Characterization of Local and Systemic Impact of Whitefly (Bemisia tabaci) Feeding and Whitefly-Transmitted Tomato Mottle Virus Infection on Tomato Leaves by Comprehensive Proteomics
Abstract
1. Introduction
2. Results
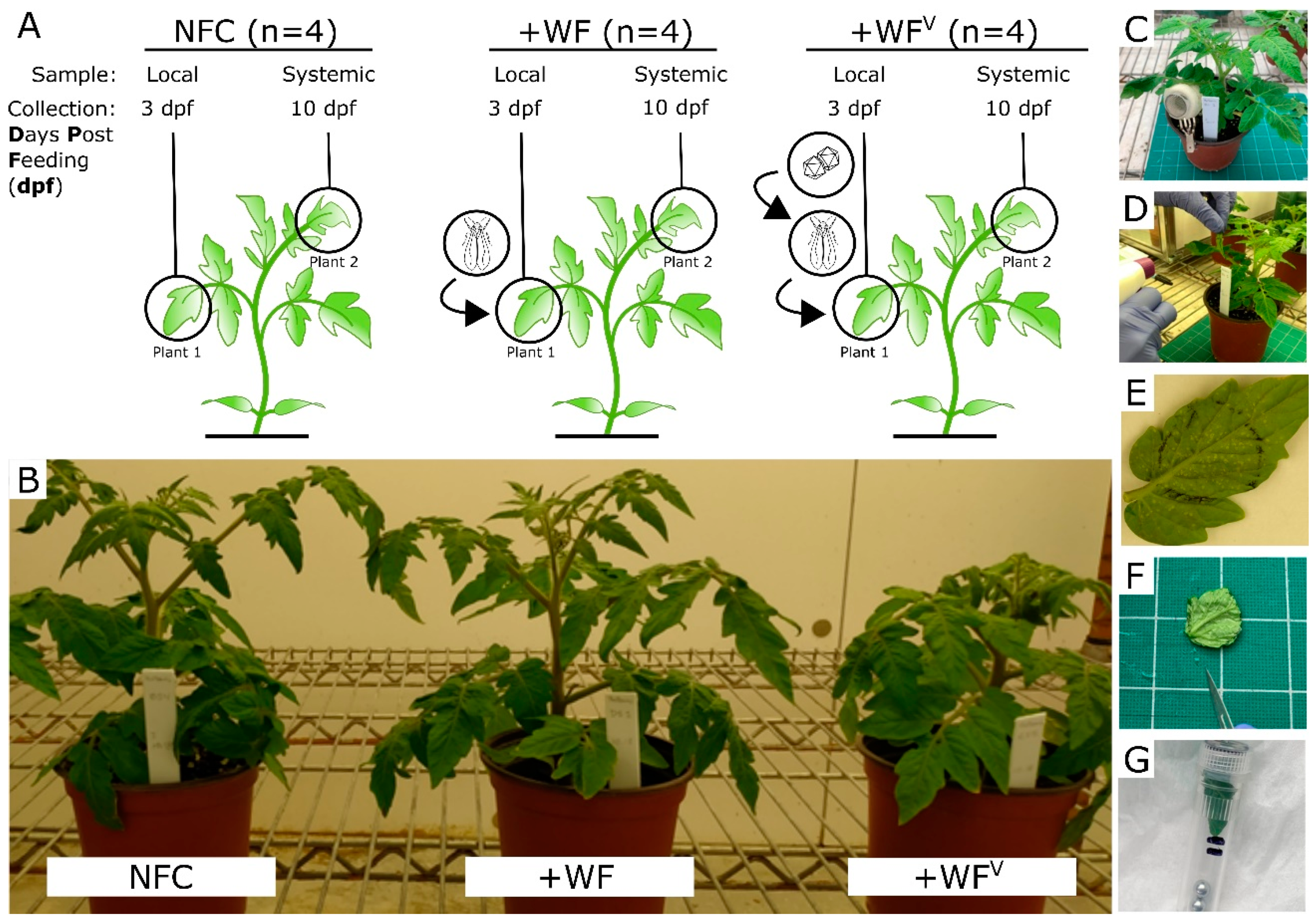
2.1. Obvious Plant Growth Defects Were Observed Only 10 Days Post Whitefly Feeding (dpf) in Tomato Subjected to Feeding by Viruliferous Whiteflies (+WFV)
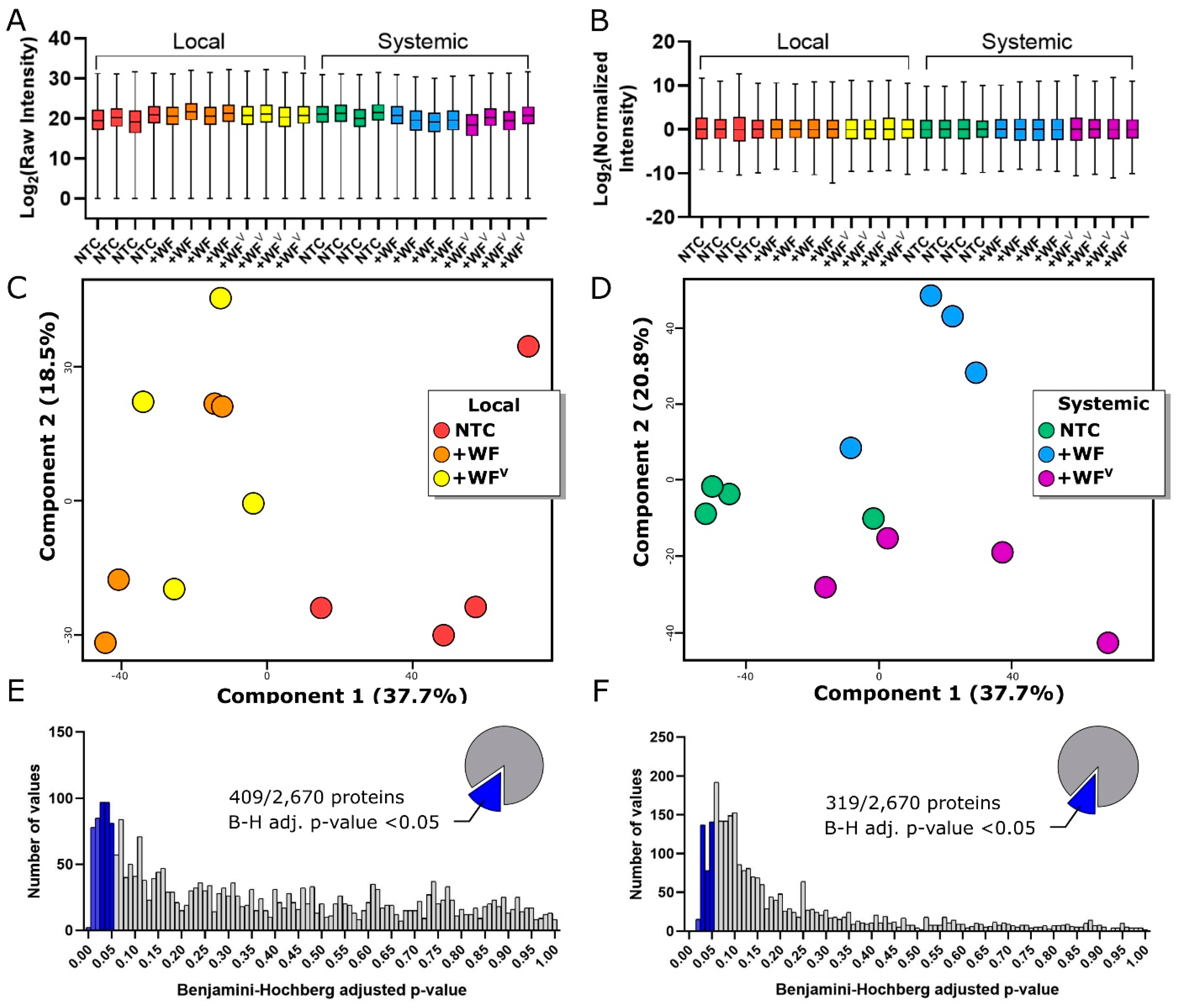
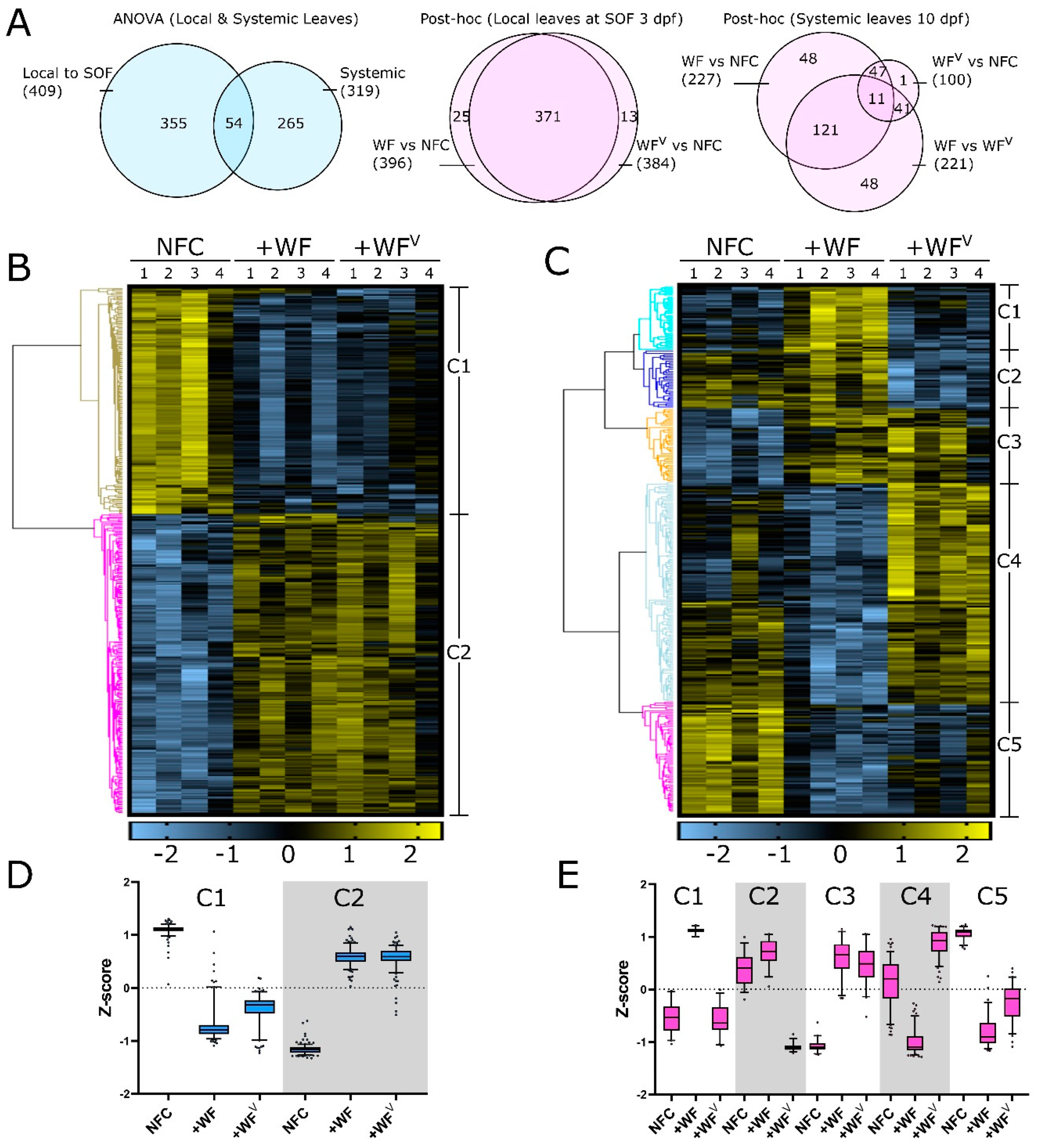
2.2. The Protein Profile of Tomato Leaves is Altered Predominantly by Whitefly Feeding at 3 dpf and Systemically by Both Whitefly and ToMoV at 10 dpf
2.3. Specific Biological Processes Are Altered by Whitefly Feeding and ToMoV Infection Both at the SOF 3 dpf and Systemically in Leaves 10 dpf
2.4. Proteins Involved in Alternative Splicing, Translation, and Plasmodesmata Dynamics Are Affected 3 dpf by +WF and +WFV in Leaves at the SOF
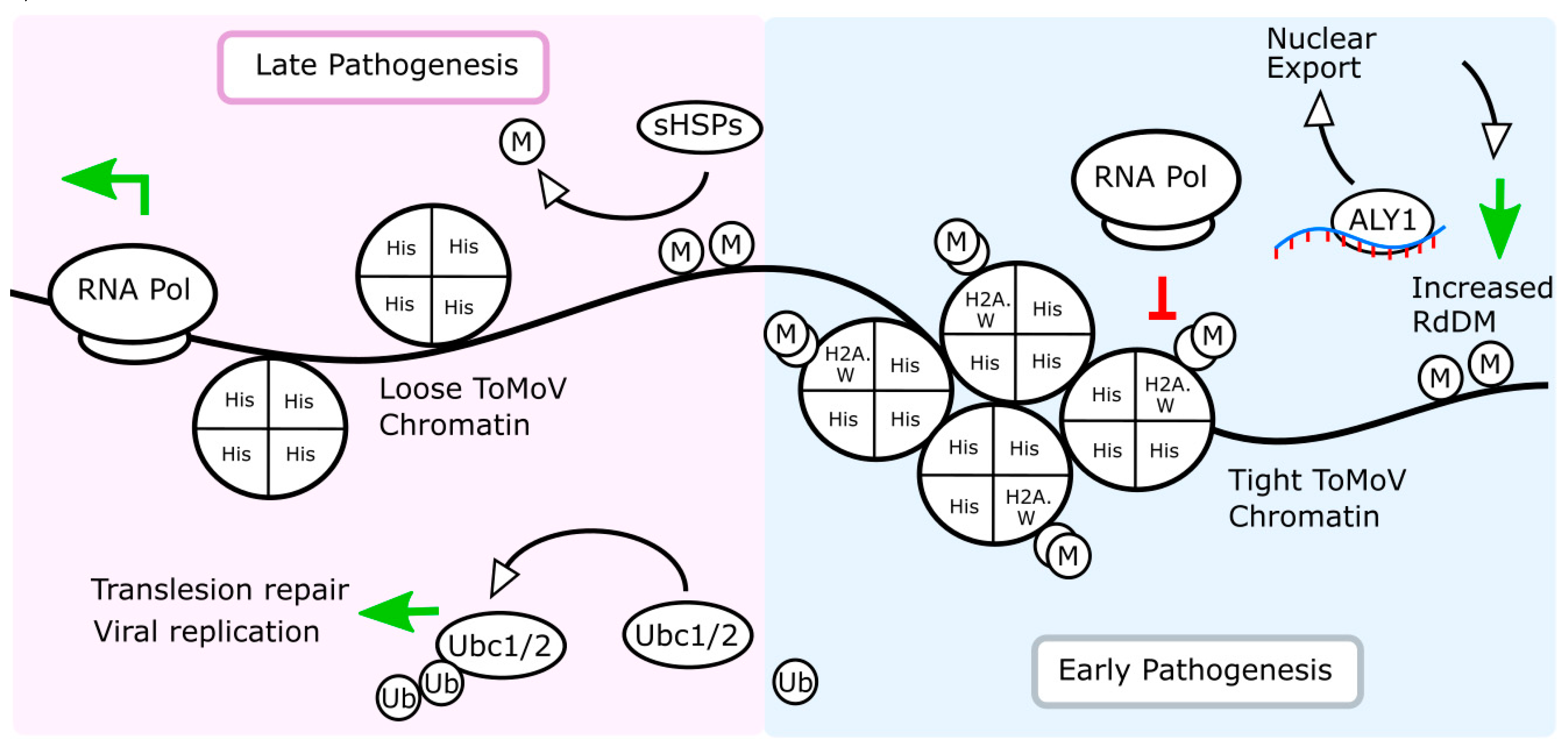
2.5. Tomato Chromatin Architecture and Alternative Splicing Are Affected by ToMoV in Systemic Leaves 10 dpf
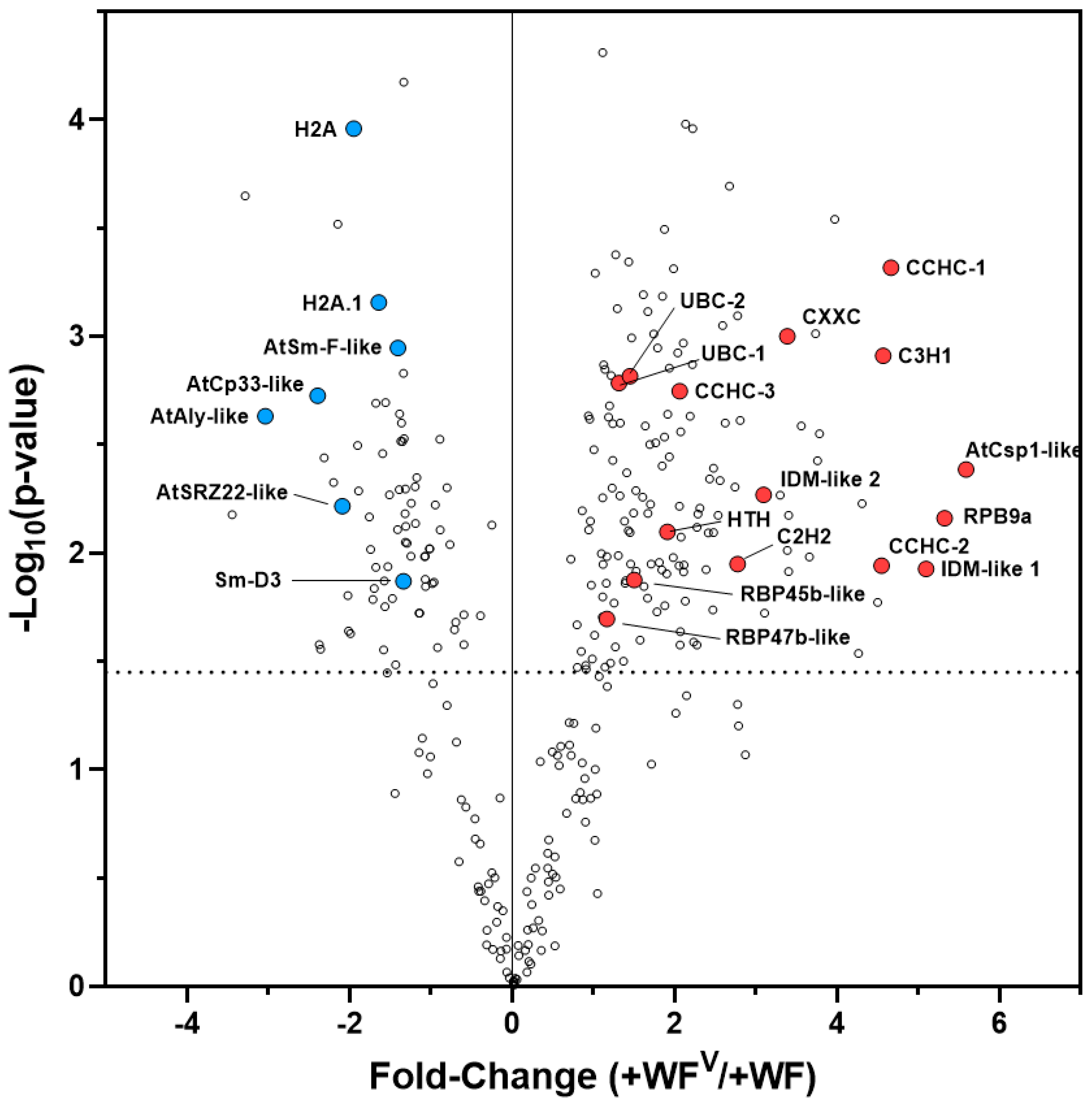
2.6. RNA-Directed DNA Methylation and Other RNA Maturation Processes Are Altered in Systemic Leaves during ToMoV Pathogenesis
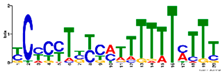
2.7. Similar DNA Motifs Are Found Upstream of Genes Regulated by ToMoV in Systemic Leaves at 10 dpf
3. Discussion
4. Materials and Methods
4.1. Plant Growth, Whitefly Colony Establishment, Feeding, and Sample Collection
4.2. Proteomics Sample Preparation
4.3. Mass Spectrometry, Spectral Search, and Data Analysis
4.4. Promoter Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Dpf | Days Post Feeding |
| GO | Gene Ontology |
| HCA | Hierarchical Cluster Analysis |
| NFC | No Feeding Control |
| PTGS | Post-Translational Gene Silencing |
| RdDM | RNA-Dependent DNA Methylation |
| SG | Stress Granule |
| SOF | Site of Feeding |
| TGS | Transcriptional Gene Silencing |
| ToMoV | Tomato Mottle Virus |
| WF | Whitefly |
References
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting Chinks in the Plant’s Armor: Evolution and Emergence of Geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef] [PubMed]
- Fauquet, C.M.; Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X. Geminivirus strain demarcation and nomenclature. Arch. Virol. 2008, 153, 783–821. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.V.; Sahu, P.P.; Prasad, M.; Praveen, S.; Pappu, H.R. Geminiviruses and Plant Hosts: A Closer Examination of the Molecular Arms Race. Viruses 2017, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A. Ictv Report Consortium. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef]
- Mubin, M.; Ijaz, S.; Nahid, N.; Hassan, M.; Younus, A.; Qazi, J.; Nawaz-Ul-Rehman, M.S. Journey of begomovirus betasatellite molecules: From satellites to indispensable partners. Virus Genes 2020, 56, 16–26. [Google Scholar] [CrossRef]
- Briddon, R.W.; Pinner, M.S.; Stanley, J.; Markham, P.G. Geminivirus coat protein gene replacement alters insect specificity. Virology 1990, 177, 85–94. [Google Scholar] [CrossRef]
- Varma, A.; Malathi, V.G. Emerging geminivirus problems: A serious threat to crop production. Ann. Appl. Biol. 2003, 142, 145–164. [Google Scholar] [CrossRef]
