Clean Grinding Technique: A Facile Synthesis and In Silico Antiviral Activity of Hydrazones, Pyrazoles, and Pyrazines Bearing Thiazole Moiety against SARS-CoV-2 Main Protease (Mpro)
Abstract
1. Introduction
2. Results and Discussion
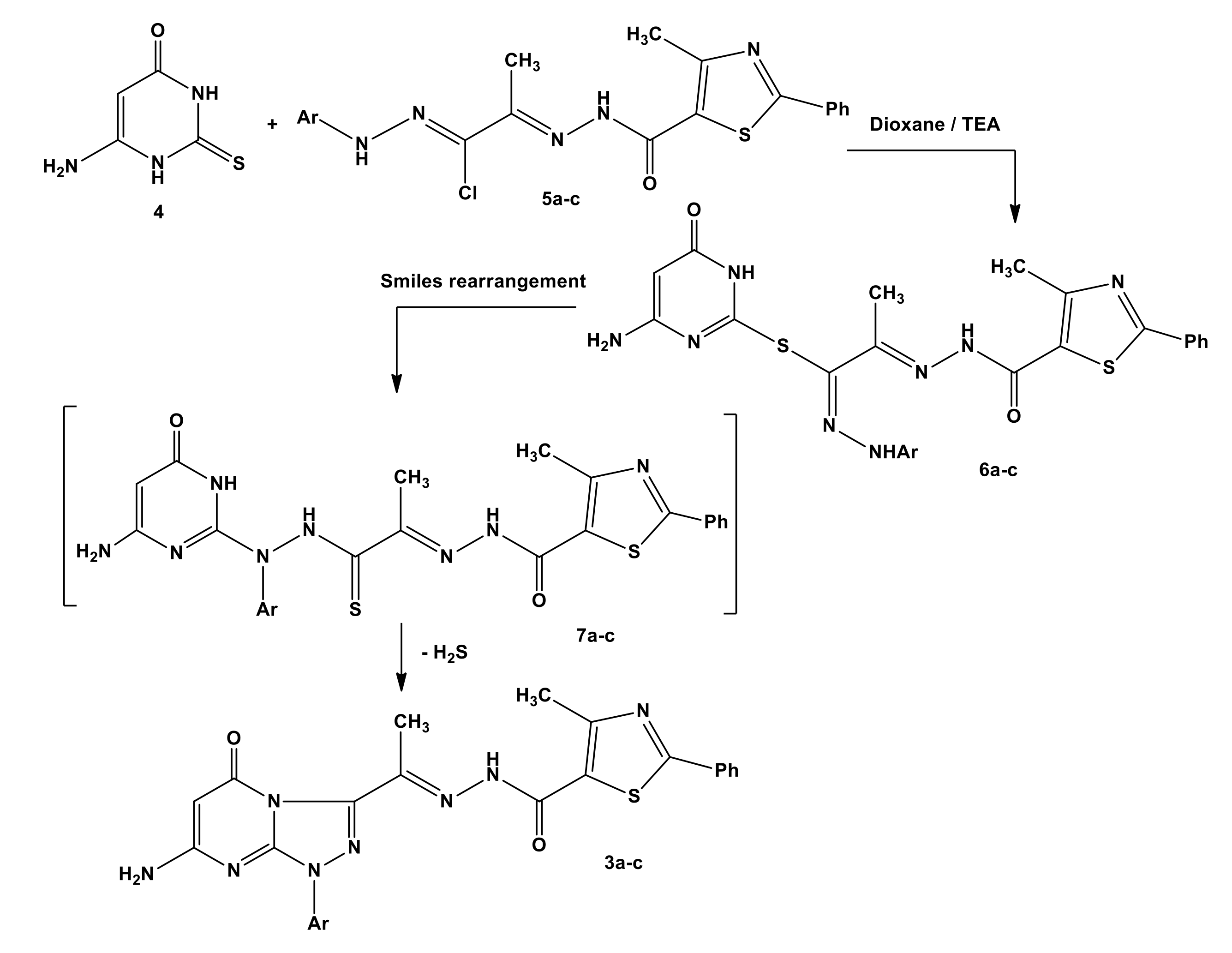
2.1. Chemistry
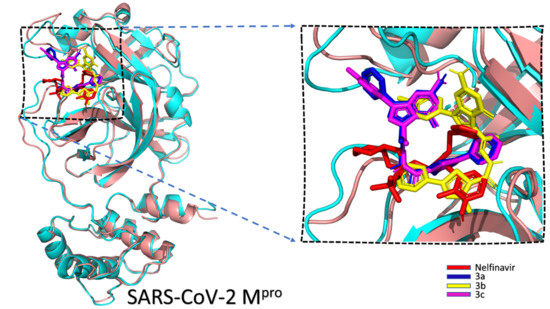
2.2. Molecular Simulations
3. Experimental
3.1. Chemistry
General Procedure for the Synthesis of Compounds 3a–c, 9, 11a–c, 16–19, 22, and 23
3.2. The In Silico Studies
3.2.1. Molecular Dynamics Simulation
3.2.2. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Horvath, I.T.; Anastas, P.T. Innovations and green chemistry. Chem. Rev. 2007, 107, 2169–2173. [Google Scholar]
- Gomha, S.M.; Riyadh, S.M.; Abdalla, M.M. Solvent-drop grinding method: Efficient synthesis, dpph radical scavenging and anti-diabetic activities of chalcones, bis-chalcones, azolines, and bis-azolines. Cur. Org. Synth. 2015, 12, 220–228. [Google Scholar]
- Amirnejad, M.; Naimi-Jamal, M.R.; Tourani, H.; Ghafuri, H. A facile solvent-free one-pot three-component method for the synthesis of 2-amino-4H-pyrans and tetrahydro-4H-chromenes at ambient temperature. Monatsh. Chem. 2013, 144, 1219–1225. [Google Scholar]
- Mashkouri, S.; Naimi-Jamal, M.R. Mechanochemical solvent-free and catalyst-free one-pot synthesis of pyrano[2,3-d]pyrimidine-2,4(1H,3H)-diones with quantitative yields. Molecules 2009, 14, 474–479. [Google Scholar]
- Vibhute, A.; Mokle, S.; Karamunge, K.; Gurav, V.; Vibhute, Y. A simple and efficient method for solvent-free iodination of hydroxylated aromatic aldehydes and ketones using iodine and iodic acid by grinding method. Chin. Chem. Lett. 2010, 21, 914–918. [Google Scholar]
- Bingul, M.; Ercan, S.; Boga, M. The design of novel 4,6-dimethoxyindole based hydrazide-hydrazones: Molecular modeling, synthesis and anticholinesterase activity. J. Mol. Str. 2020, 1213, 128202. [Google Scholar]
- Rohane, S.H.; Chauhan, A.J.; Fuloria, N.K.; Fuloria, S. Synthesis and in vitro antimycobacterial potential of novel hydrazones of eugenol. Arab. J. Chem. 2020, 13, 4495–4504. [Google Scholar]
