Electrochemical Detection and Point-of-Care Testing for Circulating Tumor Cells: Current Techniques and Future Potentials
Abstract
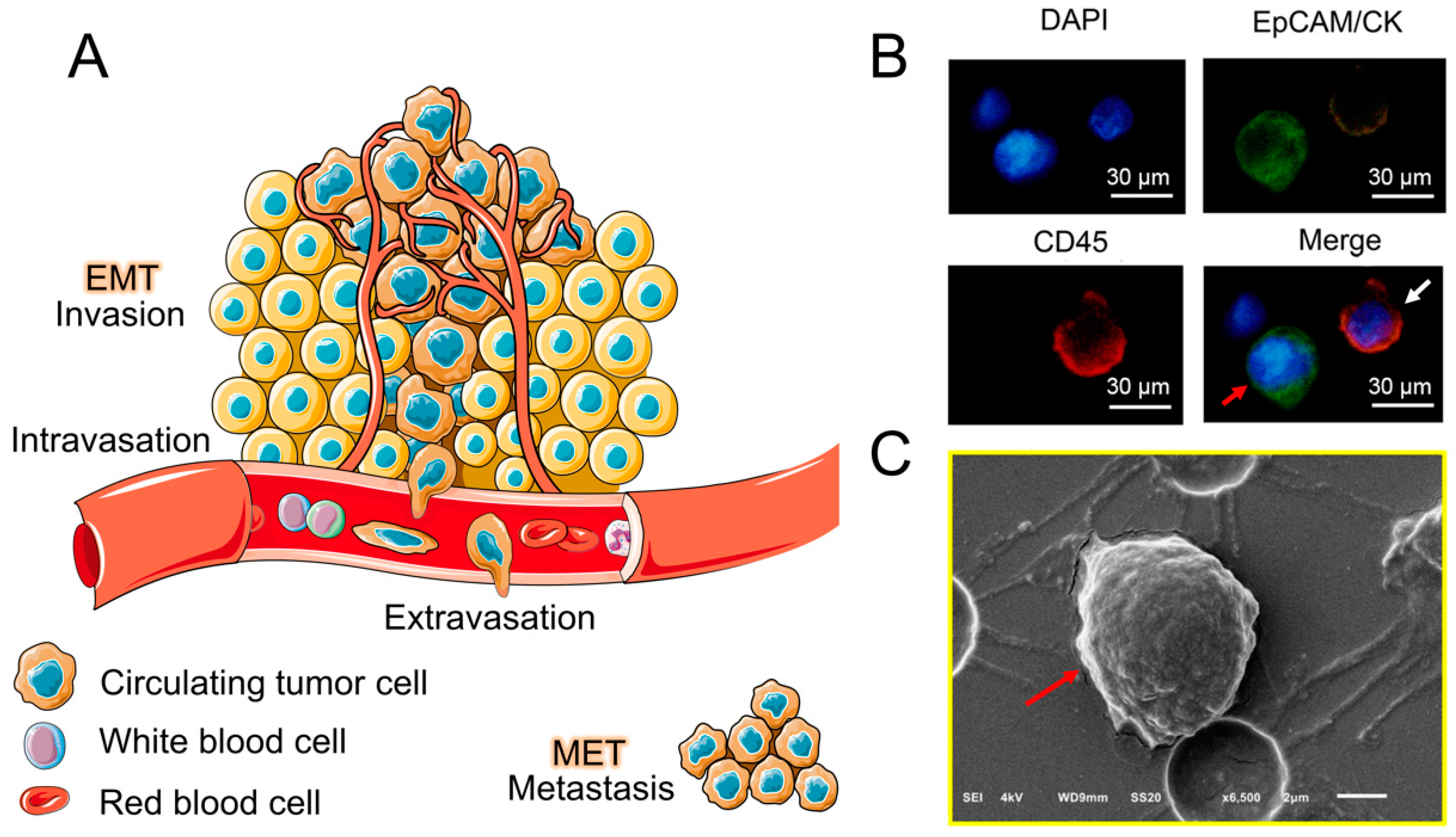
:1. Introduction
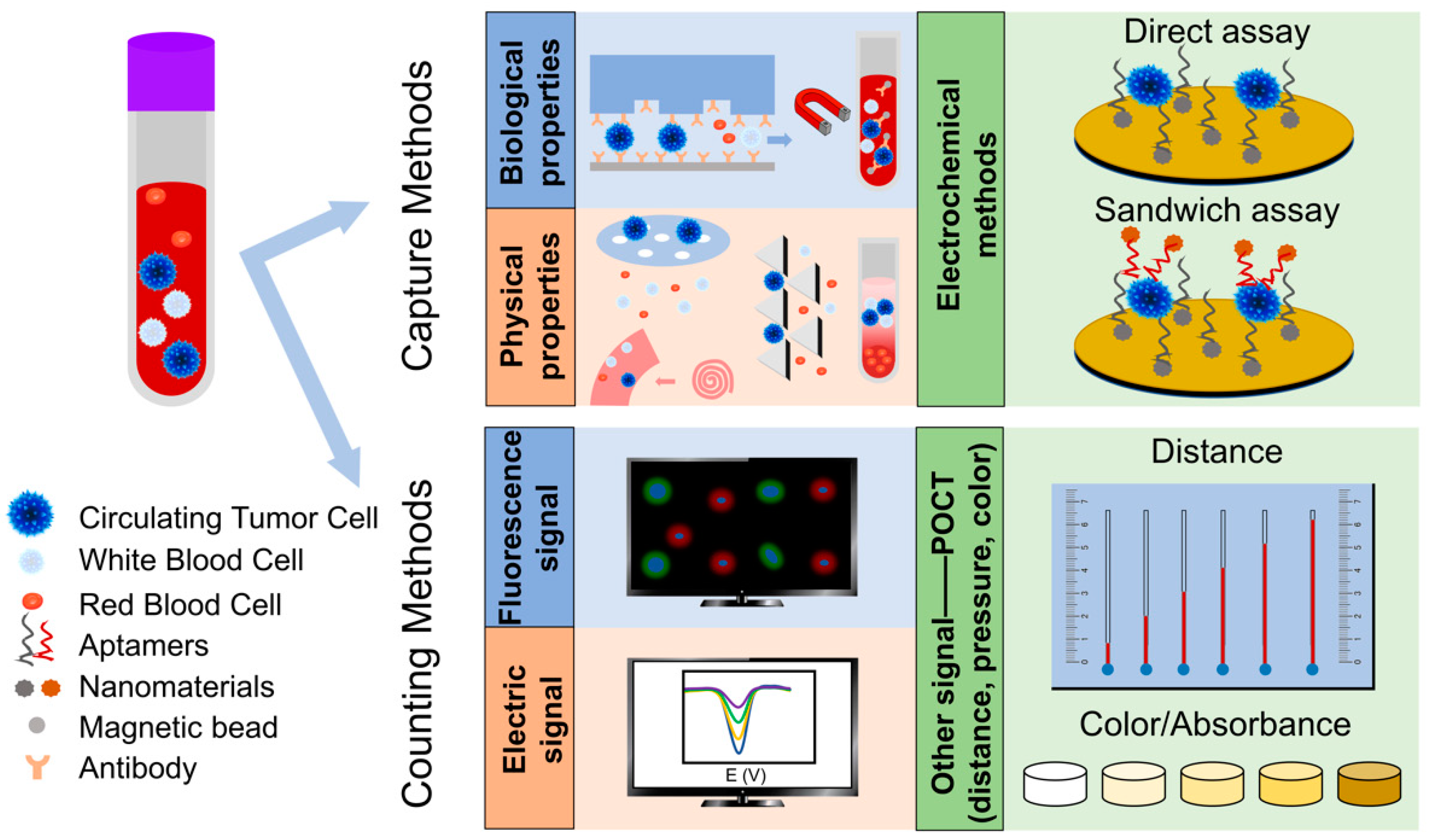
2. CTC Separation Methods Based on Biological Properties and Physical Properties
2.1. Methods Based on Biological Properties
| Separation Principles | Technology | Flow Rate (mL h−1) | Effective Flow Rate (mL h−1) | Efficiency/Sensitivity | Capture Purity | Enrichment Factor | Clinical Sample | Ref. |
|---|---|---|---|---|---|---|---|---|
| Biological properties-based methods | CellSearch | 3 | 3 | 80–82% | —— | —— | Metastatic breast, prostate or colorectal | [56] |
| CTC-chip | 1–2 | 1–2 | >60% | 49–67% | —— | Non small cell lung cancer (n = 55), prostate (n = 26), pancreatic (n = 15), breast (n = 10) and colon (n = 10) | [24] | |
| HB-chip | 1.2 | 1.2 | 91.8 ± 5.2% | 14.0 ± 0.1% | —— | Prostate (n = 15), lung (n = 4) | [25] | |
| CellCollector | 30 | 30 | 10–35% | 50–96.4% | —— | Pancreatic (n = 43) | [57] | |
| GO-chip | 1–3 | 1–3 | 84–95% | —— | —— | Breast (n = 10), pancreatic (n = 3) | [58] | |
| Physical properties-based methods | DLD | 600 | 30 | 85% | —— | 3.4 | —— | [59] |
| Vortex-chip | 240–360 | 12 | 20.7% | 89% | 3.5×104 | Breast (n = 4), lung (n = 8) | [60] | |
| Multiplexed spiral chip | 21 | 21 | 76.4–87.6% | 1CTC/30–100WBCs | ~104 | Breast (n = 5), lung (n = 5) | [61] | |
| Labyrinth-chip | 150 | 30 | >90% | WBCs number: 13,800 mL−1 (once); 663 mL−1 (twice) | —— | Breast (n = 56), pancreatic (n = 20) | [62] | |
| Micro-obstacles spiral-chip | 390 | 22 | >95% | —— | 2.29×105 | —— | [63] | |
| Spiral series-chip | 60–120 | 2.4 | 73.75–80.75% | 63.6% | —— | —— | [64] | |
| Multistage hydrodynamic focusing-chip | 0.54–0.90 | 0.90 | >90% | 85% | —— | Ovarian (n = 1) | [65] | |
| Membrane filtration | 12–60 | 12–60 | >80% | WBCs number: 1000–3000 mL−1 | —— | —— | [66] | |
| Cluster-chip | 2.5 | 2.5 | 41% (two-cell) | —— | —— | Breast (n = 27), melanoma (n = 20), prostate (n = 13) | [26] | |
| Conical-shaped holes-chip | 12 | 4 | 96% | WBC clearance efficiency: 96% | ~102 | Lung (n = 8), nasopharynx (n=2), mediastinal (n = 2), cardia (n = 1), cervical (n = 1) and breast (n = 1) | [67] | |
| Ratchets-chip | 1 | 1 | 90% | —— | ~104 | Prostate (n = 20) | [68] | |
| Parsortix-chip | 10 | 10 | 54–69% | WBCs number: 200–5000 mL−1 | —— | Breast (n = 10), colon (n = 10) and lung (n = 6) | [69] |
2.2. Methods Based on Physical Properties
3. Electrochemical Detection of CTC
| Cancer Type | Cell Line | Aptamer | DNA or RNA | Biomarker | Kd/nM | Ref. |
|---|---|---|---|---|---|---|
| Breast | MCF-7/HeLa | AS1411 | DNA | Overexpressed nucleolin on the cell surface | 100–1000 | [100] |
| MCF-7 | MUC 1 | DNA | Overexpressed mucin 1 (MUC1) glycoprotein on the cell membrane | 38.3 | [101] | |
| MCF-7/MDA-MB-231 | SYL3C | DNA | EpCAM | 38 ± 9 | [102,103] | |
| Leukemia | CCRF-CEM | Sgc8c | DNA | Tyrosine kinase-7 on the cell surface | 2.0 ± 0.2 | [104] |
| Lymphoma | Toledo | Sgd5 | DNA | —— | —— | [105] |
| Ramos | Td05 | DNA | B-cell receptor | 74.8 ± 8.7 | [105,106] | |
| Ramos | TE02 | DNA | —— | 0.76 ± 0.13 | [106] | |
| Karpas 299 | Anti-CD30 RNA aptamer | RNA | CD30 | —— | [107] | |
| Colorectal, glioblastoma and lung | —— | Anti-EGFR RNA aptamer | RNA | Epidermal growth factor receptor (EGFR) | 2.4 | [108,109] |
| Liver | Mear | TLS1c | ssDNA | —— | 9.79 ± 0.30 | [110,111] |
| HepG2/Mear | TLS11a | DNA | —— | 4.51 ± 0.39 | [110,111] | |
| Lung | —— | LC-18 | DNA | Neutrophil defensin 1 and 3 | 38 | [112] |
| Prostate | LNCaP | A10 RNA aptamer | RNA | Prostate specific membrane antigen (PSMA) | 600 | [113,114] |
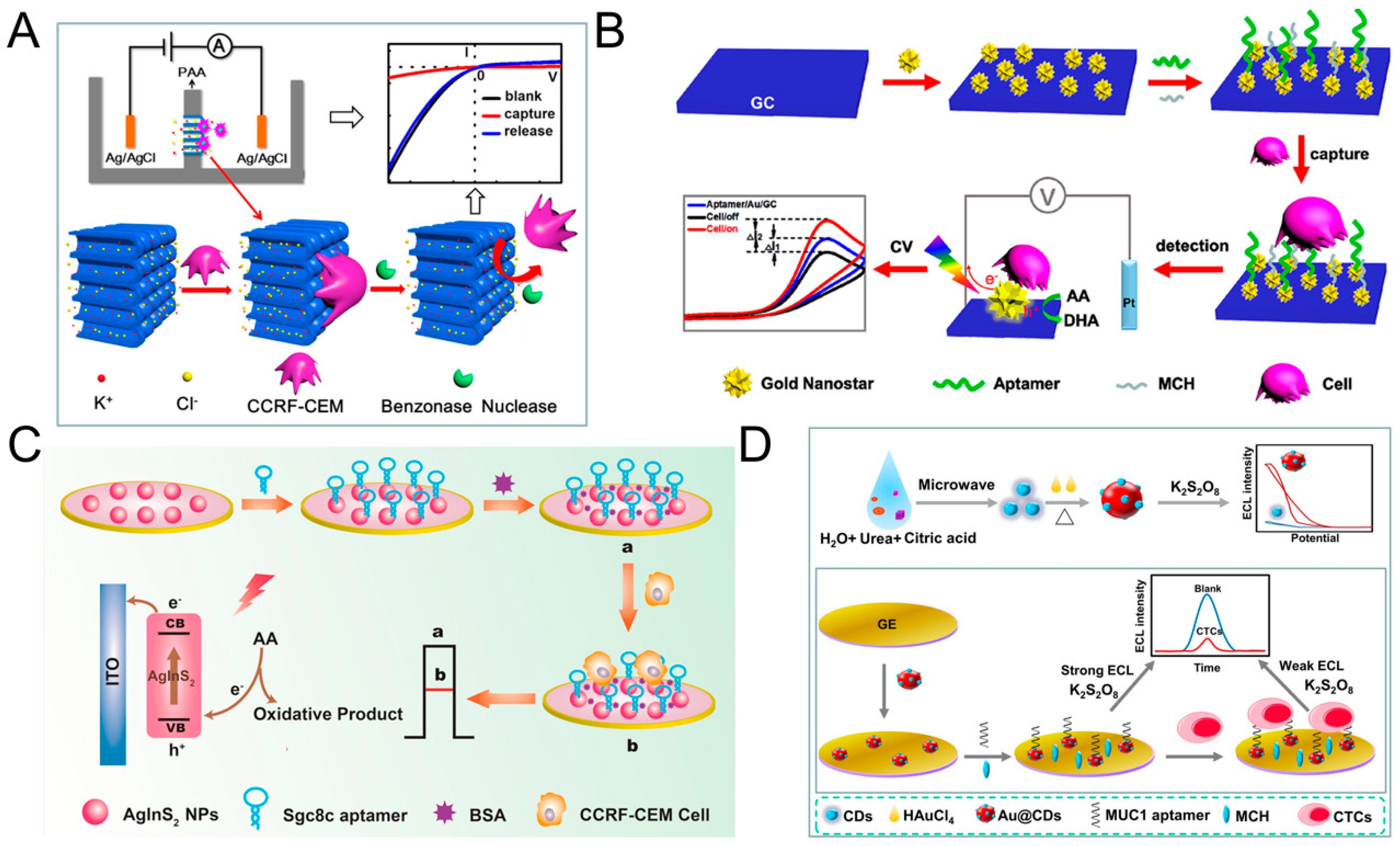
3.1. Direct Assays
| Assay Time (min) | Linear Range (Cells mL−1) | LOD (Cells mL−1) | Aptamer/Antibody | Target Cell | Capture Probe | Signal Probe | Clinical Sample | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| Direct assay | 70 | 18–1.5 × 106 | 6 | AS1411 | MCF-7 | xFe2O3-nPt (or nPt-xFe2O3)-coated graphene nanostructure+aptamer | —— | Spiked blood samples | [119] |
| 45 | 1 × 102–2 × 106 | 100 | sgc8c | CCRF-CEM | Nanochannel−ion channel +aptamer | —— | —— | [116] | |
| 90 | 100–10,000 | 34 | anti-MUC 1 aptamer | MCF-7 | Gold electrode+Au@CDs +aptamer | —— | Human serum samples | [92] | |
| 45 | 5–1 × 105 | 5 | sgc8c | CCRF-CEM | Glassy carbon electrode +AuNSs+aptamer | —— | Human serum samples and spiked blood samples | [117] | |
| 60 | 1.5 × 102–3 × 105 | 16 | sgc8c | CCRF-CEM | AgInS2 NPs-modified electrode+aptamer | —— | —— | [93] | |
| Sandwich assay | 120 | 75–5500 | 75 | E-cadherin | Not limited | CNT-AuNPs+antibody | E-cadherin antibody+QD | Fetal calf serum and mouse blood | [121] |
| 50 | 1 × 102–5 × 104 | 5 | SYL3C | MCF-7 | m-aptamer/MCH/AuE | Anti-EpCAM/HRP-AuNP | Spiked blood samples | [28] | |
| 20 | 5–500 | 4 | Td05 | Ramos | AuNPs-Fe3O4-GS+aptamer | AuNPs with the electroactive species (Fc-SH/Thi) +aptamer | Spiked blood samples and 3 leukemia patients | [98] | |
| 3 | Sgc8 | CCRF-CEM | |||||||
| 40 | 10–1 × 106 | 2 | EpCAM+vimentin | MCF-7 | Ketjen black+AuNP +antibody | PdIrBPMNS+antibody | Spiked blood samples | [29] | |
| 170 | 3–3000 | 1 | 5′-thiol modified MCF-7 binding aptamer | MCF-7 | MN+EpCAM | Cu2O+aptamer | Spiked blood samples | [30] | |
