Identification of Novel Genomic Associations and Gene Candidates for Grain Starch Content in Sorghum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotypic Data
2.3. Genotypic Data
2.4. Genome-Wide Association Analysis
2.4.1. Linear Mixed Model
2.4.2. Bayesian Sparse Linear Mixed Model
2.5. Gene Expression and Network Analysis
3. Results
3.1. Phenotypic Analysis
3.2. Genome-Wide Association
3.3. Associated Genes
3.4. Candidate Gene Expression
3.5. HSP90-6 Gene Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
Abbreviations
| GWAS | Genome-wide association study |
| LMM | Linear mixed model |
| BSLMM | Bayesian sparse linear mixed model |
| GEMMA | Genome-wide efficient mixed model association |
| BLUP | Best linear unbiased predictor |
| chr | Chromosome |
| HSP | Heat shock protein |
| CASP | Casparian membrane protein |
| CDS | Coding sequences |
| LD | Linkage disequilibrium |
| QTL | Quantitative trail loci |
| SNP | Single nucleotide polymorphism |
| DAP | Days after pollination |
| PPI | Protein–protein interaction |
| FDR | False discovery rate |
| PIP | Posterior inclusion probability |
| NIRS | Near infrared spectroscopy |
References
- Mace, E.S.; Tai, S.; Gilding, E.K.; Li, Y.; Prentis, P.J.; Bian, L.; Campbell, B.C.; Hu, W.; Innes, D.J.; Han, X.; et al. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 2013, 4, 2320. [Google Scholar] [CrossRef]
- Zhu, F. Structure, physicochemical properties, modifications, and uses of sorghum starch. Compr. Rev. Food Sci. Food Saf. 2014, 13, 597–610. [Google Scholar] [CrossRef]
- Rhodes, D.H.; Hoffmann, L.; Rooney, W.L.; Herald, T.J.; Bean, S.; Boyles, R.; Brenton, Z.W.; Kresovich, S. Genetic architecture of kernel composition in global sorghum germplasm. BMC Genom. 2017, 18, 15. [Google Scholar] [CrossRef]
- Asif, M.; Rooney, L.; Acosta-Sanchez, D.; Mack, C.; Riaz, M. Uses of sorghum grain in gluten-free products. Cereal Foods World 2010, 55, 285–291. [Google Scholar] [CrossRef]
- Pfister, B.; Zeeman, S.C. Formation of starch in plant cells. Cell. Mol. Life Sci. 2016, 73, 2781–2807. [Google Scholar] [CrossRef]
- Smith, A.M. Making starch. Curr. Opin. Plant Biol. 1999, 2, 223–229. [Google Scholar] [CrossRef]
- Sulpice, R.; Pyl, E.T.; Ishihara, H.; Trenkamp, S.; Steinfath, M.; Witucka-Wall, H.; Gibon, Y.; Usadel, B.; Poree, F.; Piques, M.C.; et al. Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 2009, 106, 10348–10353. [Google Scholar] [CrossRef]
- MacNeill, G.J.; Mehrpouyan, S.; Minow, M.A.; Patterson, J.A.; Tetlow, I.J.; Emes, M.J.; Raines, C. Starch as a source, starch as a sink: The bifunctional role of starch in carbon allocation. J. Exp. Bot. 2017, 68, 4433–4453. [Google Scholar] [CrossRef]