- Yang, X.; Baliji, S.; Buchmann, R.C.; Wang, H.; Lindbo, J.A.; Sunter, G.; Bisaro, D.M. Functional Modulation of the Geminivirus AL2 Transcription Factor and Silencing Suppressor by Self-Interaction. J. Virol. 2007, 81, 11972–11981. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 105–140. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, C. Geminiviruses and the plant cell cycle. Plant. Mol. Biol. 2000, 43, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, X.; Wang, Y.; Xie, Y.; Zhou, X. Tomato Yellow Leaf Curl Virus V2 Interacts with Host Histone Deacetylase 6 To Suppress Methylation-Mediated Transcriptional Gene Silencing in Plants. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.; Yu, F.; Gu, L.; Min, X.J. Expanding Alternative Splicing Identification by Integrating Multiple Sources of Transcription Data in Tomato. Front. Plant Sci. 2019, 10, 689. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, J.; Bang, B.; Kim, I.; Lee, H.-H.; Park, J.; Seo, Y.-S. Comparative Analyses of Tomato yellow leaf curl virus C4 Protein-Interacting Host Proteins in Healthy and Infected Tomato Tissues. Plant Pathol. J. 2016, 32, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Jackel, J.N.; Storer, J.M.; Coursey, T.; Bisaro, D.M. Arabidopsis RNA Polymerases IV and V Are Required To Establish H3K9 Methylation, but Not Cytosine Methylation, on Geminivirus Chromatin. J. Virol. 2016, 90, 7529–7540. [Google Scholar] [CrossRef]
- Zhong, X.; Wang, Z.Q.; Xiao, R.; Wang, Y.; Xie, Y.; Zhou, X. iTRAQ analysis of the tobacco leaf proteome reveals that RNA-directed DNA methylation (RdDM) has important roles in defense against geminivirus-betasatellite infection. J. Proteom. 2017, 152, 88–101. [Google Scholar] [CrossRef]
- Vinutha, T.; Kumar, G.; Garg, V.; Canto, T.; Palukaitis, P.; Ramesh, S.V.; Praveen, S. Tomato geminivirus encoded RNAi suppressor protein, AC4 interacts with host AGO4 and precludes viral DNA methylation. Gene 2018, 678, 184–195. [Google Scholar] [CrossRef]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.-J.; Chu, T.-M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global Analysis of Arabidopsis Gene Expression Uncovers a Complex Array of Changes Impacting Pathogen Response and Cell Cycle during Geminivirus Infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef]
- Góngora-Castillo, E.; Ibarra-Laclette, E.; Trejo-Saavedra, D.L.; Rivera-Bustamante, R.F. Transcriptome analysis of symptomatic and recovered leaves of geminivirus-infected pepper (Capsicum annuum). Virol. J. 2012, 9, 295. [Google Scholar] [CrossRef]
- Pierce, E.J.; Rey, M.E.C. Assessing Global Transcriptome Changes in Response to South African Cassava Mosaic Virus [ZA-99] Infection in Susceptible Arabidopsis thaliana. PLoS ONE 2013, 8, e67534. [Google Scholar] [CrossRef]