- Al-Sodies, S.A.; Aouad, M.R.; Ihmaid, S.; Aljuhani, A.; Messali, M.; Ali, I.; Rezki, N. Microwave and conventional synthesis of ester based dicationic pyridinium ionic liquids carrying hydrazone linkage: DNA binding, anticancer and docking studies. J. Mol. Str. 2020, 1207, 127756. [Google Scholar]
- Saidugari, S.; Rao, L.; Vidya, K.; Ram, B.; Balram, B. Synthesis, characterization and antibacterial activity of (E)-4-((3-methyl-4-(methylsulfonyl)pyridin-2-yl)methoxy)-N′-(substituted-benzylidene)benzohydrazide derivatives. Indian J. Chem. 2017, 56, 177–182. [Google Scholar]
- Abdelrazek, F.M.; Gomha, S.M.; Abdel-aziz, H.M.; Farghaly, M.S.; Metz, P.; Abdel-Shafy, A. Efficient Synthesis and In-Silico study of some novel pyrido[2,3-d][1,2,4]triazolo[4,3-a]pyrimidine derivatives. J. Heterocycl. Chem. 2020, 57, 1759–1769. [Google Scholar]
- Gomha, S.M.; Muhammad, Z.A.; Abdel-aziz, H.M.; Matar, I.K.; El-Sayed, A.A. Green synthesis, molecular docking and anticancer activity of novel 1,4-dihydropyridine-3,5-dicarbohydrazones under grind-stone chemistry. Green Chem. Lett. Rev. 2020, 13, 6–17. [Google Scholar]
- Belskaya, N.P.; Dehaen, W.; Bakulev, V.A. Synthesis and properties of hydrazones bearing amide, thioamide and amidine functions. Arch. Org. Chem. 2010, 1, 75–332. [Google Scholar]
- Xavier, A.J.; Thakur, M.; Marie, J.M. Synthesis and spectral characterisation of hydrazone based 14-membered octaaza macrocyclic Ni (II) complexes. J. Chem. Pharm. Res. 2012, 4, 986–990. [Google Scholar]
- Banerjee, S.; Mondal, S.; Chakraborty, W.; Sen, S.; Gachhui, R.; Butcher, R.J.; Slawin, A.M.Z.; Mandal, C.; Mitra, S. Syntheses, X-ray crystal structures, DNA binding, oxidative cleavage and antimicrobial studies of two Cu (II) hydrazone complees. Polyhedron 2009, 28, 2785–2793. [Google Scholar]
- Zheng, L.W.; Wu, L.L.; Zhao, B.X.; Dong, W.L.; Miao, J.Y. Synthesis of novel substituted pyrazole-5-carbohydrazide hydrazone derivatives and discovery of a potent apoptosis inducer in A549 lung cancer cells. Bioorg. Med. Chem. 2009, 17, 1957–1962. [Google Scholar]