- Méchin, V.; Thévenot, C.; Le Guilloux, M.; Prioul, J.L.; Damerval, C. Developmental analysis of maize endosperm proteome suggests a pivotal role for pyruvate orthophosphate dikinase. Plant Physiol. 2007, 143, 1203–1219. [Google Scholar] [CrossRef]
- Murray, S.C.; Sharma, A.; Rooney, W.L.; Klein, P.E.; Mullet, J.E.; Mitchell, S.E.; Kresovich, S. Genetic improvement of sorghum as a biofuel feedstock: I. QTL for stem sugar and grain nonstructural carbohydrates. Crop Sci. 2008, 48, 2165–2179. [Google Scholar] [CrossRef]
- Boyles, R.E.; Pfeiffer, B.K.; Cooper, E.A.; Rauh, B.L.; Zielinski, K.J.; Myers, M.T.; Brenton, Z.; Rooney, W.L.; Kresovich, S. Genetic dissection of sorghum grain quality traits using diverse and segregating populations. Theor. Appl. Genet. 2017, 130, 697–716. [Google Scholar] [CrossRef]
- Whitt, S.R.; Wilson, L.M.; Tenaillon, M.I.; Gaut, B.S.; Buckler, E.S. Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 12959–12962. [Google Scholar] [CrossRef]
- Mace, E.; Jordan, D. Location of major effect genes in sorghum (Sorghum bicolor (L.) Moench). Theor. Appl. Genet. 2010, 121, 1339–1356. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef]
- Myles, S.; Peiffer, J.; Brown, P.J.; Ersoz, E.S.; Zhang, Z.; Costich, D.E.; Buckler, E.S. Association mapping: Critical considerations shift from genotyping to experimental design. Plant Cell 2009, 21, 2194–2202. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Malosetti, M.; van der Linden, C.G.; Vosman, B.; van Eeuwijk, F.A. A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 2007, 175, 879–889. [Google Scholar] [CrossRef]
- Zhao, K.; Aranzana, M.J.; Kim, S.; Lister, C.; Shindo, C.; Tang, C.; Toomajian, C.; Zheng, H.; Dean, C.; Marjoram, P.; et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007, 3, e4. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 2014, 11, 407. [Google Scholar] [CrossRef]
- Zhou, X.; Carbonetto, P.; Stephens, M. Polygenic modeling with Bayesian sparse linear mixed models. PLoS Genet. 2013, 9, e1003264. [Google Scholar] [CrossRef]
- Sukumaran, S.; Xiang, W.; Bean, S.R.; Pedersen, J.F.; Kresovich, S.; Tuinstra, M.R.; Tesso, T.T.; Hamblin, M.T.; Yu, J. Association mapping for grain quality in a diverse sorghum collection. Plant Genome 2012, 5, 126–135. [Google Scholar] [CrossRef]
- Moose, S.P.; Dudley, J.W.; Rocheford, T.R. Maize selection passes the century mark: A unique resource for 21st century genomics. Trends Plant Sci. 2004, 9, 358–364. [Google Scholar] [CrossRef]
- Gallagher, M.D.; Chen-Plotkin, A.S. The post-GWAS era: From association to function. Am. J. Hum. Genet. 2018, 102, 717–730. [Google Scholar] [CrossRef]