| 60 | 3–1000 | 1 | anti-MUC 1 aptamer | MCF-7 | MN+EpCAM | LiFePO4/Au+aptamer | Spiked blood samples | [27] | |
| 115 | 5–3 × 104 | 1 | SYL3C-RCA primer | MCF-7 | MN+EpCAM | Aptamer+DNA amplification (RCA) | Spiked blood samples | [99] | |
| 40 | 20–1 × 106 | 3 | EpCAM | MCF-7 | Au electrode+AB +AuNP+proteinG+antibody | Pt@AgNFs+antibody | Spiked blood samples | [94] |
3.2. Sandwich Assays
3.3. Other Assays
4. Detection Method Based on Point-of-Care Testing (POCT)
4.1. POCT Based Detection of Enzyme Activity/Proteins/Compound
4.2. POCT Based Detection of CTC
5. Conclusions and Future Trends
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CTC | circulating tumor cell |
| ctDNA | circulating tumor DNA |
| EV | extracellular vesicle |
| WBC | white blood cell |
| RBC | red blood cell |
| EpCAM | epithelial cell adhesion molecule |
| CK | cytokeratin |
| POCT | point-of-care testing |
| EMT | epithelial–mesenchymal transition |
| MET | mesenchymal- epithelial transition |
| ISET | isolation by size of epithelial tumor cell |
| DLD | deterministic lateral displacement |
| ECL | electrochemiluminescence |
| PEC | photoelectrochemical |
| EIS | electrochemical impedance spectroscopy |
| LSV | linear sweep voltammetry |
| LOD | the limit of detection |
| MPA | mercaptopropionic acid |
| NP | nanoparticle |
| AuNS | Au nanostar |
| GC | glassy carbon |
| DPV | differential pulse voltammetry |
| RCA | rolling circle amplification |
| HRP | horseradish peroxidase |
| AuNP | Au nanoparticle |
| AB | acetylene black |
| HCNT | hexagonal carbon nitride tube |
| GS | graphene nanosheet |
| CV | cyclic voltammetry |
| QD | quantum dot |
| PDMS | polydimethylsiloxane |
| AuNW | Au nanowire |
| DM | daunomycin |
| TS | telomerase primer |
| CD | carbon dot |
| MN | magnetic nanosphere |
| MB | magnetic bead |
| PtNP | Pt nanoparticle |
| ALP | alkaline phosphatase |
| FDG | di-β-D-pyridine galactoside |
| FMG | fluorescein monogalactoside |
| ET | electrophoresis titration |
| MRB | moving reaction boundary |
| CEACAM5 | carcinoembryonic antigen-related cell adhesion molecules 5 |
| RI | refractive index |
| ACNP | aptamer-conjugated nanoparticle |
| GO | graphene oxide |
| EGFR | epidermal growth factor receptor |
| PSMA | prostate specific membrane antigen |
References
- Steeg, P.S. Targeting metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef]
- Sleeman, J.; Steeg, P.S. Cancer metastasis as a therapeutic target. Eur. J. Cancer 2010, 46, 1177–1180. [Google Scholar] [CrossRef]
- Vaidyanathan, R.; Soon, R.H.; Zhang, P.; Jiang, K.; Lim, C.T. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab Chip 2018, 19, 11–34. [Google Scholar] [CrossRef]
- Tadimety, A.; Closson, A.; Li, C.; Yi, S.; Shen, T.; Zhang, J.X.J. Advances in liquid biopsy on-chip for cancer management: Technologies, biomarkers, and clinical analysis. Crit. Rev. Clin. Lab. Sci. 2018, 55, 140–162. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snow, A.; Chen, D.; Lang, J.E. The current status of the clinical utility of liquid biopsies in cancer. Expert Rev. Mol. Diagn. 2019, 19, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Tellez-Gabriel, M.; Heymann, M.F.; Heymann, D. Circulating Tumor Cells as a Tool for Assessing Tumor Heterogeneity. Theranostics 2019, 9, 4580–4594. [Google Scholar] [CrossRef]
- O’Flaherty, J.D.; Gray, S.; Richard, D.; Fennell, D.; O’Leary, J.J.; Blackhall, F.H.; O’Byrne, K.J. Circulating tumour cells, their role in metastasis and their clinical utility in lung cancer. Lung Cancer 2012, 76, 19–25. [Google Scholar] [CrossRef]
- Samlowski, W.E.; McGregor, J.R.; Samlowski, S.T.; Tharkar, S.; Shen, S.; Bentz, J.S. Growth of Circulating Tumor Cell-Derived Colonies from Peripheral Blood of Melanoma Patients: Preliminary Characterization of Colony Composition. Health 2014, 6, 1467–1481. [Google Scholar] [CrossRef] [Green Version]
- Cheung, K.J.; Ewald, A.J. A collective route to metastasis: Seeding by tumor cell clusters. Science 2016, 352, 167–169. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Mu, Z.; Chervoneva, I.; Austin, L.; Ye, Z.; Rossi, G.; Palazzo, J.P.; Sun, C.; Abu-Khalaf, M.; Myers, R.E.; et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res. Treat. 2017, 161, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Grenci, G.; Lim, Y.B.; Lee, S.C.; Han, J.; Lim, C.T. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat. Protoc. 2018, 13, 34–58. [Google Scholar] [CrossRef] [PubMed]
- Yin, S.; Xi, R.; Wu, A.; Wang, S.; Li, Y.; Wang, C.; Tang, L.; Xia, Y.; Yang, D.; Li, J.; et al. Patient-derived tumor-like cell clusters for drug testing in cancer therapy. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tu, C.; Liang, Y.; Huang, B.; Fang, Y.; Liang, X.; Ye, X. The label-free separation and culture of tumor cells in a microfluidic biochip. Analyst 2020, 145, 1706–1715. [Google Scholar] [CrossRef]
- Cheng, Y.H.; Chen, Y.C.; Lin, E.; Brien, R.; Jung, S.; Chen, Y.T.; Lee, W.; Hao, Z.; Sahoo, S.; Min Kang, H.; et al. Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat. Commun. 2019, 10, 2163. [Google Scholar] [CrossRef]
- Hou, H.W.; Warkiani, M.E.; Khoo, B.L.; Li, Z.R.; Soo, R.A.; Tan, D.S.; Lim, W.T.; Han, J.; Bhagat, A.A.; Lim, C.T. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci. Rep. 2013, 3, 1259. [Google Scholar] [CrossRef] [Green Version]
- Alix-Panabieres, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef]
