 1971, 2, 971–972. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Lemskaya, N.A.; Kulemzina, A.I.; Beklemisheva, V.R.; Biltueva, L.S.; Proskuryakova, A.A.; Hallenbeck, J.M.; Perelman, P.L.; Graphodatsky, A.S. A combined banding method that allows the reliable identification of chromosomes as well as differentiation of AT- and GC-rich heterochromatin. Chromosome Res. 2018, 26, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; O’Brien, P.C.; Milne, B.S.; Graphodatsky, A.S.; Solanky, N.; Trifonov, V.; Rens, W.; Sargan, D.; Ferguson-Smith, M.A. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 1999, 62, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Ferguson-Smith, M.A. Genetic analysis by chromosome sorting and painting: Phylogenetic and diagnostic applications. Eur. J. Hum. Genet. 1997, 5, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.; Wang, J.; O’Brien, P.C.M.; Fu, B.; Ying, T.; Ferguson-Smith, M.A.; Yang, F. The genome phylogeny of domestic cat, red panda and five mustelid species revealed by comparative chromosome painting and G-banding. Chromosome Res. 2002, 10, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Maden, B.E.; Dent, C.L.; Farrell, T.E.; Garde, J.; McCallum, F.S.; Wakeman, J.A. Clones of human ribosomal DNA containing the complete 18 S-rRNA and 28 S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochem. J. 1987, 246, 519–527. [Google Scholar] [CrossRef] [PubMed]
- IJdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)ngenerated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef]
- Graphodatsky, A.S.; Yang, F.; O’Brien, P.C.; Perelman, P.; Milne, B.S.; Serdukova, N.; Kawada, S.I.; Ferguson-Smith, M.A. Phylogenetic implications of the 38 putative ancestral chromosome segments for four canid species. Cytogenet. Cell Genet. 2001, 92, 243–247. [Google Scholar] [CrossRef]
- Arnason, U. Localization of nucleolar organizing regions in pinniped karyotypes. Hereditas 1981, 94, 29–34. [Google Scholar] [CrossRef]
- Perelman, P.L.; Graphodatsky, A.S.; Dragoo, J.W.; Serdyukova, N.A.; Stone, G.; Cavagna, P.; Menotti, A.; Nie, W.; O’Brien, P.C.M.; Wang, J.; et al. Chromosome painting shows that skunks (Mephitidae, Carnivora) have highly rearranged karyotypes. Chromosome Res. 2008, 16, 1215–1231. [Google Scholar] [CrossRef]
- Nie, W.; Wang, J.; Su, W.; Wang, D.; Tanomtong, A.; Perelman, P.L.; Graphodatsky, A.S.; Yang, F. Chromosomal rearrangements and karyotype evolution in carnivores revealed by chromosome painting. Heredity 2012, 108, 17–27. [Google Scholar] [CrossRef]
- Dumas, F.; Cuttaia, H.; Sineo, L. Chromosomal distribution of interstitial telomeric sequences in nine neotropical primates (Platyrrhini): Possible implications in evolution and phylogeny. J. Zool. Syst. Evol. Res. 2016, 54, 226–236. [Google Scholar] [CrossRef]
- Fay, F.H.; Rausch, V.R.; Feltz, E.T. Cyntogenic comparison of some Pinnipeds (Mammalia: Eutheria). Can. J. Zool. 1967, 45, 773–778. [Google Scholar] [CrossRef]
- Arnason, U. The relationship between the four principal pinniped karyotypes. Hereditas 1977, 87, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Robinson, T.J.; Condy, P.R. The chromosomes of the southern elephant seal, Mirounga leonina (Phocidae: Mammalia). Cytogenet. Cell Genet. 1979, 23, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Graphodatsky, A.S. Integrated Comparative Genome Maps and Their Implications for Karyotype Evolution of Carnivores. Chrom. Today 2004, 215–224. [Google Scholar] [CrossRef]
- Nash, W.G.; Wienberg, J.; Ferguson-Smith, M.A.; Menninger, J.C.; O’Brien, S.J. Comparative genomics: Tracking chromosome evolution in the family ursidae using reciprocal chromosome painting. Cytogenet. Cell Genet. 1998, 83, 182–192. [Google Scholar] [CrossRef]
- Tian, Y.; Nie, W.; Wang, J.; Ferguson-Smith, M.A.; Yang, F. Chromosome evolution in bears: Reconstructing phylogenetic relationships by cross-species chromosome painting. Chromosome Res. 2004, 12, 55–63. [Google Scholar] [CrossRef]
- Vondrak, T.; Ávila Robledillo, L.; Novák, P.; Koblížková, A.; Neumann, P.; Macas, J. Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant J. 2020, 101, 484–500. [Google Scholar] [CrossRef]
- Miga, K.H.; Koren, S.; Rhie, A.; Vollger, M.R.; Gershman, A.; Bzikadze, A.; Brooks, S.; Howe, E.; Porubsky, D.; Logsdon, G.A.; et al. Telomere-to-telomere assembly of a complete human X chromosome. Nature 2020, 585, 79–84. [Google Scholar] [CrossRef]
- Perelman, P.L.; Graphodatsky, A.S.; Serdukova, N.A.; Nie, W.; Alkalaeva, E.Z.; Fu, B.; Robinson, T.J.; Yang, F. Karyotypic conservatism in the suborder Feliformia (Order carnivora). Cytogenet. Genome Res. 2005, 108, 348–354. [Google Scholar] [CrossRef]
- Graphodatsky, A.S.; Yang, F.; Perelman, P.L.; O’Brien, P.C.M.; Serdukova, N.A.; Milne, B.S.; Biltueva, L.S.; Fu, B.; Vorobieva, N.V.; Kawada, S.-I.; et al. Comparative molecular cytogenetic studies in the order Carnivora: Mapping chromosomal rearrangements onto the phylogenetic tree. Cytogenet. Genome Res. 2002, 96, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.L.; Bradshaw, C.J.A.; Goldsworthy, S.D.; Sunnucks, P. Lower reproductive success in hybrid fur seal males indicates fitness costs to hybridization. Mol. Ecol. 2007, 16, 3187–3197. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Mishra, R.K. Heterochromatic hues of transcription-the diverse roles of noncoding transcripts from constitutive heterochromatin. FEBS J. 2019, 286, 4626–4641. [Google Scholar] [CrossRef] [PubMed]