- Casa, A.M.; Pressoir, G.; Brown, P.J.; Mitchell, S.E.; Rooney, W.L.; Tuinstra, M.R.; Franks, C.D.; Kresovich, S. Community resources and strategies for association mapping in sorghum. Crop Sci. 2008, 48, 30–40. [Google Scholar] [CrossRef]
- Boyles, R.E.; Cooper, E.A.; Myers, M.T.; Brenton, Z.; Rauh, B.L.; Morris, G.P.; Kresovich, S. Genome-wide association studies of grain yield components in diverse sorghum germplasm. Plant Genome 2016, 9. [Google Scholar] [CrossRef]
- Sapkota, S.; Boyles, R.; Cooper, E.; Brenton, Z.; Myers, M.; Kresovich, S. Impact of sorghum racial structure and diversity on genomic prediction of grain yield components. Crop Sci. 2020, 60, 132–148. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D.; Riera-Lizarazu, O.; Brown, P.J.; Acharya, C.B.; Mitchell, S.E.; et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA 2013, 110, 453–458. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Swarts, K.; Li, H.; Romero Navarro, J.A.; An, D.; Romay, M.C.; Hearne, S.; Acharya, C.; Glaubitz, J.C.; Mitchell, S.; Elshire, R.J.; et al. Novel methods to optimize genotypic imputation for low-coverage, next-generation sequence data in crop plants. Plant Genome 2014, 7. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Davidson, R.M.; Gowda, M.; Moghe, G.; Lin, H.; Vaillancourt, B.; Shiu, S.H.; Jiang, N.; Robin Buell, C. Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 2012, 71, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Mu, H.H.; Mu-Forster, C.; Wasserman, B.P. Polypeptides of the maize amyloplast stroma: Stromal localization of starch-biosynthetic enzymes and identification of an 81-kilodalton amyloplast stromal heat-shock cognate. Plant Physiol. 1998, 116, 1451–1460. [Google Scholar] [CrossRef]
- Khan, A.; Ali, M.; Khattak, A.M.; Gai, W.X.; Zhang, H.X.; Wei, A.M.; Gong, Z.H.; Gong, Z.-H. Heat Shock Proteins: Dynamic Biomolecules to Counter Plant Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2019, 20, 5321. [Google Scholar]
- Woldesemayat, A.A.; Van Heusden, P.; Ndimba, B.K.; Christoffels, A. An integrated and comparative approach towards identification, characterization and functional annotation of candidate genes for drought tolerance in sorghum (Sorghum bicolor (L.) Moench). BMC Genet. 2017, 18, 119. [Google Scholar] [CrossRef]
- Woldesemayat, A.A.; Ntwasa, M. Pathways and Network Based Analysis of Candidate Genes to Reveal Cross-Talk and Specificity in the Sorghum (Sorghum bicolor (L.) Moench) Responses to Drought and It’s Co-occurring Stresses. Front. Genet. 2018, 9, 557. [Google Scholar] [CrossRef]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef]