- Weissenstein, U.; Schumann, A.; Reif, M.; Link, S.; Toffol-Schmidt, U.D.; Heusser, P. Detection of circulating tumor cells in blood of metastatic breast cancer patients using a combination of cytokeratin and EpCAM antibodies. BMC Cancer 2012, 12, 206. [Google Scholar] [CrossRef] [Green Version]
- Banko, P.; Lee, S.Y.; Nagygyorgy, V.; Zrinyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Luo, G.; Du, W.; Kong, T.; Liu, C.; Liu, Z. Recent Advances in Microfluidic Platforms Applied in Cancer Metastasis: Circulating Tumor Cells’ (CTCs) Isolation and Tumor-On-A-Chip. Small 2020, 16, e1903899. [Google Scholar] [CrossRef] [PubMed]
- Nasiri, R.; Shamloo, A.; Ahadian, S.; Amirifar, L.; Akbari, J.; Goudie, M.J.; Lee, K.; Ashammakhi, N.; Dokmeci, M.R.; Di Carlo, D.; et al. Microfluidic-Based Approaches in Targeted Cell/Particle Separation Based on Physical Properties: Fundamentals and Applications. Small 2020, 16, e2000171. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Lu, J.; Zhang, L.; Chen, Z. Aptamer-based electrochemical cytosensors for tumor cell detection in cancer diagnosis: A review. Anal. Chim. Acta 2019, 1082, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Huang, J.; He, H.; Ma, C.; Wang, K. Contributing to liquid biopsy: Optical and electrochemical methods in cancer biomarker analysis. Coord. Chem. Rev. 2020, 415. [Google Scholar] [CrossRef]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stott, S.L.; Hsu, C.H.; Tsukrov, D.I.; Yu, M.; Miyamoto, D.T.; Waltman, B.A.; Rothenberg, S.M.; Shah, A.M.; Smas, M.E.; Korir, G.K.; et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 2010, 107, 18392–18397. [Google Scholar] [CrossRef] [Green Version]
- Sarioglu, A.F.; Aceto, N.; Kojic, N.; Donaldson, M.C.; Zeinali, M.; Hamza, B.; Engstrom, A.; Zhu, H.; Sundaresan, T.K.; Miyamoto, D.T.; et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 2015, 12, 685–691. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, H.; Yang, M.; Liao, L. Electrochemical assay for detection of circulating tumor cells based on LiFePO4 as electrochemical probe. Mater. Lett. 2020, 276. [Google Scholar] [CrossRef]
- Yang, J.; Li, X.; Jiang, B.; Yuan, R.; Xiang, Y. In Situ-Generated Multivalent Aptamer Network for Efficient Capture and Sensitive Electrochemical Detection of Circulating Tumor Cells in Whole Blood. Anal. Chem. 2020, 92, 7893–7899. [Google Scholar] [CrossRef]
- Peng, Y.; Peng, Y.; Tang, S.; Shen, H.; Sheng, S.; Wang, Y.; Wang, T.; Cai, J.; Xie, G.; Feng, W. PdIrBP mesoporous nanospheres combined with superconductive carbon black for the electrochemical determination and collection of circulating tumor cells. Mikrochim. Acta 2020, 187, 216. [Google Scholar] [CrossRef]
- Luo, J.; Liang, D.; Zhao, D.; Yang, M. Photoelectrochemical detection of circulating tumor cells based on aptamer conjugated Cu2O as signal probe. Biosens. Bioelectron. 2020, 151, 111976. [Google Scholar] [CrossRef]
- Park, J.; Han, D.H.; Park, J.K. Towards practical sample preparation in point-of-care testing: User-friendly microfluidic devices. Lab Chip 2020, 20, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Tian, T.; Chen, X.; Lei, Z.; Song, Y.; Shi, Y.; Ji, T.; Zhu, Z.; Yang, L.; Yang, C. Gas-generating reactions for point-of-care testing. Analyst 2018, 143, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Manz, A.; Graber, N.; Widmer, H.M. Miniaturized Total Chemical-Analysis Systems—A Novel Concept for Chemical Sensing. Sens. Actuator B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Goetz, J.G. Metastases go with the flow. Science 2018, 362, 999–1000. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Kyprianou, N. Targeting anoikis resistance in prostate cancer metastasis. Mol. Asp. Med. 2010, 31, 205–214. [Google Scholar] [CrossRef] [Green Version]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef] [Green Version]
- Yeung, K.T.; Yang, J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Yang, H.; Zhang, M.; Zhang, D.; Liu, Y.; Liu, Y.; Song, Y.; Zhang, X.; Li, H.; Ma, W.; et al. Circulating tumor cells (CTCs) detected by triple-marker EpCAM, CK19, and hMAM RT-PCR and their relation to clinical outcome in metastatic breast cancer patients. Cell Biochem. Biophys. 2013, 65, 263–273. [Google Scholar] [CrossRef]
- Yoon, S.O.; Kim, Y.T.; Jung, K.C.; Jeon, Y.K.; Kim, B.H.; Kim, C.W. TTF-1 mRNA-positive circulating tumor cells in the peripheral blood predict poor prognosis in surgically resected non-small cell lung cancer patients. Lung Cancer 2011, 71, 209–216. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [Green Version]
- Yates, D.R.; Roupret, M.; Drouin, S.J.; Comperat, E.; Ricci, S.; Lacave, R.; Sebe, P.; Cancel-Tassin, G.; Bitker, M.O.; Cussenot, O. Quantitative RT-PCR analysis of PSA and prostate-specific membrane antigen mRNA to detect circulating tumor cells improves recurrence-free survival nomogram prediction after radical prostatectomy. Prostate 2012, 72, 1382–1388. [Google Scholar] [CrossRef] [PubMed]
- Shimada, R.; Iinuma, H.; Akahane, T.; Horiuchi, A.; Watanabe, T. Prognostic significance of CTCs and CSCs of tumor drainage vein blood in Dukes’ stage B and C colorectal cancer patients. Oncol. Rep. 2012, 27, 947–953. [Google Scholar] [CrossRef] [Green Version]