- Nicetto, D.; Zaret, K.S. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 2019, 55, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nishibuchi, G.; Déjardin, J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res. 2017, 25, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jia, S.T.; Jia, S. New insights into the regulation of Heterochromatin. Trends Genet. 2016, 32, 284–294. [Google Scholar] [CrossRef]
- Enukashvily, N.I.; Ponomartsev, N.V. Mammalian satellite DNA: A speaking dumb. Adv. Protein Chem. Struct. Biol. 2013, 90, 31–65. [Google Scholar]
- Penagos-Puig, A.; Furlan-Magaril, M. Heterochromatin as an important driver of genome organization. Front. Cell Dev. Biol. 2020, 8, 579137. [Google Scholar] [CrossRef]
- Finlan, L.E.; Sproul, D.; Thomson, I.; Boyle, S.; Kerr, E.; Perry, P.; Ylstra, B.; Chubb, J.R.; Bickmore, W.A. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008, 4, e1000039. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M.; Hübner, B.; Strickfaden, H.; Smeets, D.; Popken, J.; Sterr, M.; Markaki, Y.; Rippe, K.; Cremer, C. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015, 589, 2931–2943. [Google Scholar] [CrossRef]
- Cremer, T.; Kreth, G.; Koester, H.; Fink, R.H.A.; Heintzmann, R.; Cremer, M.; Solovei, I.; Zink, D.; Cremer, C. Chromosome territories, interchromatin domain compartment, and nuclear matrix: An integrated view of the functional nuclear architecture. Crit. Rev. Eukaryot. Gene Expr. 2000, 10, 38. [Google Scholar] [CrossRef]
- Bickmore, W.A.; van Steensel, B. Genome architecture: Domain organization of interphase chromosomes. Cell 2013, 152, 1270–1284. [Google Scholar] [CrossRef] [PubMed]
- Page, B.; Goldsworthy, S.D.; Hindell, M.A.; Mckenzie, J. Interspecific differences in male vocalizations of three sympatric fur seals (Arctocephalus spp.). J. Zool. 2002, 258, 49–56. [Google Scholar] [CrossRef]
- Reisinger, R.R.; Landman, M.; Mgibantaka, N.; Smale, M.J.; Bester, M.N.; De Bruyn, P.J.N.; Pistorius, P.A. Overlap and temporal variation in the diets of sympatric Antarctic and Subantarctic fur seals (Arctocephalus spp.) at Marion Island, Prince Edward Islands. Polar Res. 2018, 37, 1451142. [Google Scholar] [CrossRef]
- Kerley, G.I.H. Relative population sizes and trends, and hybridization of fur seals Arctocephalus tropicalisandA. gazellaat the Prince Edward Islands, Southern Ocean. S. Afr. J. Zool. 1983, 18, 388–392. [Google Scholar]
- Graphodatsky, A.S.; Yang, F.; Serdukova, N.; Perelman, P.; Zhdanova, N.S.; Ferguson-Smith, M.A. Dog chromosome-specific paints reveal evolutionary inter- and intrachromosomal rearrangements in the American mink and human. Cytogenet. Cell Genet. 2000, 90, 275–278. [Google Scholar] [CrossRef]
- Yang, F.; Graphodatsky, A.S.; O’Brien, P.C.; Colabella, A.; Solanky, N.; Squire, M.; Sargan, D.R.; Ferguson-Smith, M.A. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res. 2000, 8, 393–404. [Google Scholar] [CrossRef]
- Tian, Y.; Nie, W.-H.; Wang, J.-H.; Yang, Y.-F.; Yang, F.-T. Comparative chromosome painting shows the red panda (Ailurus fulgens) has a highly conserved karyotype. Yi Chuan Xue Bao 2002, 29, 124–127. [Google Scholar]
- Marshall, O.J.; Chueh, A.C.; Wong, L.H.; Choo, K.H.A. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 2008, 82, 261–282. [Google Scholar] [CrossRef]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67. [Google Scholar] [CrossRef]
- Capozzi, O.; Purgato, S.; D’Addabbo, P.; Archidiacono, N.; Battaglia, P.; Baroncini, A.; Capucci, A.; Stanyon, R.; Della Valle, G.; Rocchi, M. Evolutionary descent of a human chromosome 6 neocentromere: A jump back to 17 million years ago. Genome Res. 2009, 19, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Wurster-Hill, D.H.; Gray, C.W. The interrelationships of chromosome banding patterns in procyonids, viverrids, and felids. Cytogenet. Cell Genet. 1975, 15, 306–331. [Google Scholar] [CrossRef] [PubMed]
- Dutrillaux, B.; Couturier, J. The ancestral karyotype of Carnivora: Comparison with that of platyrrhine monkeys. Cytogenet. Genome Res. 1983, 35, 200–208. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.; Nash, W. Genetic mapping in mammals: Chromosome map of domestic cat. Science 1982, 216, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Farré, M.; Auvil, L.; Capitanu, B.; Larkin, D.M.; Ma, J.; Lewin, H.A. Reconstruction and evolutionary history of eutherian chromosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E5379–E5388. [Google Scholar] [CrossRef]
- Lewin, H.A.; Graves, J.A.M.; Ryder, O.A.; Graphodatsky, A.S.; O’Brien, S.J. Precision nomenclature for the new genomics. Gigascience 2019, 8. [Google Scholar] [CrossRef]
- TimeTree. The Timescale of Life. Available online: http://www.timetree.org/ (accessed on 30 April 2020).