- Miozzi, L.; Napoli, C.; Sardo, L.; Accotto, G.P. Transcriptomics of the Interaction between the Monopartite Phloem-Limited Geminivirus Tomato Yellow Leaf Curl Sardinia Virus and Solanum lycopersicum Highlights a Role for Plant Hormones, Autophagy and Plant Immune System Fine Tuning during Infection. PLoS ONE 2014, 9, e89951. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chi, Y.; Zhang, X.-J.; Lei, T.; Wang, X.-W.; Liu, S.-S. Comparative proteomic analysis provides new insight into differential transmission of two begomoviruses by a whitefly. Virol. J. 2019, 16, 32. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Saurabh, S.; Maurya, R.; Mudawal, A.; Parmar, D.; Singh, P.K. Proteome analysis of Bemisia tabaci suggests specific targets for RNAi mediated control. J. Proteom. 2016, 132, 93–102. [Google Scholar] [CrossRef]
- Su, Q.; Peng, Z.; Tong, H.; Xie, W.; Wang, S.; Wu, Q.; Zhang, J.; Li, C.; Zhang, Y. A salivary ferritin in the whitefly suppresses plant defenses and facilitates host exploitation. J. Exp. Bot. 2019, 70, 3343–3355. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yan, J.; Wu, Y.; Zhang, H.; Mo, S.; Xu, X.; Zhou, F.; Ding, H. Proteomic analysis by iTRAQ-PRM provides integrated insight into mechanisms of resistance in pepper to Bemisia tabaci (Gennadius). BMC Plant Biol. 2019, 19, 270. [Google Scholar] [CrossRef]
- Abouzid, A.M.; Polston, J.E.; Hiebert, E. The nucleotide sequence of tomato mottle virus, a new geminivirus isolated from tomatoes in Florida. J. Gen. Virol. 1992, 73, 3225–3229. [Google Scholar] [CrossRef]
- Polston, J.E.; Hiebert, E.; McGovern, R.J.; Stansly, P.A.; Schuster, D.J. Host range of tomato mottle virus, a new geminivirus infecting tomato in Florida. Plant Disease 1993, 77, 1181–1184. [Google Scholar] [CrossRef]
- Rajabu, C.A.; Kennedy, G.G.; Ndunguru, J.; Ateka, E.M.; Tairo, F.; Hanley-Bowdoin, L.; Ascencio-Ibáñez, J.T. Lanai: A small, fast growing tomato variety is an excellent model system for studying geminiviruses. J. Virol. Methods 2018, 256, 89–99. [Google Scholar] [CrossRef]
- McKenzie, C.L. Effect of tomato mottle virus (ToMoV) on Bemisia tabaci biotype B (Homoptera: Aleyrodidae) oviposition and adult survivorship on healthy tomato. Fla. Entomol. 2002, 85, 367–368. [Google Scholar] [CrossRef]
- Hao, L.; Wang, H.; Sunter, G.; Bisaro, D.M. Geminivirus AL2 and L2 Proteins Interact with and Inactivate SNF1 Kinase. Plant Cell 2003, 15, 1034–1048. [Google Scholar] [CrossRef]
- Raja, P.; Sanville, B.C.; Buchmann, R.C.; Bisaro, D.M. Viral Genome Methylation as an Epigenetic Defense against Geminiviruses. J. Virol. 2008, 82, 8997–9007. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, R.C.; Asad, S.; Wolf, J.N.; Mohannath, G.; Bisaro, D.M. Geminivirus AL2 and L2 Proteins Suppress Transcriptional Gene Silencing and Cause Genome-Wide Reductions in Cytosine Methylation. J. Virol. 2009, 83, 5005–5013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, H.; Huang, X.; Xia, R.; Zhao, Q.; Lai, J.; Teng, K.; Li, Y.; Liang, L.; Du, Q.; et al. BSCTV C2 Attenuates the Degradation of SAMDC1 to Suppress DNA Methylation-Mediated Gene Silencing in Arabidopsis. Plant Cell 2011, 23, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Bruns, A.N.; Li, S.; Mohannath, G.; Bisaro, D.M. Phosphorylation of Arabidopsis eIF4E and eIFiso4E by SnRK1 inhibits translation. FEBS J. 2019, 286, 3778–3796. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2018, 47, D419–D426. [Google Scholar] [CrossRef]