- Iinuma, H.; Watanabe, T.; Mimori, K.; Adachi, M.; Hayashi, N.; Tamura, J.; Matsuda, K.; Fukushima, R.; Okinaga, K.; Sasako, M.; et al. Clinical significance of circulating tumor cells, including cancer stem-like cells, in peripheral blood for recurrence and prognosis in patients with Dukes’ stage B and C colorectal cancer. J. Clin. Oncol. 2011, 29, 1547–1555. [Google Scholar] [CrossRef]
- Aktas, B.; Kasimir-Bauer, S.; Heubner, M.; Kimmig, R.; Wimberger, P. Molecular profiling and prognostic relevance of circulating tumor cells in the blood of ovarian cancer patients at primary diagnosis and after platinum-based chemotherapy. Int. J. Gynecol. Cancer 2011, 21, 822–830. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Li, P.; He, S.; Long, T.; Zhang, N.; Fang, J.; Yu, Z. Detection of circulating tumour cells with the CellSearch system in patients with advanced-stage head and neck cancer: Preliminary results. J. Laryngol. Otol. 2013, 127, 788–793. [Google Scholar] [CrossRef]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J. Oncol. 2010, 2010, 617421. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Wen, C.Y.; Wu, L.L.; Hong, S.L.; Hu, J.; Xu, C.M.; Pang, D.W.; Zhang, Z.L. A chip assisted immunomagnetic separation system for the efficient capture and in situ identification of circulating tumor cells. Lab Chip 2016, 16, 1214–1223. [Google Scholar] [CrossRef]
- Talasaz, A.H.; Powell, A.A.; Huber, D.E.; Berbee, J.G.; Roh, K.H.; Yu, W.; Xiao, W.; Davis, M.M.; Pease, R.F.; Mindrinos, M.N.; et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. USA 2009, 106, 3970–3975. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.; Dubash, T.D.; Edd, J.F.; Jewett, M.K.; Garre, S.G.; Karabacak, N.M.; Rabe, D.C.; Mutlu, B.R.; Walsh, J.R.; Kapur, R.; et al. Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc. Natl. Acad. Sci. USA 2020, 117, 16839–16847. [Google Scholar] [CrossRef]
- Liu, H.; Sun, N.; Ding, P.; Chen, C.; Wu, Z.; Zhu, W.; Liu, L.; Wang, Z.; Pei, R. Fabrication of aptamer modified TiO2 nanofibers for specific capture of circulating tumor cells. Colloids Surf. B Biointerfaces 2020, 191, 110985. [Google Scholar] [CrossRef]
- Lou, H.Y.; Zhao, W.; Hanson, L.; Zeng, C.; Cui, Y.; Cui, B. Dual-Functional Lipid Coating for the Nanopillar-Based Capture of Circulating Tumor Cells with High Purity and Efficiency. Langmuir 2017, 33, 1097–1104. [Google Scholar] [CrossRef]
- Jolly, M.K. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front. Oncol. 2015, 5. [Google Scholar] [CrossRef] [Green Version]
- Diepenbruck, M.; Christofori, G. Epithelial–mesenchymal transition (EMT) and metastasis: Yes, no, maybe? Curr. Opin. Cell Biol. 2016, 43, 7–13. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Zhao, H.; Shu, W.; Tian, J.; Huang, Y.; Song, Y.; Wang, R.; Li, E.; Slamon, D.; Hou, D.; et al. An integrated microfluidic device for rapid and high-sensitivity analysis of circulating tumor cells. Sci. Rep. 2017, 7, 42612. [Google Scholar] [CrossRef] [Green Version]
- Nanou, A.; Crespo, M.; Flohr, P.; De Bono, J.S.; Terstappen, L. Scanning Electron Microscopy of Circulating Tumor Cells and Tumor-Derived Extracellular Vesicles. Cancers 2018, 10, 416. [Google Scholar] [CrossRef] [Green Version]
- Riethdorf, S.; Fritsche, H.; Muller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Janicke, F.; et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: A validation study of the CellSearch system. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef] [Green Version]
- Theil, G.; Fischer, K.; Weber, E.; Medek, R.; Hoda, R.; Lucke, K.; Fornara, P. The Use of a New CellCollector to Isolate Circulating Tumor Cells from the Blood of Patients with Different Stages of Prostate Cancer and Clinical Outcomes—A Proof-of-Concept Study. PLoS ONE 2016, 11, e0158354. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.J.; Shanker, A.; Wang, Y.; Kozminsky, M.; Jin, Q.; Palanisamy, N.; Burness, M.L.; Azizi, E.; Simeone, D.M.; Wicha, M.S.; et al. Tunable Thermal-Sensitive Polymer-Graphene Oxide Composite for Efficient Capture and Release of Viable Circulating Tumor Cells. Adv. Mater. 2016, 28, 4891–4897. [Google Scholar] [CrossRef] [Green Version]
- Loutherback, K.; D’Silva, J.; Liu, L.Y.; Wu, A.; Austin, R.H.; Sturm, J.C. Deterministic separation of cancer cells from blood at 10 mL/min. AIP Adv. 2012, 2. [Google Scholar] [CrossRef] [Green Version]
- Sollier, E.; Go, D.E.; Che, J.; Gossett, D.R.; O’Byrne, S.; Weaver, W.M.; Kummer, N.; Rettig, M.; Goldman, J.; Nickols, N.; et al. Size-selective collection of circulating tumor cells using Vortex technology. Lab Chip 2014, 14, 63–77. [Google Scholar] [CrossRef]
- Warkiani, M.E.; Khoo, B.L.; Tan, D.S.W.; Bhagat, A.A.S.; Lim, W.T.; Yap, Y.S.; Lee, S.C.; Soo, R.A.; Han, J.; Lim, C.T. An ultra-high-throughput spiral microfluidic biochip for the enrichment of circulating tumor cells. Analyst 2014, 139, 3245–3255. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.; Rivera-Báez, L.; Fouladdel, S.; Yoon, H.J.; Guthrie, S.; Wieger, J.; Deol, Y.; Keller, E.; Sahai, V.; Simeone, D.M.; et al. High-Throughput Microfluidic Labyrinth for the Label-free Isolation of Circulating Tumor Cells. Cell Syst. 2017, 5, 295–304.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, S.; Tian, C.; Li, T.; Xu, J.; Chen, S.-W.; Tu, Q.; Yuan, M.-S.; Liu, W.; Wang, J. Spiral microchannel with ordered micro-obstacles for continuous and highly-efficient particle separation. Lab Chip 2017, 17, 3578–3591. [Google Scholar] [CrossRef]