- Nyakatura, K.; Bininda-Emonds, O.R.P. Updating the evolutionary history of Carnivora (Mammalia): A new species-level supertree complete with divergence time estimates. BMC Biol. 2012, 10, 12. [Google Scholar] [CrossRef]
- Yonezawa, T.; Kohno, N.; Hasegawa, M. The monophyletic origin of sea lions and fur seals (Carnivora; Otariidae) in the Southern Hemisphere. Gene 2009, 441, 89–99. [Google Scholar] [CrossRef]
- Arnason, U.; Gullberg, A.; Janke, A.; Kullberg, M.; Lehman, N.; Petrov, E.A.; Väinölä, R. Pinniped phylogeny and a new hypothesis for their origin and dispersal. Mol. Phylogenet. Evol. 2006, 41, 345–354. [Google Scholar] [CrossRef]
- Kulemzina, A.I.; Proskuryakova, A.A.; Beklemisheva, V.R.; Lemskaya, N.A.; Perelman, P.L.; Graphodatsky, A.S. Comparative chromosome map and heterochromatin features of the gray whale Karyotype (Cetacea). Cytogenet. Genome Res. 2016, 148, 25–34. [Google Scholar] [CrossRef]
- Kulemzina, A.I.; Trifonov, V.A.; Perelman, P.L.; Rubtsova, N.V.; Volobuev, V.; Ferguson-Smith, M.A.; Stanyon, R.; Yang, F.; Graphodatsky, A.S. Cross-species chromosome