- Sanfaçon, H. Plant Translation Factors and Virus Resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef] [PubMed]
- Peal, L.; Jambunathan, N.; Mahalingam, R. Phylogenetic and expression analysis of RNA-binding proteins with triple RNA recognition motifs in plants. Mol. Cells 2011, 31, 55–64. [Google Scholar] [CrossRef]
- Muthuramalingam, M.; Wang, Y.; Li, Y.; Mahalingam, R. Interacting protein partners of Arabidopsis RNA-binding protein AtRBP45b. Plant Biol. 2017, 19, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, A.; Lorković, Z.J.; Kobayashi, W.; Tachiwana, H.; Yelagandula, R.; Kurumizaka, H.; Berger, F. Histone H2A variants confer specific properties to nucleosomes and impact on chromatin accessibility. Nucleic Acids Res. 2018, 46, 7675–7685. [Google Scholar] [CrossRef] [PubMed]
- Choudury, S.G.; Shahid, S.; Cuerda-Gil, D.; Panda, K.; Cullen, A.; Ashraf, Q.; Sigman, M.J.; McCue, A.D.; Slotkin, R.K. The RNA Export Factor ALY1 Enables Genome-Wide RNA-Directed DNA Methylation. Plant Cell 2019, 31, 759–774. [Google Scholar] [CrossRef]
- Stankovic, N.; Schloesser, M.; Joris, M.; Sauvage, E.; Hanikenne, M.; Motte, P. Dynamic Distribution and Interaction of the Arabidopsis SRSF1 Subfamily Splicing Factors. Plant Physiol. 2016, 170, 1000–1013. [Google Scholar] [CrossRef] [PubMed]
- Swaraz, A.M.; Park, Y.-D.; Hur, Y. Knock-out mutations of Arabidopsis SmD3-b induce pleotropic phenotypes through altered transcript splicing. Plant Sci. 2011, 180, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Kanno, T.; Lin, W.-D.; Fu, J.L.; Matzke, A.J.M.; Matzke, M. A genetic screen implicates a CWC16/Yju2/CCDC130 protein and SMU1 in alternative splicing in Arabidopsis thaliana. RNA 2017, 23, 1068–1079. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Ohta, M.; Sugiura, M.; Sugita, M. Chloroplast Ribonucleoproteins Function as a Stabilizing Factor of Ribosome-free mRNAs in the Stroma. J. Biol. Chem. 2001, 276, 147–152. [Google Scholar] [CrossRef]
- Teubner, M.; Fuß, J.; Kühn, K.; Krause, K.; Schmitz-Linneweber, C. The RNA recognition motif protein CP33A is a global ligand of chloroplast mRNAs and is essential for plastid biogenesis and plant development. Plant J. 2017, 89, 472–485. [Google Scholar] [CrossRef]
- Tan, E.H.; Blevins, T.; Ream, T.S.; Pikaard, C.S. Functional Consequences of Subunit Diversity in RNA Polymerases II and V. Cell Rep. 2012, 1, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Torres-Acosta, J.A.; Pastushok, L.; Lai, X.; Pelzer, L.; Wang, H.; Xiao, W. Arabidopsis UEV1D Promotes Lysine-63–Linked Polyubiquitination and Is Involved in DNA Damage Response. Plant Cell 2008, 20, 213–227. [Google Scholar] [CrossRef]
- Zhao, Y.; Xie, S.; Li, X.; Wang, C.; Chen, Z.; Lai, J.; Gong, Z. REPRESSOR OF SILENCING5 Encodes a Member of the Small Heat Shock Protein Family and Is Required for DNA Demethylation in Arabidopsis. Plant Cell 2014, 26, 2660–2675. [Google Scholar] [CrossRef]
- Qian, W.; Miki, D.; Lei, M.; Zhu, X.; Zhang, H.; Liu, Y.; Li, Y.; Lang, Z.; Wang, J.; Tang, K.; et al. Regulation of Active DNA Demethylation by an α-Crystallin Domain Protein in Arabidopsis. Mol. Cell 2014, 55, 361–371. [Google Scholar] [CrossRef]