- Dong, S.; Beckles, D.M. Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response. J. Plant Physiol. 2019, 234, 80–93. [Google Scholar] [CrossRef]
- Rami, J.F.; Dufour, P.; Trouche, G.; Fliedel, G.; Mestres, C.; Davrieux, F.; Blanchard, P.; Hamon, P. Quantitative trait loci for grain quality, productivity, morphological and agronomical traits in sorghum (Sorghum bicolor L. Moench). Theor. Appl. Genet. 1998, 97, 605–616. [Google Scholar] [CrossRef]
- Shen, X.; Carlborg, Ö. Beware of risk for increased false positive rates in genome-wide association studies for phenotypic variability. Front. Genet. 2013, 4, 93. [Google Scholar] [CrossRef]
- Johnson, R.C.; Nelson, G.W.; Troyer, J.L.; Lautenberger, J.A.; Kessing, B.D.; Winkler, C.A.; O’Brien, S.J. Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genom. 2010, 11, 724. [Google Scholar] [CrossRef]
- Roppolo, D.; Boeckmann, B.; Pfister, A.; Boutet, E.; Rubio, M.C.; Dénervaud-Tendon, V.; Vermeer, J.E.; Gheyselinck, J.; Xenarios, I.; Geldner, N. Functional and evolutionary analysis of the CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN family. Plant Physiol. 2014, 165, 1709–1722. [Google Scholar] [CrossRef]
- Yang, J.; Ding, C.; Xu, B.; Chen, C.; Narsai, R.; Whelan, J.; Hu, Z.; Zhang, M. A Casparian strip domain-like gene, CASPL, negatively alters growth and cold tolerance. Sci. Rep. 2015, 5, 14299. [Google Scholar] [CrossRef]
- Vierling, E. The roles of heat shock proteins in plants. Annu. Rev. Plant Biol. 1991, 42, 579–620. [Google Scholar] [CrossRef]
- Boston, R.S.; Viitanen, P.V.; Vierling, E. Molecular chaperones and protein folding in plants. In Post-Transcriptional Control of Gene Expression in Plants; Springer: Berlin/Heidelberg, Germany, 1996; pp. 191–222. [Google Scholar]
- Kamara, J.S.; Hoshino, M.; Satoh, Y.; Nayar, N.; Takaoka, M.; Sasanuma, T.; Abe, T. Japanese sake-brewing rice cultivars show high levels of globulin-like protein and a chloroplast stromal HSP70. Crop Sci. 2009, 49, 2198–2206. [Google Scholar] [CrossRef]
- Duan, H.; Tong, H.; Zhu, A.; Zhang, H.; Liu, L. Effects of heat, drought and their combined effects on morphological structure and physicochemical properties of rice (Oryza sativa L.) starch. J. Cereal Sci. 2020, 95, 103059. [Google Scholar] [CrossRef]
- Yang, H.; Gu, X.; Ding, M.; Lu, W.; Lu, D. Heat stress during grain filling affects activities of enzymes involved in grain protein and starch synthesis in waxy maize. Sci. Rep. 2018, 8, 15665. [Google Scholar] [CrossRef]