painting in Cetartiodactyla: Reconstructing the karyotype evolution in key phylogenetic lineages. Chromosome Res. 2009, 17, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Bielec, P.E.; Gallagher, D.S.; Womack, J.E.; Busbee, D.L. Homologies between human and dolphin chromosomes detected by heterologous chromosome painting. Cytogenet. Cell Genet. 1998, 81, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, J.A.; Arnason, U.; Widegren, B. Sequence organization and evolution, in all extant whalebone whales, of a DNA satellite with terminal chromosome localization. Chromosoma 1993, 102, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Arnason, U. Karyotype stability in marine mammals. Cytogenet. Cell Genet. 1982, 33, 274–276. [Google Scholar]
- Widegren, B.; Arnason, U.; Akusjärvi, G. Characteristics of a conserved 1,579-bp highly repetitive component in the killer whale, Orcinus orca. Mol. Biol. Evol. 1985, 2, 411–419. [Google Scholar]
- Kapitonov, V.V.; Holmquist, G.P.; Jurka, J. L1 repeat is a basic unit of heterochromatin satellites in cetaceans. Mol. Biol. Evol. 1998, 15, 611–612. [Google Scholar] [CrossRef]
- Arnason, U.; Widegren, B. Composition and chromosomal localization of cetacean highly repetitive DNA with special reference to the blue whale, Balaenoptera musculus. Chromosoma 1989, 98, 323–329. [Google Scholar] [CrossRef]
- Arnason, U.; Widegren, B. Different rates of divergence in highly repetitive DNA of cetaceans. Hereditas 1984, 101, 171–177. [Google Scholar] [CrossRef]
- Arnason, U. Comparative chromosome studies in Cetacea. Hereditas 1974, 77, 1–36. [Google Scholar] [CrossRef]
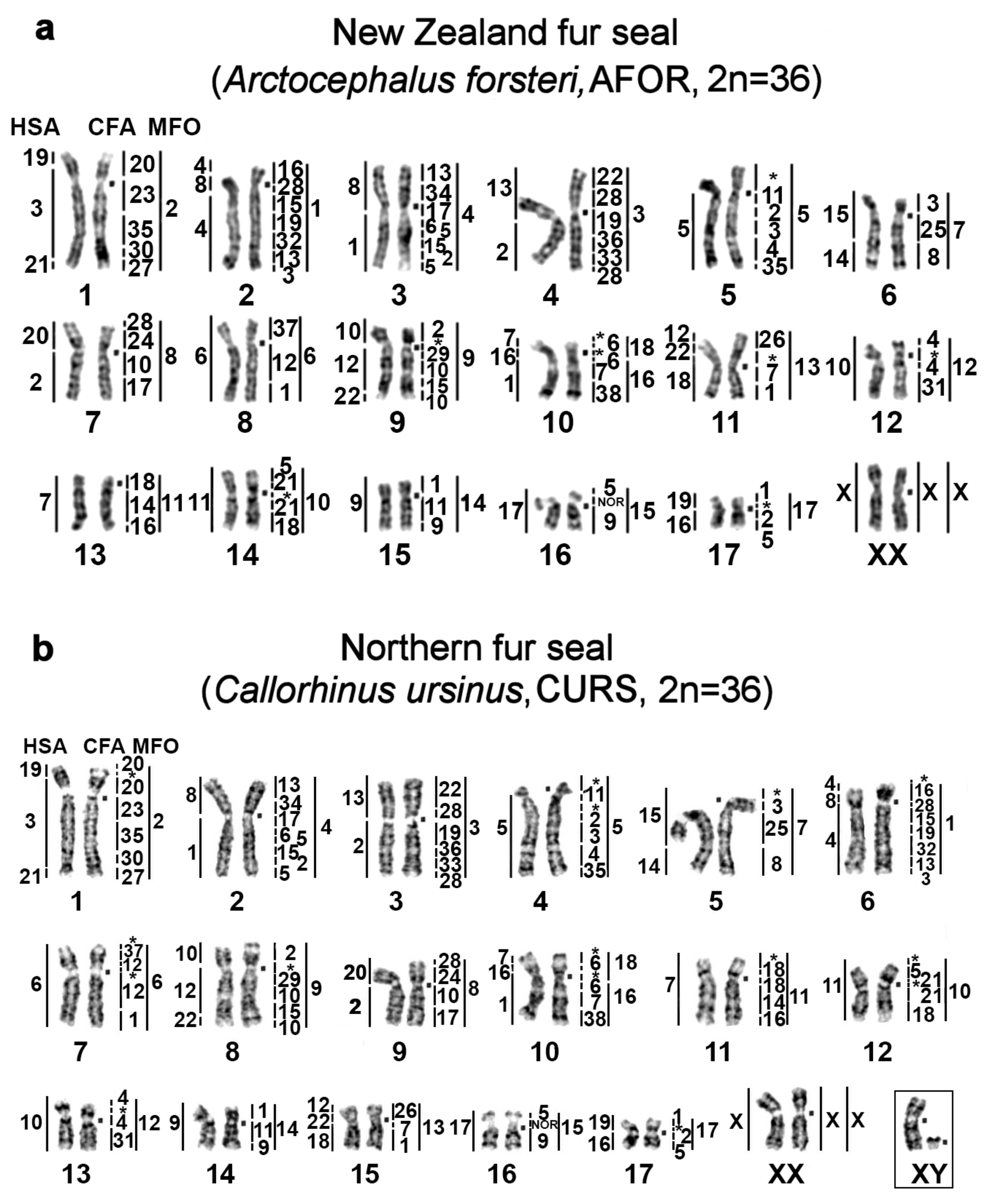
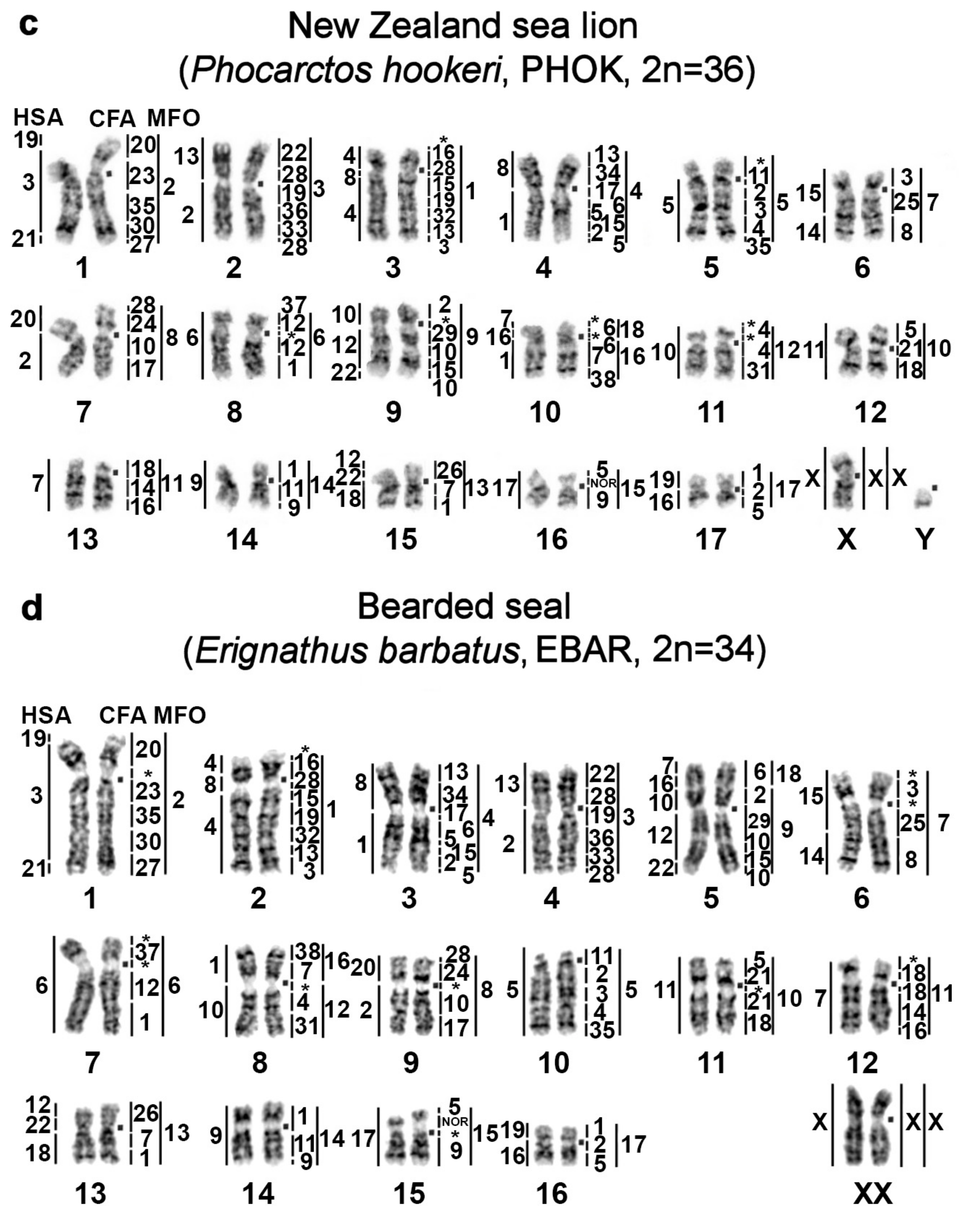
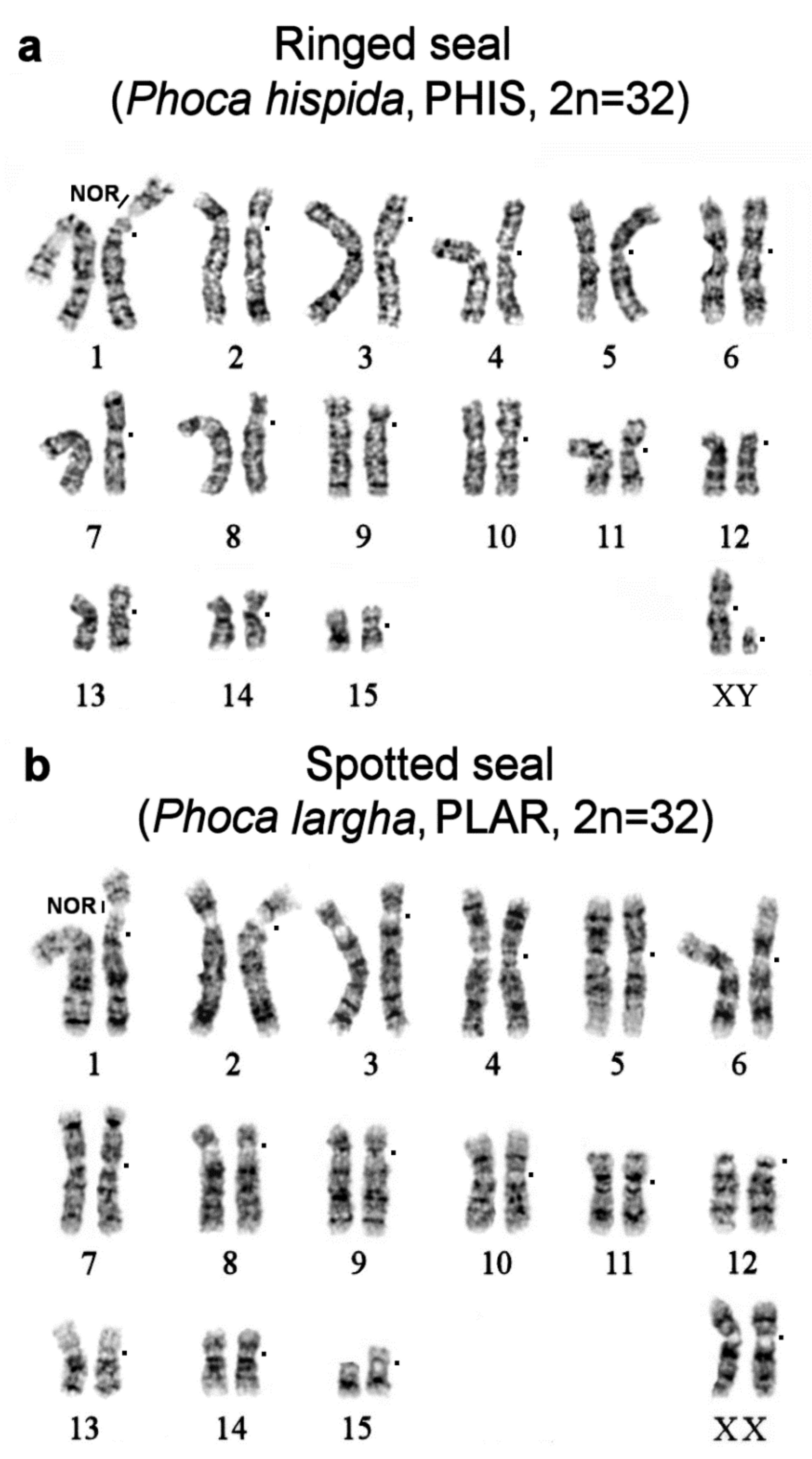
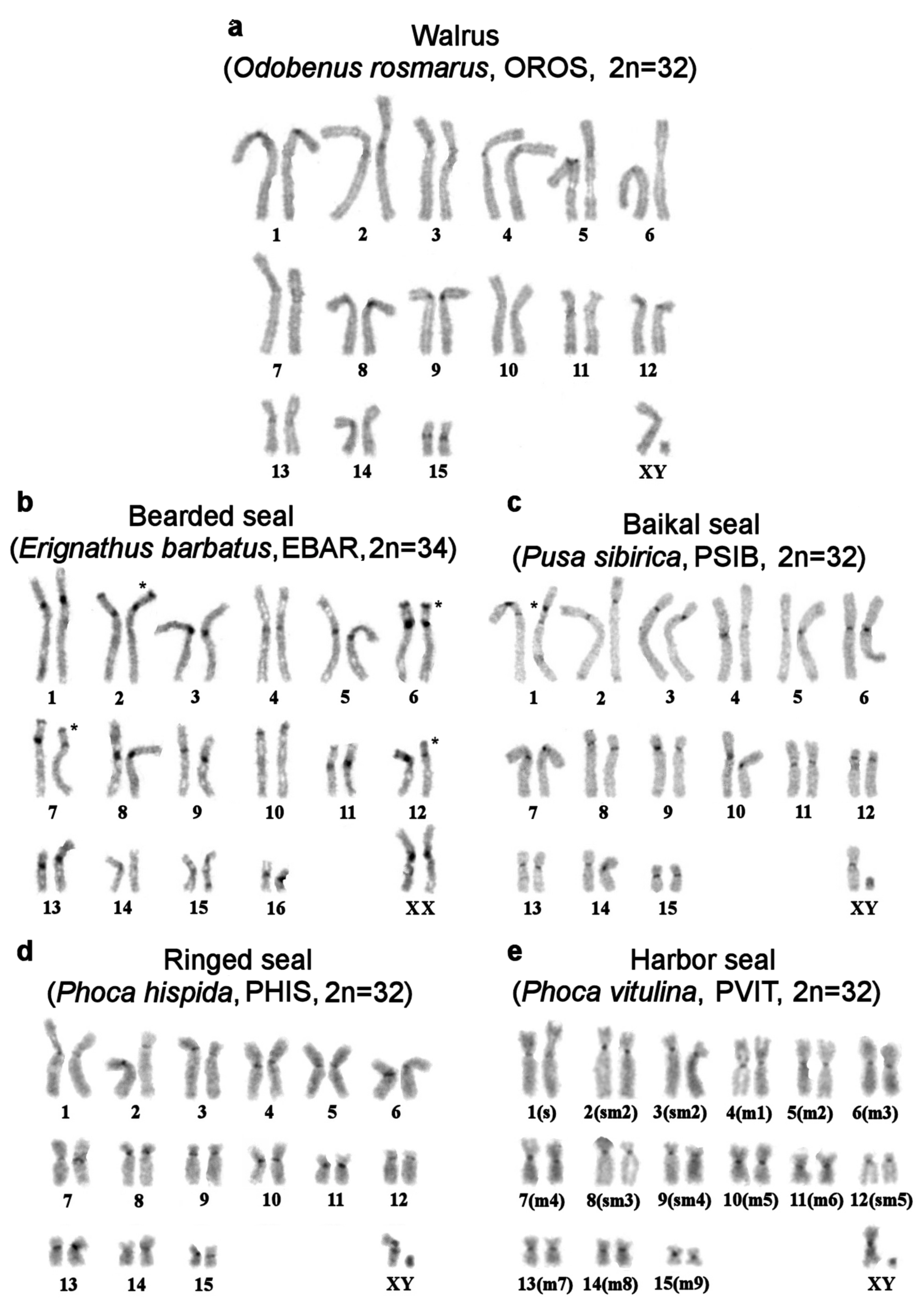
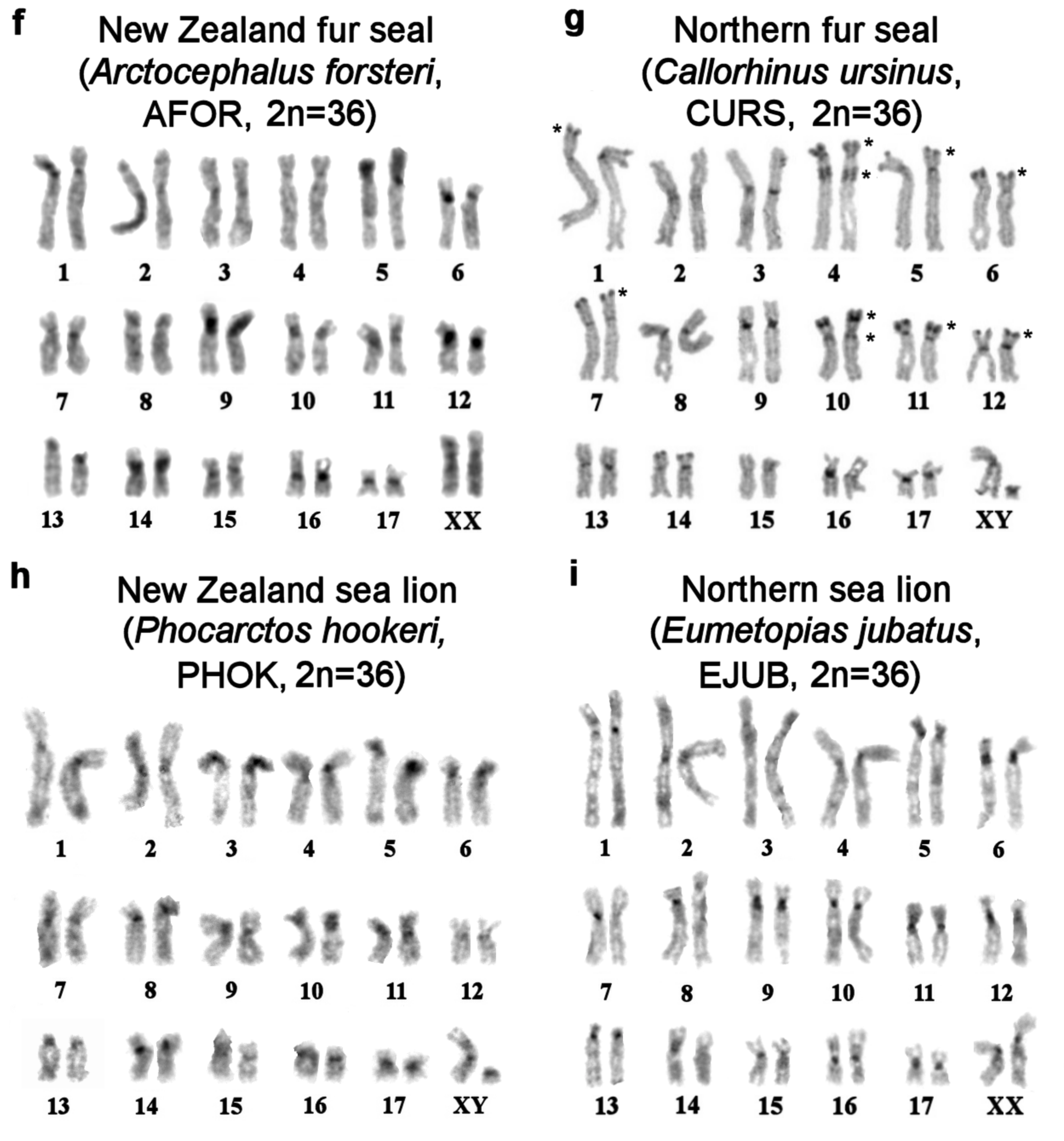
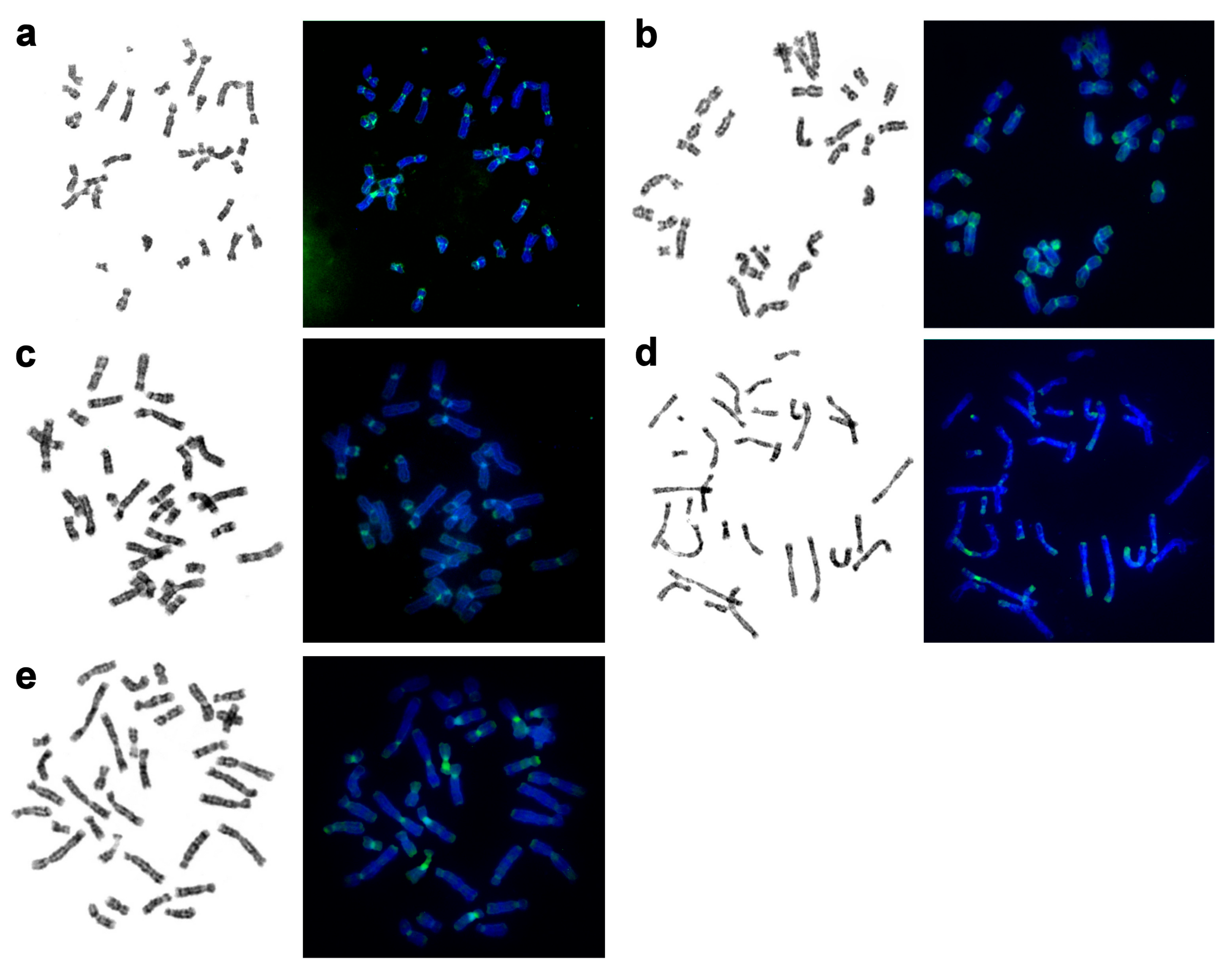
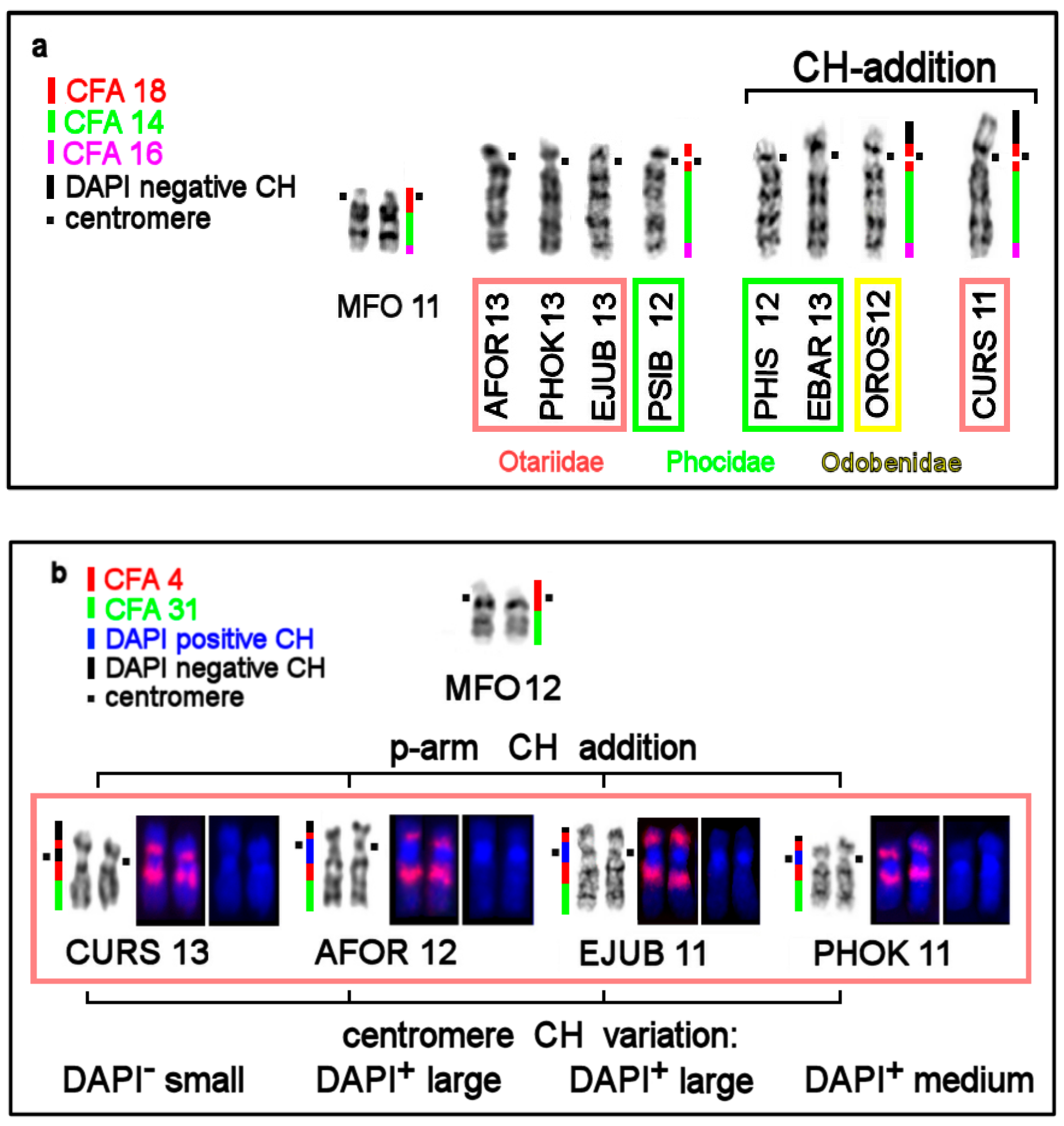
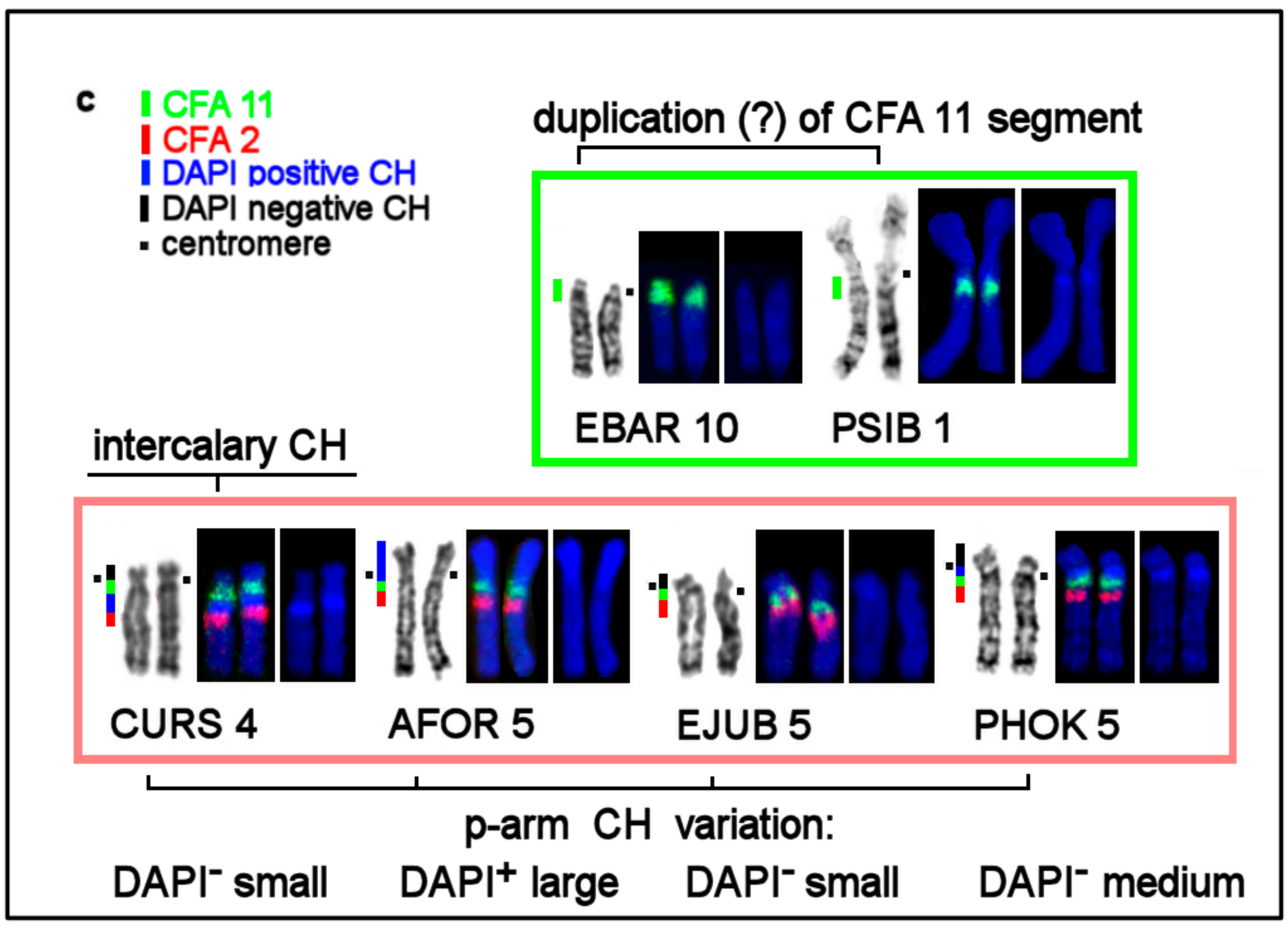
| No | Family | Latin Names | Code | 2n | Sex | Common Names | Reference for Fluorescence In Situ Hybridization (FISH) Data |
|---|---|---|---|---|---|---|---|
| 1 | Odobenidae Walruses | Odobenus rosmarus | OROS | 32 | M | walrus | [9] |