- Abdulla, A.; Liu, W.; Gholamipour-Shirazi, A.; Sun, J.; Ding, X. High-Throughput Isolation of Circulating Tumor Cells Using Cascaded Inertial Focusing Microfluidic Channel. Anal. Chem. 2018, 90, 4397–4405. [Google Scholar] [CrossRef]
- Gao, R.; Cheng, L.; Wang, S.; Bi, X.; Wang, X.; Wang, R.; Chen, X.; Zha, Z.; Wang, F.; Xu, X.; et al. Efficient separation of tumor cells from untreated whole blood using a novel multistage hydrodynamic focusing microfluidics. Talanta 2020, 207, 120261. [Google Scholar] [CrossRef]
- Hosokawa, M.; Hayata, T.; Fukuda, Y.; Arakaki, A.; Yoshino, T.; Tanaka, T.; Matsunaga, T. Size-Selective Microcavity Array for Rapid and Efficient Detection of Circulating Tumor Cells. Anal. Chem. 2010, 82, 6629–6635. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Shi, J.; Li, S.; Wang, L.; Cayre, Y.E.; Chen, Y. Microfluidic device with integrated microfilter of conical-shaped holes for high efficiency and high purity capture of circulating tumor cells. Sci. Rep. 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- Park, E.S.; Jin, C.; Guo, Q.; Ang, R.R.; Duffy, S.P.; Matthews, K.; Azad, A.; Abdi, H.; Todenhöfer, T.; Bazov, J.; et al. Continuous Flow Deformability-Based Separation of Circulating Tumor Cells Using Microfluidic Ratchets. Small 2016, 12, 1909–1919. [Google Scholar] [CrossRef]
- Hvichia, G.E.; Parveen, Z.; Wagner, C.; Janning, M.; Quidde, J.; Stein, A.; Muller, V.; Loges, S.; Neves, R.P.; Stoecklein, N.H.; et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. Cancer 2016, 138, 2894–2904. [Google Scholar] [CrossRef]
- Hao, S.-J.; Wan, Y.; Xia, Y.-Q.; Zou, X.; Zheng, S.-Y. Size-based separation methods of circulating tumor cells. Adv. Drug Deliv. Rev. 2018, 125, 3–20. [Google Scholar] [CrossRef]
- Park, S.; Ang, R.R.; Duffy, S.P.; Bazov, J.; Chi, K.N.; Black, P.C.; Ma, H. Morphological differences between circulating tumor cells from prostate cancer patients and cultured prostate cancer cells. PLoS ONE 2014, 9, e85264. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Iwata, M. Stiffness of cancer cells measured with an AFM indentation method. J. Mech. Behav. Biomed. Mater. 2015, 49, 105–111. [Google Scholar] [CrossRef]
- Wu, P.H.; Aroush, D.R.; Asnacios, A.; Chen, W.C.; Dokukin, M.E.; Doss, B.L.; Durand-Smet, P.; Ekpenyong, A.; Guck, J.; Guz, N.V.; et al. A comparison of methods to assess cell mechanical properties. Nat. Methods 2018, 15, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Sajeesh, P.; Raj, A.; Doble, M.; Sen, A.K. Characterization and sorting of cells based on stiffness contrast in a microfluidic channel. RSC Adv. 2016, 6, 74704–74714. [Google Scholar] [CrossRef]
- Zahalak, G.I.; McConnaughey, W.B.; Elson, E.L. Determination of cellular mechanical properties by cell poking, with an application to leukocytes. J. Biomech. Eng. 1990, 112, 283–294. [Google Scholar] [CrossRef]
- Zhou, Z.L.; Hui, T.H.; Tang, B.; Ngan, A.H.W. Accurate measurement of stiffness of leukemia cells and leukocytes using an optical trap by a rate-jump method. RSC Adv. 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- Raab, M.; Gentili, M.; de Belly, H.; Thiam, H.R.; Vargas, P.; Jimenez, A.J.; Lautenschlaeger, F.; Voituriez, R.; Lennon-Dumenil, A.M.; Manel, N.; et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 2016, 352, 359–362. [Google Scholar] [CrossRef]
- Zhou, M.D.; Hao, S.; Williams, A.J.; Harouaka, R.A.; Schrand, B.; Rawal, S.; Ao, Z.; Brenneman, R.; Gilboa, E.; Lu, B.; et al. Separable bilayer microfiltration device for viable label-free enrichment of circulating tumour cells. Sci. Rep. 2014, 4, 7392. [Google Scholar] [CrossRef]
- Shim, J.-E.; Bu, J.; Lee, M.-K.; Cho, Y.-H.; Kim, T.-H.; Bu, J.-U.; Han, S.-W. Viable and high-throughput isolation of heterogeneous circulating tumor cells using tapered-slit filters. Sens. Actuators B Chem. 2020, 321. [Google Scholar] [CrossRef]
- Farace, F.; Massard, C.; Vimond, N.; Drusch, F.; Jacques, N.; Billiot, F.; Laplanche, A.; Chauchereau, A.; Lacroix, L.; Planchard, D.; et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br. J. Cancer 2011, 105, 847–853. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, R.; Gertler, R.; Friederichs, J.; Fuehrer, K.; Dahm, M.; Phelps, R.; Thorban, S.; Nekarda, H.; Siewert, J.R. Comparison of two density gradient centrifugation systems for the enrichment of disseminated tumor cells in blood. Cytometry 2002, 49, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Kulasinghe, A.; Kenny, L.; Perry, C.; Thiery, J.P.; Jovanovic, L.; Vela, I.; Nelson, C.; Punyadeera, C. Impact of label-free technologies in head and neck cancer circulating tumour cells. Oncotarget 2016, 7, 71223–71234. [Google Scholar] [CrossRef] [Green Version]
- Yoon, Y.; Kim, S.; Lee, J.; Choi, J.; Kim, R.K.; Lee, S.J.; Sul, O.; Lee, S.B. Clogging-free microfluidics for continuous size-based separation of microparticles. Sci. Rep. 2016, 6, 26531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Huang, F.; Du, J.; Shu, W.; Feng, H.; Xu, X.; Chen, Y. Rapid isolation of cancer cells using microfluidic deterministic lateral displacement structure. Biomicrofluidics 2013, 7, 11801. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, W.; Huang, F.; Feng, H.; Shu, W.; Xu, X.; Chen, Y. High throughput capture of circulating tumor cells using an integrated microfluidic system. Biosens. Bioelectron. 2013, 47, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Warkiani, M.E.; Tan, D.S.; Bhagat, A.A.; Irwin, D.; Lau, D.P.; Lim, A.S.; Lim, K.H.; Krisna, S.S.; Lim, W.T.; et al. Clinical validation of an ultra high-throughput spiral microfluidics for the detection and enrichment of viable circulating tumor cells. PLoS ONE 2014, 9, e99409. [Google Scholar] [CrossRef] [Green Version]