- Růžička, K.; Zhang, M.; Campilho, A.; Bodi, Z.; Kashif, M.; Saleh, M.; Eeckhout, D.; El-Showk, S.; Li, H.; Zhong, S.; et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017, 215, 157–172. [Google Scholar] [CrossRef]
- Weber, C.; Nover, L.; Fauth, M. Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J. 2008, 56, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Kosmacz, M.; Luzarowski, M.; Kerber, O.; Leniak, E.; Gutiérrez-Beltrán, E.; Moreno, J.C.; Gorka, M.; Szlachetko, J.; Veyel, D.; Graf, A.; et al. Interaction of 2′,3′-cAMP with Rbp47b Plays a Role in Stress Granule Formation. Plant Physiol. 2018, 177, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Lorković, Z.J.; Kirk, D.A.W.; Klahre, U.; Hemmings-Mieszczak, M.; Filipowicz, W. RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 2000, 6, 1610–1624. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Protter, D.S.W.; Parker, R. Principles and Properties of Stress Granules. Trends Cell Biol. 2016, 26, 668–679. [Google Scholar] [CrossRef]
- Kosmacz, M.; Gorka, M.; Schmidt, S.; Luzarowski, M.; Moreno, J.C.; Szlachetko, J.; Leniak, E.; Sokolowska, E.M.; Sofroni, K.; Schnittger, A.; et al. Protein and metabolite composition of Arabidopsis stress granules. New Phytol. 2019, 222, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Carmo, L.S.T.; Resende, R.O.; Silva, L.P.; Ribeiro, S.G.; Mehta, A. Identification of host proteins modulated by the virulence factor AC2 of Tomato chlorotic mottle virus in Nicotiana benthamiana. Proteomics 2013, 13, 1947–1960. [Google Scholar] [CrossRef]
- Zhang, P.-J.; He, Y.-C.; Zhao, C.; Ye, Z.-H.; Yu, X.-P. Jasmonic Acid-Dependent Defenses Play a Key Role in Defending Tomato Against Bemisia tabaci Nymphs, but Not Adults. Front. Plant Sci. 2018, 9, 1065. [Google Scholar]
- Simpson, C.; Thomas, C.; Findlay, K.; Bayer, E.; Maule, A.J. An Arabidopsis GPI-Anchor Plasmodesmal Neck Protein with Callose Binding Activity and Potential to Regulate Cell-to-Cell Trafficking. Plant Cell 2009, 21, 581–594. [Google Scholar] [CrossRef]
- Ernst, A.M.; Jekat, S.B.; Zielonka, S.; Müller, B.; Neumann, U.; Rüping, B.; Twyman, R.M.; Krzyzanek, V.; Prüfer, D.; Noll, G.A. Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proc. Natl. Acad. Sci. USA 2012, 109, 1980–1989. [Google Scholar] [CrossRef]
- Froelich, D.R.; Mullendore, D.L.; Jensen, K.H.; Ross-Elliott, T.J.; Anstead, J.A.; Thompson, G.A.; Pélissier, H.C.; Knoblauch, M. Phloem Ultrastructure and Pressure Flow: Sieve-Element-Occlusion-Related Agglomerations Do Not Affect Translocation. Plant Cell 2011, 23, 4428–4445. [Google Scholar] [CrossRef]
- Noueiry, A.O.; Lucas, W.J.; Gilbertson, R.L. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 1994, 76, 925–932. [Google Scholar] [CrossRef]
- Lazarowitz, S.G.; Beachy, R.N. Viral Movement Proteins as Probes for Intracellular and Intercellular Trafficking in Plants. Plant Cell 1999, 11, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Aitken, C.E.; Lorsch, J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012, 19, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. The Scanning Mechanism of Eukaryotic Translation Initiation. Annu. Rev. Biochem. 2014, 83, 779–812. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef]