- Ozdemir, Z.; Kandilici, H.B.; Gumusel, B.; Calis, U.; Bilgin, A.A. Synthesis and studies on antidepressant and anticonvulsant activities of some 3-(2-furyl)-pyrazoline derivatives. Eur. J. Med. Chem. 2007, 42, 373–379. [Google Scholar]
- Rajendra, P.Y.; Lakshmana, R.A.; Prasoona, L.; Murali, K.; Ravi, K.P. Synthesis and antidepressant activity of some 1,3,5-triphenyl-2-pyrazolines and 3-(2″-hydroxy naphthalen-1″-yl)-1,5-diphenyl-2- pyrazolines. Bioorg. Med. Chem. Lett. 2005, 15, 5030–5034. [Google Scholar]
- Gomha, S.M.; Riyadh, S.M. Synthesis of triazolo[4,3-b][1,2,4,5]tetrazines and triazolo[3,4-b] [1,3,4]thiadiazines using chitosan as ecofriendly catalyst under microwave irradiation. Arkivoc 2009, XI, 58–68. [Google Scholar]
- Gomha, S.M.; Riyadh, S.M.; Mahmmoud, E.A.; Elaasser, M.M. Synthesis and anticancer activity of arylazothiazoles and 1,3,4-thiadiazoles using chitosan-grafted-poly(4-vinylpyridine) as a novel copolymer basic catalyst. Chem. Heterocycl. Compds. 2015, 51, 1030–1038. [Google Scholar]
- Gomha, S.M.; Abdel-aziz, H.M.; Abolibda, T.Z.; Hassan, S.A.; Abdalla, M.M. Green synthesis, molecular docking and pharmacological evaluation of new triazolo-thiadiazepinylcoumarine derivatives as sedative-hypnotic scaffold. J. Heterocycl. Chem. 2020, 57, 1034–1043. [Google Scholar]
- Gomha, S.M.; Badrey, M.G.; Arafa, W.A.A. DABCO-catalyzed green synthesis of thiazole and 1,3-thiazine derivatives linked to benzofuran. Heterocycles 2016, 92, 1450–1461. [Google Scholar]
- Gomha, S.M.; Khalil, K.D.; El-Zanate, A.M.; Riyadh, S.M. A Facile green synthesis and anti-cancer activity of bis-arylhydrazononitriles, triazolo[5,1-c][1,2,4]triazine, and 1,3,4-thiadiazoline. Heterocycles 2013, 87, 1109–1120. [Google Scholar]
- Gomha, S.M.; Eldebss, T.M.A.; Badrey, M.G.; Abdulla, M.M.; Mayhoub, A.S. Novel 4-heteroaryl- antipyrines as DPP-IV Inhibitors. Chem. Biol. Drug Des. 2015, 86, 1292–1303. [Google Scholar]
- World Health Organization Situation Report 72. April 2020. Available online: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200401-sitrep-72-covid-19.pdf?sfvrsn=3dd8971b_2 (accessed on 5 October 2020).