- Boya, M.; Chu, C.H.; Liu, R.; Ozkaya-Ahmadov, T.; Sarioglu, A.F. Circulating Tumor Cell Enrichment Technologies. Recent Results Cancer Res. 2020, 215, 25–55. [Google Scholar] [CrossRef]
- Wu, M.; Huang, P.H.; Zhang, R.; Mao, Z.; Chen, C.; Kemeny, G.; Li, P.; Lee, A.V.; Gyanchandani, R.; Armstrong, A.J.; et al. Circulating Tumor Cell Phenotyping via High-Throughput Acoustic Separation. Small 2018, 14, e1801131. [Google Scholar] [CrossRef]
- Li, P.; Mao, Z.; Peng, Z.; Zhou, L.; Chen, Y.; Huang, P.H.; Truica, C.I.; Drabick, J.J.; El-Deiry, W.S.; Dao, M.; et al. Acoustic separation of circulating tumor cells. Proc. Natl. Acad. Sci. USA 2015, 112, 4970–4975. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Li, Q.; Du, X.; Liu, M. Application of electrochemical biosensors in tumor cell detection. Thorac. Cancer 2020, 11, 840–850. [Google Scholar] [CrossRef]
- Safarpour, H.; Dehghani, S.; Nosrati, R.; Zebardast, N.; Alibolandi, M.; Mokhtarzadeh, A.; Ramezani, M. Optical and electrochemical-based nano-aptasensing approaches for the detection of circulating tumor cells (CTCs). Biosens. Bioelectron. 2020, 148, 111833. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, L.; Zhao, K.; Liu, Z.; Cao, H.; Ye, S.; Liang, G. High luminous efficiency Au@CDs for sensitive and label-free electrochemiluminescent detection of circulating tumor cells in serum. Sens. Actuators B Chem. 2020, 316. [Google Scholar] [CrossRef]
- Li, J.; Lin, X.; Zhang, Z.; Tu, W.; Dai, Z. Red light-driven photoelectrochemical biosensing for ultrasensitive and scatheless assay of tumor cells based on hypotoxic AgInS2 nanoparticles. Biosens. Bioelectron. 2019, 126, 332–338. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Shen, H.; Hao, Y.; Huang, Z.; Tao, Y.; Peng, Y.; Guo, Y.; Xie, G.; Feng, W. A novel cytosensor based on Pt@Ag nanoflowers and AuNPs/Acetylene black for ultrasensitive and highly specific detection of Circulating Tumor Cells. Biosens. Bioelectron. 2018, 104, 72–78. [Google Scholar] [CrossRef]
- Peng, Y.; Pan, Y.; Han, Y.; Sun, Z.; Jalalah, M.; Al-Assiri, M.S.; Harraz, F.A.; Yang, J.; Li, G. Direct Analysis of Rare Circulating Tumor Cells in Whole Blood Based on Their Controlled Capture and Release on Electrode Surface. Anal. Chem. 2020. [Google Scholar] [CrossRef]
- Cao, H.-X.; Liu, P.-F.; Wang, L.; Liu, Z.-J.; Ye, S.-Y.; Liang, G.-X. Nonenzymatic chemiluminescence detection of circulating tumor cells in blood based on Au@luminol nanoparticles, hybridization chain reaction and magnetic isolation. Sens. Actuators B Chem. 2020, 318. [Google Scholar] [CrossRef]
- Vajhadin, F.; Ahadian, S.; Travas-Sejdic, J.; Lee, J.; Mazloum-Ardakani, M.; Salvador, J.; Aninwene, G.E., 2nd; Bandaru, P.; Sun, W.; Khademhossieni, A. Electrochemical cytosensors for detection of breast cancer cells. Biosens. Bioelectron. 2020, 151, 111984. [Google Scholar] [CrossRef]
- Dou, B.; Xu, L.; Jiang, B.; Yuan, R.; Xiang, Y. Aptamer-Functionalized and Gold Nanoparticle Array-Decorated Magnetic Graphene Nanosheets Enable Multiplexed and Sensitive Electrochemical Detection of Rare Circulating Tumor Cells in Whole Blood. Anal. Chem. 2019, 91, 10792–10799. [Google Scholar] [CrossRef]
- Shen, C.; Liu, S.; Li, X.; Yang, M. Electrochemical Detection of Circulating Tumor Cells Based on DNA Generated Electrochemical Current and Rolling Circle Amplification. Anal. Chem. 2019, 91, 11614–11619. [Google Scholar] [CrossRef]
- Figueiredo, J.; Lopes-Nunes, J.; Carvalho, J.; Antunes, F.; Ribeiro, M.; Campello, M.P.C.; Paulo, A.; Paiva, A.; Salgado, G.F.; Queiroz, J.A.; et al. AS1411 derivatives as carriers of G-quadruplex ligands for cervical cancer cells. Int. J. Pharm. 2019, 568. [Google Scholar] [CrossRef]
- Hu, Y.; Duan, J.H.; Zhan, Q.M.; Wang, F.D.; Lu, X.; Yang, X.D. Novel MUC1 Aptamer Selectively Delivers Cytotoxic Agent to Cancer Cells In Vitro. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Y.L.; Zhu, Z.; An, Y.; Zhang, W.T.; Zhang, H.M.; Liu, D.; Yu, C.D.; Duan, W.; Yang, C.J. Selection of DNA Aptamers against Epithelial Cell Adhesion Molecule for Cancer Cell Imaging and Circulating Tumor Cell Capture. Anal. Chem. 2013, 85, 4141–4149. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Zhang, Q.; Feng, S.; Zhu, J.J.; Wang, Q.; Wang, H. Robust nonenzymatic hybrid nanoelectrocatalysts for signal amplification toward ultrasensitive electrochemical cytosensing. J. Am. Chem. Soc. 2014, 136, 2288–2291. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.F.; Shangguan, D.; Liu, H.; Phillips, J.A.; Zhang, X.; Chen, Y.; Tan, W. Molecular assembly of an aptamer-drug conjugate for targeted drug delivery to tumor cells. ChemBioChem 2009, 10, 862–868. [Google Scholar] [CrossRef] [Green Version]
- Zheng, F.Y.; Cheng, Y.; Wang, J.; Lu, J.; Zhang, B.; Zhao, Y.J.; Gu, Z.Z. Aptamer-Functionalized Barcode Particles for the Capture and Detection of Multiple Types of Circulating Tumor Cells. Adv. Mater. 2014, 26, 7333–7338. [Google Scholar] [CrossRef]