- Hébrard, E.; Poulicard, N.; Gérard, C.; Traoré, O.; Wu, H.-C.; Albar, L.; Fargette, D.; Bessin, Y.; Vignols, F. Direct Interaction Between the Rice yellow mottle virus (RYMV) VPg and the Central Domain of the Rice eIF(iso)4G1 Factor Correlates with Rice Susceptibility and RYMV Virulence. Mol. Plant-Microbe Interact. 2010, 23, 1506–1513. [Google Scholar]
- Hinnebusch, A.G. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006, 31, 553–562. [Google Scholar] [CrossRef]
- Thiébeauld, O.; Schepetilnikov, M.; Park, H.-S.; Geldreich, A.; Kobayashi, K.; Keller, M.; Hohn, T.; Ryabova, L.A. A new plant protein interacts with eIF3 and 60S to enhance virus-activated translation re-initiation. EMBO J. 2009, 28, 3171–3184. [Google Scholar] [CrossRef]
- Truniger, V.; Aranda, M.A. Chapter 4-Recessive Resistance to Plant Viruses. In Advances in Virus Research; Natural and Engineered Resistance to Plant Viruses, Part I.; Loebenstein, G., Carr, J.P., Eds.; Academic Press: Cambridge, MA, USA, 2009; Volume 75, pp. 119–231. [Google Scholar]
- Wang, A.; Krishnaswamy, S. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 2012, 13, 795–803. [Google Scholar] [CrossRef]
- Julio, E.; Cotucheau, J.; Decorps, C.; Volpatti, R.; Sentenac, C.; Candresse, T.; Dorlhac de Borne, F. A Eukaryotic Translation Initiation Factor 4E (eIF4E) is Responsible for the “va” Tobacco Recessive Resistance to Potyviruses. Plant Mol. Biol. Rep. 2015, 33, 609–623. [Google Scholar] [CrossRef]
- Albar, L.; Bangratz-Reyser, M.; Hébrard, E.; Ndjiondjop, M.-N.; Jones, M.; Ghesquière, A. Mutations in the eIF(iso)4G translation initiation factor confer high resistance of rice to Rice yellow mottle virus. Plant J. 2006, 47, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sridhar, V.V.; Zhu, J.; Kapoor, A.; Zhu, J.-K. Distinctive Core Histone Post-Translational Modification Patterns in Arabidopsis thaliana. PLoS ONE 2007, 2, e1210. [Google Scholar] [CrossRef] [PubMed]
- Yelagandula, R.; Stroud, H.; Holec, S.; Zhou, K.; Feng, S.; Zhong, X.; Muthurajan, U.M.; Nie, X.; Kawashima, T.; Groth, M.; et al. The Histone Variant H2A.W Defines Heterochromatin and Promotes Chromatin Condensation in Arabidopsis. Cell 2014, 158, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Piquerez, S.J.M.; Bendahmane, A.; Hirt, H.; Raynaud, C.; Benhamed, M. Modify the Histone to Win the Battle: Chromatin Dynamics in Plant–Pathogen Interactions. Front Plant Sci. 2018, 9, 355. [Google Scholar] [CrossRef]
- Gallego-Bartolomé, J. DNA methylation in plants: Mechanisms and tools for targeted manipulation. New Phytol. 2020, 227, 38–44. [Google Scholar] [CrossRef]
- Heath, C.G.; Viphakone, N.; Wilson, S.A. The role of TREX in gene expression and disease. Biochem. J. 2016, 473, 2911–2935. [Google Scholar] [CrossRef]
- Pfaff, C.; Ehrnsberger, H.F.; Flores-Tornero, M.; Sørensen, B.B.; Schubert, T.; Längst, G.; Griesenbeck, J.; Sprunck, S.; Grasser, M.; Grasser, K.D. ALY RNA-Binding Proteins Are Required for Nucleocytosolic mRNA Transport and Modulate Plant Growth and Development. Plant Physiol. 2018, 177, 226–240. [Google Scholar] [CrossRef]