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [PubMed]
- Leach, A. Molecular Modelling: Principles and Applications, 2nd ed.; Prentice Hall: Upper New Jersey River, NJ, USA, 2001. [Google Scholar]
- Elfiky, A.A. Reply to a letter to the editor. Life Sci. 2020, 252, 117715. [Google Scholar]
- Ganesan, A.; Barakat, K. Applications of computer-aided approaches in the development of hepatitis c antiviral agents. Expert Opin. Drug Discov. 2017, 12, 407–425. [Google Scholar] [PubMed]
- Saleh, N.A.; Elhaes, H.; Ibrahim, M. Design and Development of Some Viral Protease Inhibitors by QSAR and Molecular Modeling Studies, in Viral Proteases and Their Inhibitors; Elsevier: Amsterdam, The Netherlands, 2017; pp. 25–58. [Google Scholar]
- Saleh, N.A.; Ezat, A.A.; ElFiky, A.A.; Elshemey, W.M.; Bayoumy, A.M. Theoretical Study on Modified Boceprevir Compounds as NS3 Protease Inhibitors. J. Comput. Theor. Nanosci. 2015, 12, 371–375. [Google Scholar]
- Elfiky, A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. J. Biomol. Struct. Dyn. 2020, 1–9. [Google Scholar] [CrossRef]
- Elfiky, A.A. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [PubMed]
- Elfiky, A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020, 248, 117477. [Google Scholar] [PubMed]
- Elfiky, A.A. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn. 2020, 1–10. [Google Scholar] [CrossRef]
- Mosselhi, M.A.M. A convenient synthesis of novel derivatives of pyrido[2,3-d][1,2,4]triazolo[4,3-a] pyrimidine-5,6-dione. Monatsh. Chem. 2002, 133, 1297–1304. [Google Scholar]
- Araniciu, C.; Oniga, S.D.; Benedec, D.; Crisan, O.; Vlase, L.; Palage, M.; Oniga, O. New 5-thiazolyl- carbohydrazon-n-allyl-thiazolines. Synthesis, characterization and antioxidant activity. Rev. Chim. 2019, 70, 2340–2343. [Google Scholar]
- Gomha, S.M.; Abdelaziz, M.R.; Kheder, N.A.; Abdelaziz, H.M.; Alterary, S.; Mabkhot, Y.N. A facile access and evaluation of some novel thiazole and 1,3,4-thiadiazole derivatives incorporating thiazole moiety as potent anticancer agents. Chem. Cent. J. 2017, 11, 105. [Google Scholar]
- Shawali, A.S.; Abdallah, M.A.; Mosselhi, M.A.N.; Farghaly, T.A. A Facile one-pot regioselective synthesis of [1,2,4]triazolo[4,3-a]5(1H)-pyrimidinones via tandem Japp–Klingemann, Smiles rearrangement, and cyclization reactions. Heteroatom Chem. 2002, 13, 136–140. [Google Scholar]
- Tazoo, D.; Oniga, O.; Bohle, D.S.; Chua, Z.; Dongo, E. General two--step preparation of chalcones containing thiazole. J. Heterocycl. Chem. 2012, 49, 768–773. [Google Scholar]
- Min, Z.-L.; Zhang, Q.; Hong, X.; Cao, X.-L.; Hu, X.-M. A Green protocol for catalyst-free synthesis of pyrazole in glycerol-water solution. Asian J. Chem. 2015, 27, 3205–3207. [Google Scholar]
- Ailawadi, S.; Jyoti, A.S.; Yadav, M.; Pathak, D. Synthesis and characterization of some substituted pyrazoles as analgesics and anti-inflammatory agents. Der Pharm. Chem. 2011, 3, 215–222. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein–ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar]
- van Dijk, A.D.; Bonvin, A.M. Solvated docking: Introducing water into the modelling of biomolecular complexes. Bioinformatics 2006, 22, 2340–2347. [Google Scholar]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar]
- Schrödinger, LLC. The PyMOL Molecular Graphics System; Version 1.7.6; Schrödinger, LLC: New York, NY, USA.