| 2 | Otariidae Eared seals | Arctocephalus forsteri | AFOR | 36 | F | New Zealand fur seal (South Australian fur seal) | this article |
| 3 | Callorhinus ursinus | CURS | 36 | M | northern fur seal | this article | |
| 4 | Phocarctos hookeri | PHOK | 36 | M | New Zealand sea lion | this article | |
| 5 | Eumetopias jubatus | EJUB | 36 | M | northern sea lion (Steller’s sea lion) | [9] | |
| 6 | Phocidae True seals | Erignathus barbatus | EBAR | 34 | F | bearded seal (square flipper seal) | this article |
| 7 | Phoca hispida | PHIS | 32 | M | ringed seal | ||
| 8 | Phoca largha | PLAR | 32 | F | spotted seal (largha seal) | ||
| 9 | Phoca vitulina | PVIT | 32 | M | harbor seal (common seal) | [8] | |
| 10 | Pusa sibirica | PSIB | 32 | M | Baikal seal | [9] | |
| 11 | Canidae | Canis familiaris | CFA | 78 | domestic dog | ||
| 12 | Mustelidae | Martes foina | MFO | 38 | stone marten | ||
| 13 | Hominidae | Homo sapiens | HSA | 46 | human |
| Location of CH | AT/GC Composition of CH Detected by Fluorochromes DAPI and CMA3 | Notes |
|---|---|---|