- Komor, E.; Liegl, I.; Schobert, C. Loading and translocation of various cytokinins in phloem and xylem of the seedlings of Ricinus communis L. Planta 1993, 191, 252–255. [Google Scholar] [CrossRef]
- Canto, T.; Uhrig, J.F.; Swanson, M.; Wright, K.M.; MacFarlane, S.A. Translocation of Tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. J. Virol. 2006, 80, 9064–9072. [Google Scholar] [CrossRef]
- Haag, J.R.; Pikaard, C.S. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat. Rev. Mol. Cell Biol. 2011, 12, 483–492. [Google Scholar] [CrossRef]
- Ream, T.S.; Haag, J.R.; Wierzbicki, A.T.; Nicora, C.D.; Norbeck, A.D.; Zhu, J.-K.; Hagen, G.; Guilfoyle, T.J.; Pasa-Tolić, L.; Pikaard, C.S. Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol. Cell 2009, 33, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Woychik, N.A.; Lane, W.S.; Young, R.A. Yeast RNA polymerase II subunit RPB9 is essential for growth at temperature extremes. J. Biol. Chem. 1991, 266, 19053–19055. [Google Scholar] [PubMed]
- Paul, A.; Rao, S.; Mathur, S. The α-Crystallin Domain Containing Genes: Identification, Phylogeny and Expression Profiling in Abiotic Stress, Phytohormone Response and Development in Tomato (Solanum lycopersicum). Front. Plant Sci. 2016, 7, 426. [Google Scholar] [CrossRef]
- Whitham, S.A.; Anderberg, R.J.; Chisholm, S.T.; Carrington, J.C. Arabidopsis RTM2 Gene Is Necessary for Specific Restriction of Tobacco Etch Virus and Encodes an Unusual Small Heat Shock–like Protein. Plant Cell 2000, 12, 569–582. [Google Scholar] [PubMed]
- Castillo-González, C.; Liu, X.; Huang, C.; Zhao, C.; Ma, Z.; Hu, T.; Sun, F.; Zhou, Y.; Zhou, X.; Wang, X.-J.; et al. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. eLife 2015, 4, e06671. [Google Scholar] [CrossRef]
- Saunders, K.; Lucy, A.; Stanley, J. RNA-primed complementary-sense DNA synthesis of the geminivirus African cassava mosaic virus. Nucleic Acids Res. 1992, 20, 6311–6315. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.S.; Serra, H.; White, C.I.; Jeske, H. The recombination mediator RAD51D promotes geminiviral infection. Virology 2016, 493, 113–127. [Google Scholar] [CrossRef]
- Jeske, H.; Lütgemeier, M.; Preiß, W. DNA forms indicate rolling circle and recombination-dependent replication of Abutilon mosaic virus. EMBO J. 2001, 20, 6158–6167. [Google Scholar] [CrossRef]
- Mäkinen, K.; Lõhmus, A.; Pollari, M. Plant RNA Regulatory Network and RNA Granules in Virus Infection. Front Plant Sci. 2017, 8, 2093. [Google Scholar] [CrossRef]
- Krapp, S.; Greiner, E.; Amin, B.; Sonnewald, U.; Krenz, B. The stress granule component G3BP is a novel interaction partner for the nuclear shuttle proteins of the nanovirus pea necrotic yellow dwarf virus and geminivirus abutilon mosaic virus. Virus Res. 2017, 227, 6–14. [Google Scholar] [CrossRef]