- Liu, G.D.; Mao, X.; Phillips, J.A.; Xu, H.; Tan, W.H.; Zeng, L.W. Aptamer-Nanoparticle Strip Biosensor for Sensitive Detection of Cancer Cells. Anal. Chem. 2009, 81, 10013–10018. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Z.; Tung, C.H.; Zu, Y. A cancer cell-activatable aptamer-reporter system for one-step assay of circulating tumor cells. Mol. Ther. Nucleic Acids 2014, 3, e184. [Google Scholar] [CrossRef]
- Wan, Y.; Tan, J.F.; Asghar, W.; Kim, Y.T.; Liu, Y.L.; Iqbal, S.M. Velocity Effect on Aptamer-Based Circulating Tumor Cell Isolation in Microfluidic Devices. J. Phys. Chem. B 2011, 115, 13891–13896. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, Y.L.; Allen, P.B.; Asghar, W.; Mahmood, M.A.I.; Tan, J.F.; Duhon, H.; Kim, Y.T.; Ellington, A.D.; Iqbal, S.M. Capture, isolation and release of cancer cells with aptamer-functionalized glass bead array. Lab Chip 2012, 12, 4693–4701. [Google Scholar] [CrossRef] [Green Version]
- Shangguan, D.H.; Meng, L.; Cao, Z.H.C.; Xiao, Z.Y.; Fang, X.H.; Li, Y.; Cardona, D.; Witek, R.P.; Liu, C.; Tan, W.H. Identification of liver cancer-specific aptamers using whole live cells. Anal. Chem. 2008, 80, 721–728. [Google Scholar] [CrossRef]
- Qu, L.; Xu, J.; Tan, X.; Liu, Z.; Xu, L.; Peng, R. Dual-aptamer modification generates a unique interface for highly sensitive and specific electrochemical detection of tumor cells. ACS Appl. Mater. Interfaces 2014, 6, 7309–7315. [Google Scholar] [CrossRef] [PubMed]
- Frank, L.A.; Bashmakova, E.E.; Goncharova, N.S.; Krasitskaya, V.V. Bioluminescent Binding Microassay Using Aptamers as Biospecific Elements. J. Sib. Fed. Univ. Biol. 2019, 244–252. [Google Scholar] [CrossRef]
- Bagalkot, V.; Farokhzad, O.C.; Langer, R.; Jon, S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. 2006, 45, 8149–8152. [Google Scholar] [CrossRef] [PubMed]
- Min, K.; Song, K.M.; Cho, M.; Chun, Y.S.; Shim, Y.B.; Ku, J.K.; Ban, C. Simultaneous electrochemical detection of both PSMA (+) and PSMA (-) prostate cancer cells using an RNA/peptide dual-aptamer probe. Chem. Commun. 2010, 46, 5566–5568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, H.; Yang, J.; Chen, Z.; Chen, X.; Wang, L.; Hu, J.; Ji, F.; Xie, G.; Feng, W. A novel label-free and reusable electrochemical cytosensor for highly sensitive detection and specific collection of CTCs. Biosens. Bioelectron. 2016, 81, 495–502. [Google Scholar] [CrossRef]
- Cao, J.; Zhao, X.P.; Younis, M.R.; Li, Z.Q.; Xia, X.H.; Wang, C. Ultrasensitive Capture, Detection, and Release of Circulating Tumor Cells Using a Nanochannel-Ion Channel Hybrid Coupled with Electrochemical Detection Technique. Anal. Chem. 2017, 89, 10957–10964. [Google Scholar] [CrossRef]
- Wang, S.S.; Zhao, X.P.; Liu, F.F.; Younis, M.R.; Xia, X.H.; Wang, C. Direct Plasmon-Enhanced Electrochemistry for Enabling Ultrasensitive and Label-Free Detection of Circulating Tumor Cells in Blood. Anal. Chem. 2019, 91, 4413–4420. [Google Scholar] [CrossRef]
- Cordaro, A.; Neri, G.; Sciortino, M.T.; Scala, A.; Piperno, A. Graphene-Based Strategies in Liquid Biopsy and in Viral Diseases Diagnosis. Nanomaterials 2020, 10, 1014. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, F.; Wu, X.; Liu, Y.; Chen, A. Recognition and sensitive detection of CTCs using a controllable label-free electrochemical cytosensor. Mikrochim. Acta 2020, 187, 487. [Google Scholar] [CrossRef]
- Bábelová, L.; Sohová, M.E.; Poturnayová, A.; Buríková, M.; Bizík, J.; Hianik, T. Label-free Electrochemical Aptasensor for Jurkat Cells Detection as a Potential Diagnostic Tool for Leukemia. Electroanalysis 2018, 30, 1487–1495. [Google Scholar] [CrossRef]
- Du, X.; Zhang, Z.; Zheng, X.; Zhang, H.; Dong, D.; Zhang, Z.; Liu, M.; Zhou, J. An electrochemical biosensor for the detection of epithelial-mesenchymal transition. Nat. Commun. 2020, 11, 192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Li, Y.; Wu, H.; Huang, W.; Ju, H.; Ding, S. A amperometric immunosensor for sensitive detection of circulating tumor cells using a tyramide signal amplification-based signal enhancement system. Biosens. Bioelectron. 2019, 130, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Lu, J.; Luo, Z.; Zhang, L.; Liu, P.; Chen, Z. Competitive electrochemical platform for ultrasensitive cytosensing of liver cancer cells by using nanotetrahedra structure with rolling circle amplification. Biosens. Bioelectron. 2018, 120, 8–14. [Google Scholar] [CrossRef] [PubMed]
- An, L.; Wang, G.; Han, Y.; Li, T.; Jin, P.; Liu, S. Electrochemical biosensor for cancer cell detection based on a surface 3D micro-array. Lab Chip 2018, 18, 335–342. [Google Scholar] [CrossRef]
- Yan, S.; Chen, P.; Zeng, X.; Zhang, X.; Li, Y.; Xia, Y.; Wang, J.; Dai, X.; Feng, X.; Du, W.; et al. Integrated Multifunctional Electrochemistry Microchip for Highly Efficient Capture, Release, Lysis, and Analysis of Circulating Tumor Cells. Anal. Chem. 2017, 89, 12039–12044. [Google Scholar] [CrossRef]
- Zhai, T.T.; Ye, D.; Zhang, Q.W.; Wu, Z.Q.; Xia, X.H. Highly Efficient Capture and Electrochemical Release of Circulating Tumor Cells by Using Aptamers Modified Gold Nanowire Arrays. ACS Appl. Mater. Interfaces 2017, 9, 34706–34714. [Google Scholar] [CrossRef]
- Zhang, Q.W.; Ouyang, J.; Wang, Y.; Zhai, T.T.; Wang, C.; Wu, Z.Q.; Zhang, T.Q.; Wang, K.; Xia, X.H. Specific cell capture and noninvasive release via moderate electrochemical oxidation of boronic ester linkage. Biosens. Bioelectron. 2019, 138, 111316. [Google Scholar] [CrossRef]