| Centromeric region | varies but GC-rich in most species: CMA3+ | |
| DAPI+ | near-centromeric blocks on some autosomes of walrus and true seals (but not in bearded seal | |
| CMA3+/DAPI+ | 4–7 small and mid-sized autosomal pairs in eared seals | |
| Pericentromeric region in q-arms | AT-rich: DAPI+ | |
| Terminal CH in p-arms | GC-rich in most species: CMA3+; DAPI+ on AFOR 5p | absent in Phoca and Pusa |
| Interstitial CH | AT-rich: DAPI+ | in eared seals |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beklemisheva, V.R.; Perelman, P.L.; Lemskaya, N.A.; Proskuryakova, A.A.; Serdyukova, N.A.; Burkanov, V.N.; Gorshunov, M.B.; Ryder, O.; Thompson, M.; Lento, G.; et al. Karyotype Evolution in 10 Pinniped Species: Variability of Heterochromatin versus High Conservatism of Euchromatin as Revealed by Comparative Molecular Cytogenetics. Genes 2020, 11, 1485. https://doi.org/10.3390/genes11121485
Beklemisheva VR, Perelman PL, Lemskaya NA, Proskuryakova AA, Serdyukova NA, Burkanov VN, Gorshunov MB, Ryder O, Thompson M, Lento G, et al. Karyotype Evolution in 10 Pinniped Species: Variability of Heterochromatin versus High Conservatism of Euchromatin as Revealed by Comparative Molecular Cytogenetics. Genes. 2020; 11(12):1485. https://doi.org/10.3390/genes11121485
Chicago/Turabian StyleBeklemisheva, Violetta R., Polina L. Perelman, Natalya A. Lemskaya, Anastasia A. Proskuryakova, Natalya A. Serdyukova, Vladimir N. Burkanov, Maksim B. Gorshunov, Oliver Ryder, Mary Thompson, Gina Lento, and et al. 2020. "Karyotype Evolution in 10 Pinniped Species: Variability of Heterochromatin versus High Conservatism of Euchromatin as Revealed by Comparative Molecular Cytogenetics" Genes 11, no. 12: 1485. https://doi.org/10.3390/genes11121485
APA StyleBeklemisheva, V. R., Perelman, P. L., Lemskaya, N. A., Proskuryakova, A. A., Serdyukova, N. A., Burkanov, V. N., Gorshunov, M. B., Ryder, O., Thompson, M., Lento, G., O’Brien, S. J., & Graphodatsky, A. S. (2020). Karyotype Evolution in 10 Pinniped Species: Variability of Heterochromatin versus High Conservatism of Euchromatin as Revealed by Comparative Molecular Cytogenetics. Genes, 11(12), 1485. https://doi.org/10.3390/genes11121485