- Kamal, N.M.; Gorafi, Y.S.A.; Abdelrahman, M.; Abdellatef, E.; Tsujimoto, H. Stay-green trait: A prospective approach for yield potential, and drought and heat stress adaptation in globally important cereals. Int. J. Mol. Sci. 2019, 20, 5837. [Google Scholar] [CrossRef]
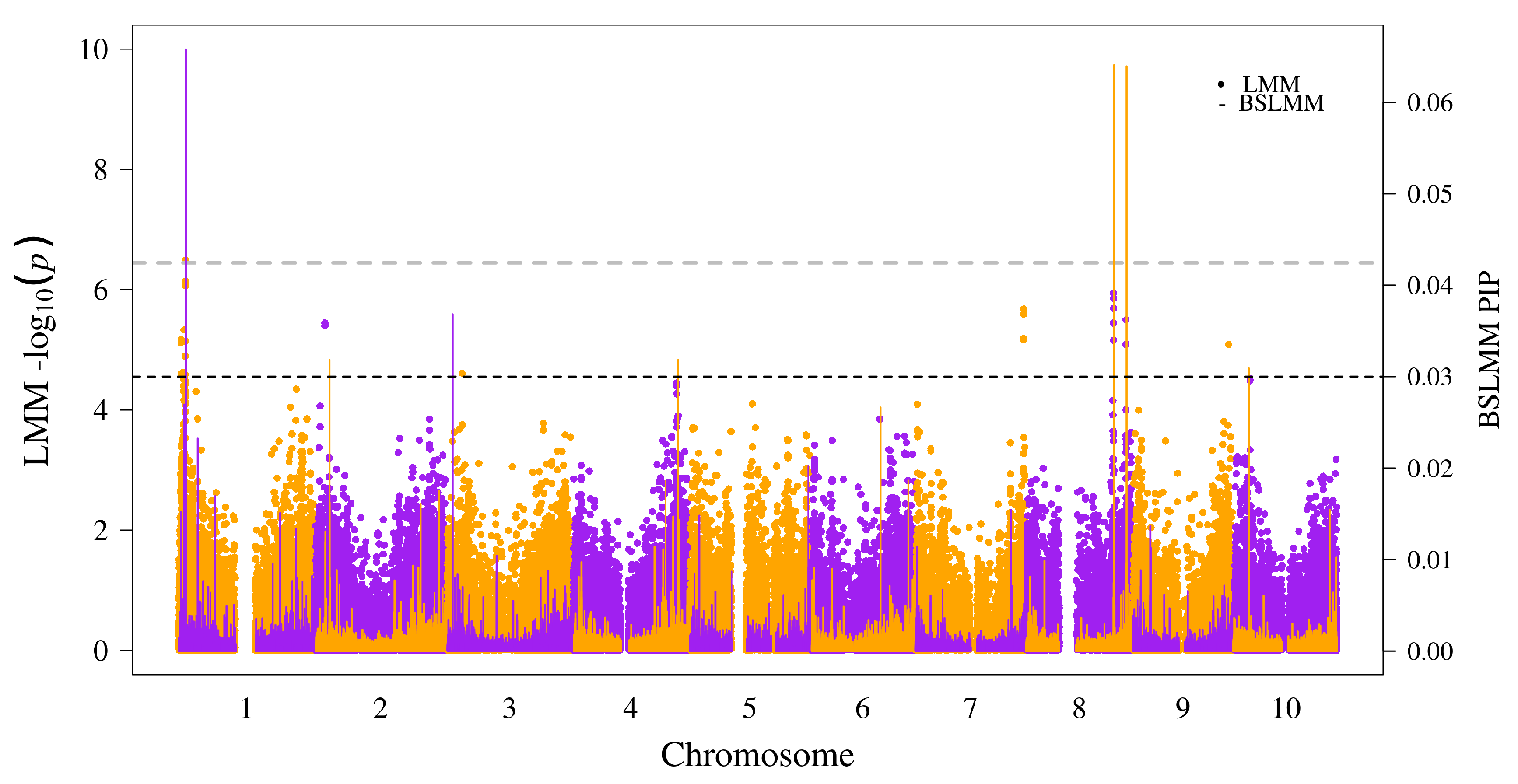



| Chr | Position | −log10(p) (LMM) | PIP (BSLMM) | Rank (LMM,BSLMM) | GeneID | Annotation | Impact |
|---|---|---|---|---|---|---|---|
| 1 | 4067535 | 6.812 | 0.066 | 1,1 | Sobic.001G054500 | Missense | Moderate |
| 1 | 4067364 | 6.412 | 0.064 | 2,4 | Sobic.001G054500 | Missense | Moderate |
| 1 | 4067377 | 6.332 | 0.053 | 4,5 | Sobic.001G054500 | Missense | Moderate |
| 8 | 51715166 | 5.350 | 0.022 | 20,20 | Sobic.008G111500 | Missense | Moderate |
| 8 | 51719704 | 5.682 | 0.047 | 12,8 | Sobic.008G111600 | Synonymous | Low |
| 8 | 51720767 | 6.171 | 0.064 | 6,2 | Sobic.008G111600 | Intron | Modifier |
| 8 | 51721062 | 6.009 | 0.053 | 7,6 | Sobic.008G111600 | Intron | Modifier |
| 8 | 51721065 | 5.728 | 0.039 | 10,9 | Sobic.008G111600 | Intron | Modifier |
| 8 | 51726098 | 6.229 | 0.051 | 5,7 | Sobic.008G111600 | Synonymous | Low |
| 8 | 59121722 | 5.260 | 0.064 | 23,3 | Sobic.008G158332 | Synonymous | Low |
| Gene | Name | Chromosome | Start | End | Maize Homolog |
|---|---|---|---|---|---|
| Sobic.001G054500 | Uncharacterized protein | 1 | 4066711 | 4067588 | Zm00001d034482 |
| Sobic.008G111500 | CASP-like protein 8 | 8 | 51714673 | 51715254 | Zm00001d023936 |
| Sobic.008G111600 | Heat shock protein 90-6 | 8 | 51719209 | 51726960 | Zm00001d041719 |
| Sobic.008G158332 | Lipid transfer protein 1 | 8 | 59121190 | 59129810 | Zm00001d027290 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sapkota, S.; Boatwright, J.L.; Jordan, K.; Boyles, R.; Kresovich, S. Identification of Novel Genomic Associations and Gene Candidates for Grain Starch Content in Sorghum. Genes 2020, 11, 1448. https://doi.org/10.3390/genes11121448
Sapkota S, Boatwright JL, Jordan K, Boyles R, Kresovich S. Identification of Novel Genomic Associations and Gene Candidates for Grain Starch Content in Sorghum. Genes. 2020; 11(12):1448. https://doi.org/10.3390/genes11121448
Chicago/Turabian StyleSapkota, Sirjan, J. Lucas Boatwright, Kathleen Jordan, Richard Boyles, and Stephen Kresovich. 2020. "Identification of Novel Genomic Associations and Gene Candidates for Grain Starch Content in Sorghum" Genes 11, no. 12: 1448. https://doi.org/10.3390/genes11121448
APA StyleSapkota, S., Boatwright, J. L., Jordan, K., Boyles, R., & Kresovich, S. (2020). Identification of Novel Genomic Associations and Gene Candidates for Grain Starch Content in Sorghum. Genes, 11(12), 1448. https://doi.org/10.3390/genes11121448