Sample Availability: Samples of the compounds are not available from the authors. |





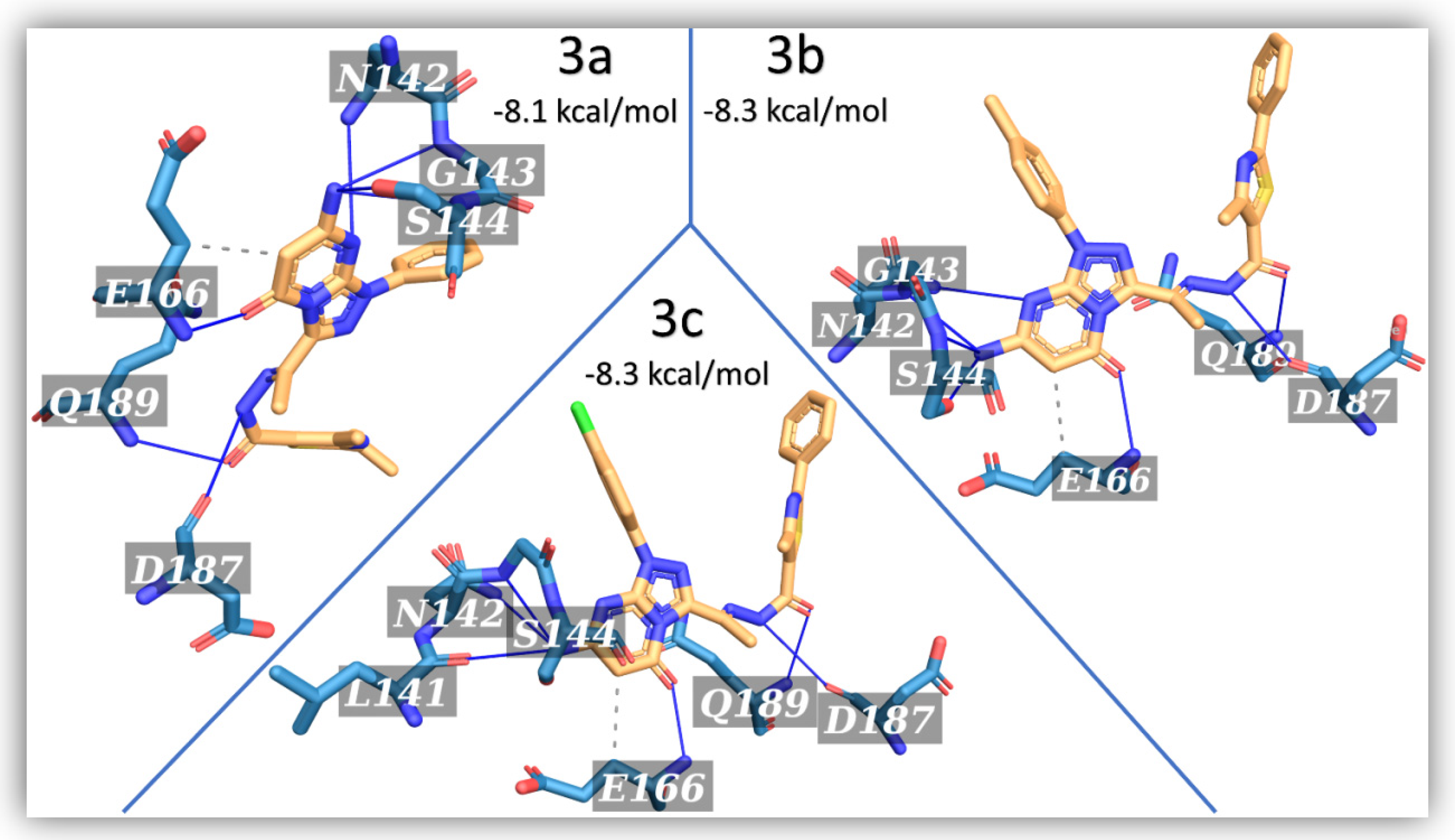

| Compound | AutoDock Vina Score | H-Bonding | Hydrophobic Interaction | ||
|---|---|---|---|---|---|
| Number | Residues from SARS-CoV-2 Mpro Take Part in the Interaction | Number | Residues from SARS-CoV-2 Mpro Take Part in the Interaction | ||
| Nelfinavir | −6.7 | 1 | E166 | 3 | T25, L27, and M165 |
| 3a | −8.1 | 7 | N142, G143, S144(2), E166, D187, Q189 | 1 | E166 |
| 3b | −8.3 | 7 | N142, G143, S144(2), E166, D187, Q189 | 1 | E166 |
| 3c | −8.3 | 7 | L141, N142, G143, S144, E166, D187, Q189 | 1 | E166 |
| 9 | −7.3 | 1 | D187 | 3 | T25, L27, Q189 |
| 11a | −6.6 | 1 | E166 | 4 | L27, N142, M165, E166 |
| 11b | −6.7 | 1 | E166 | 3 | T25, L27, E166 |
| 11c | −6.4 | 1 | G143 | 2 | L27, E166 |
| 13 | −6.4 | 1 | Q189 | 0 | N/A |
| 15 | −6.9 | 3 | G143, S144, C145 | 2 | E166, Q189 |
| 17 | −7.0 | 0 | N/A | 2 | T25, Q189 |
| 19 | −7.0 | 6 | N142, G143, S144, C145, E166, Q189 | 1 | Q189 |
| 21 | −7.0 | 3 | G143, S144, E166 | 4 | T25, L27(2), E166 |
| 23 | −7.3 | 3 | G143, S144, C145 | 1 | E166 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abu-Melha, S.; Edrees, M.M.; Riyadh, S.M.; Abdelaziz, M.R.; Elfiky, A.A.; Gomha, S.M. Clean Grinding Technique: A Facile Synthesis and In Silico Antiviral Activity of Hydrazones, Pyrazoles, and Pyrazines Bearing Thiazole Moiety against SARS-CoV-2 Main Protease (Mpro). Molecules 2020, 25, 4565. https://doi.org/10.3390/molecules25194565
Abu-Melha S, Edrees MM, Riyadh SM, Abdelaziz MR, Elfiky AA, Gomha SM. Clean Grinding Technique: A Facile Synthesis and In Silico Antiviral Activity of Hydrazones, Pyrazoles, and Pyrazines Bearing Thiazole Moiety against SARS-CoV-2 Main Protease (Mpro). Molecules. 2020; 25(19):4565. https://doi.org/10.3390/molecules25194565
Chicago/Turabian StyleAbu-Melha, Sraa, Mastoura M. Edrees, Sayed M. Riyadh, Mohamad R. Abdelaziz, Abdo A. Elfiky, and Sobhi M. Gomha. 2020. "Clean Grinding Technique: A Facile Synthesis and In Silico Antiviral Activity of Hydrazones, Pyrazoles, and Pyrazines Bearing Thiazole Moiety against SARS-CoV-2 Main Protease (Mpro)" Molecules 25, no. 19: 4565. https://doi.org/10.3390/molecules25194565
APA StyleAbu-Melha, S., Edrees, M. M., Riyadh, S. M., Abdelaziz, M. R., Elfiky, A. A., & Gomha, S. M. (2020). Clean Grinding Technique: A Facile Synthesis and In Silico Antiviral Activity of Hydrazones, Pyrazoles, and Pyrazines Bearing Thiazole Moiety against SARS-CoV-2 Main Protease (Mpro). Molecules, 25(19), 4565. https://doi.org/10.3390/molecules25194565