- Gurudatt, N.G.; Chung, S.; Kim, J.M.; Kim, M.H.; Jung, D.K.; Han, J.Y.; Shim, Y.B. Separation detection of different circulating tumor cells in the blood using an electrochemical microfluidic channel modified with a lipid-bonded conducting polymer. Biosens. Bioelectron. 2019, 146, 111746. [Google Scholar] [CrossRef]
- Ferreira, C.E.S.; Guerra, J.C.C.; Slhessarenko, N.; Scartezini, M.; Franca, C.N.; Colombini, M.P.; Berlitz, F.; Machado, A.M.O.; Campana, G.A.; Faulhaber, A.C.L.; et al. Point-of-Care Testing: General Aspects. Clin. Lab 2018, 64, 1–9. [Google Scholar] [CrossRef]
- Xu, L.; Wang, A.; Li, X.; Oh, K.W. Passive micropumping in microfluidics for point-of-care testing. Biomicrofluidics 2020, 14, 031503. [Google Scholar] [CrossRef]
- Wang, J.; Li, D.; Qiu, Y.; Liu, X.; Huang, L.; Wen, H.; Hu, J. An europium functionalized carbon dot-based fluorescence test paper for visual and quantitative point-of-care testing of anthrax biomarker. Talanta 2020, 220, 121377. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.Y.; Kong, F.Z.; Zhang, Q.; Liu, W.W.; Liu, X.P.; Li, G.Q.; Zhong, R.; Fan, L.Y.; Xiao, H.; Cao, C.X. iPhone-imaged and cell-powered electrophoresis titration chip for the alkaline phosphatase assay in serum by the moving reaction boundary. Lab Chip 2018, 18, 1758–1766. [Google Scholar] [CrossRef]
- Abate, M.F.; Jia, S.; Ahmed, M.G.; Li, X.; Lin, L.; Chen, X.; Zhu, Z.; Yang, C. Visual Quantitative Detection of Circulating Tumor Cells with Single-Cell Sensitivity Using a Portable Microfluidic Device. Small 2019, 15, e1804890. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, L.; Li, B.; Yang, C.J.; Jin, Y. Point-of-Care Assay of Telomerase Activity at Single-Cell Level via Gas Pressure Readout. Anal. Chem. 2017, 89, 8311–8318. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Z.; Wang, Y.; Wu, C.; Li, Q.; Tang, B. Rapid and Sensitive Detection of Cancer Cells Based on the Photothermal Effect of Graphene Functionalized Magnetic Microbeads. ACS Appl. Mater. Interfaces 2016, 8, 29933–29938. [Google Scholar] [CrossRef]
- Ma, Y.; Mao, G.; Wu, G.; He, Z.; Huang, W. Magnetic bead-enzyme assemble for triple-parameter telomerase detection at single-cell level. Anal. Bioanal. Chem. 2020, 412, 5283–5289. [Google Scholar] [CrossRef]
- Xiao, A.; Huang, Y.; Zheng, J.; Chen, P.; Guan, B.O. An Optical Microfiber Biosensor for CEACAM5 Detection in Serum: Sensitization by a Nanosphere Interface. ACS Appl. Mater. Interfaces 2020, 12, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Ainla, A.; Mousavi, M.P.S.; Tsaloglou, M.N.; Redston, J.; Bell, J.G.; Fernandez-Abedul, M.T.; Whitesides, G.M. Open-Source Potentiostat for Wireless Electrochemical Detection with Smartphones. Anal. Chem. 2018, 90, 6240–6246. [Google Scholar] [CrossRef] [Green Version]
- Guo, J. Smartphone-Powered Electrochemical Dongle for Point-of-Care Monitoring of Blood beta-Ketone. Anal. Chem. 2017, 89, 8609–8613. [Google Scholar] [CrossRef] [Green Version]
- Marrinucci, D.; Bethel, K.; Bruce, R.H.; Curry, D.N.; Hsieh, B.; Humphrey, M.; Krivacic, R.T.; Kroener, J.; Kroener, L.; Ladanyi, A.; et al. Case study of the morphologic variation of circulating tumor cells. Hum. Pathol. 2007, 38, 514–519. [Google Scholar] [CrossRef]
- Keller, L.; Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 2019, 19, 553–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Huang, X.; Gan, C.; Yuan, R.; Xiang, Y. Highly specific and sensitive point-of-care detection of rare circulating tumor cells in whole blood via a dual recognition strategy. Biosens. Bioelectron. 2019, 143. [Google Scholar] [CrossRef]
- Xia, N.; Wu, D.; Yu, H.; Sun, W.; Yi, X.; Liu, L. Magnetic bead-based electrochemical and colorimetric assays of circulating tumor cells with boronic acid derivatives as the recognition elements and signal probes. Talanta 2020, 221. [Google Scholar] [CrossRef]
- Liu, J.; Geng, Z.; Fan, Z.; Liu, J.; Chen, H. Point-of-care testing based on smartphone: The current state-of-the-art (2017–2018). Biosens. Bioelectron. 2019, 132, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Knowlton, S.; Joshi, A.; Syrrist, P.; Coskun, A.F.; Tasoglu, S. 3D-printed smartphone-based point of care tool for fluorescence- and magnetophoresis-based cytometry. Lab Chip 2017, 17, 2839–2851. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Han, J.; Sun, X.; Yang, G. Electrochemical Detection and Point-of-Care Testing for Circulating Tumor Cells: Current Techniques and Future Potentials. Sensors 2020, 20, 6073. https://doi.org/10.3390/s20216073
Lu C, Han J, Sun X, Yang G. Electrochemical Detection and Point-of-Care Testing for Circulating Tumor Cells: Current Techniques and Future Potentials. Sensors. 2020; 20(21):6073. https://doi.org/10.3390/s20216073
Chicago/Turabian StyleLu, Chunyang, Jintao Han, Xiaoyi Sun, and Gen Yang. 2020. "Electrochemical Detection and Point-of-Care Testing for Circulating Tumor Cells: Current Techniques and Future Potentials" Sensors 20, no. 21: 6073. https://doi.org/10.3390/s20216073
APA StyleLu, C., Han, J., Sun, X., & Yang, G. (2020). Electrochemical Detection and Point-of-Care Testing for Circulating Tumor Cells: Current Techniques and Future Potentials. Sensors, 20(21), 6073. https://doi.org/10.3390/s20216073