- Leene, J.V.; Stals, H.; Eeckhout, D.; Persiau, G.; Slijke, E.V.D.; Isterdael, G.V.; Clercq, A.D.; Bonnet, E.; Laukens, K.; Remmerie, N.; et al. A Tandem Affinity Purification-based Technology Platform to Study the Cell Cycle Interactome in Arabidopsis thaliana. Mol. Cell. Proteom. 2007, 6, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Skirycz, A.; Claeys, H.; Bodt, S.D.; Oikawa, A.; Shinoda, S.; Andriankaja, M.; Maleux, K.; Eloy, N.B.; Coppens, F.; Yoo, S.-D.; et al. Pause-and-Stop: The Effects of Osmotic Stress on Cell Proliferation during Early Leaf Development in Arabidopsis and a Role for Ethylene Signaling in Cell Cycle Arrest. Plant Cell 2011, 23, 1876–1888. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.-L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Baliji, S.; Lacatus, G.; Sunter, G. The Interaction between geminivirus pathogenicity proteins and adenosine kinase leads to increased expression of primary cytokinin-responsive genes. Virology 2010, 402, 238–247. [Google Scholar] [CrossRef]
- Polston, J.E.; Capobianco, H. Transmitting Plant Viruses Using Whiteflies. JoVE J. of Vis. Exp. 2013, e4332. [Google Scholar] [CrossRef]
- Nakayasu, E.S.; Nicora, C.D.; Sims, A.C.; Burnum-Johnson, K.E.; Kim, Y.-M.; Kyle, J.E.; Matzke, M.M.; Shukla, A.K.; Chu, R.K.; Schepmoes, A.A.; et al. MPLEx: A Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses. mSystems 2016, 1. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367. [Google Scholar] [CrossRef]
- Consortium, U. UniProt: A hub for protein information. Nucleic Acids Res. 2014, 43, D204–D212. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Bandeira, N.; Sharma, V.; Perez-Riverol, Y.; Carver, J.J.; Kundu, D.J.; García-Seisdedos, D.; Jarnuczak, A.F.; Hewapathirana, S.; Pullman, B.S.; et al. The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 2020, 48, D1145–D1152. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote) omics data. Nat. Methods 2016, 13, 731. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Ochs, M.F., Casagrande, J.T., Davuluri, R.V., Eds.; Springer US: Boston, MA, USA, 2010; pp. 267–277. ISBN 978-1-4419-5714-6. [Google Scholar]
- Yeung, K.Y.; Haynor, D.R.; Ruzzo, W.L. Validating clustering for gene expression data. Bioinformatics 2001, 17, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
| Tissue | Clust | Select GO Terms (GO Term) | # of Proteins | F.E. | B. adj. p Value |
|---|---|---|---|---|---|
| Local | 1 | demethylation (GO:0070988) | 2 | 32.77 | 2.16 × 10−3 |
| 1 | mRNA splice site selection (GO:0006376) | 1 | 25.49 | 4.26 × 10−2 | |
| 1 | defense response (GO:0006952) | 3 | 6.37 | 1.26 × 10−2 | |
| 1 | translational elongation (GO:0006414) | 5 | 3.39 | 1.72 × 10−2 | |
| 2 | chaperone-mediated protein folding (GO:0061077) | 3 | 7.81 | 7.60 × 10−3 | |
| 2 | translational elongation (GO:0006414) | 7 | 3.29 | 6.26 × 10−3 | |
| 2 | mRNA processing (GO:0006397) | 4 | 3.21 | 3.85 × 10−2 | |
| 2 | proteolysis (GO:0006508) | 10 | 2.77 | 3.95 × 10−3 | |
| Systemic | 1 | Golgi to vacuole transport (GO:0006896) | 1 | 66.87 | 1.59 × 10−2 |
| 1 | regulation of actin polymerization or depolymerization (GO:0008064) | 1 | 44.58 | 2.32 × 10−2 | |
| 1 | chromatin silencing (GO:0006342) | 2 | 36.01 | 1.53 × 10−3 | |
| 2 | RNA metabolic process (GO:0016070) | 5 | 3.51 | 1.32 × 10−2 | |
| 3 | Golgi to plasma membrane transport (GO:0006893) | 1 | 37.49 | 2.76 × 10−2 | |
| 3 | defense response to bacterium (GO:0042742) | 1 | 22.49 | 4.47 × 10−2 | |
| 3 | proteolysis (GO:0006508) | 3 | 4.11 | 3.67 × 10−2 | |
| 4 | DNA demethylation (GO:0070988) | 2 | 38.96 | 1.54 × 10−3 | |
| 4 | cellular response to oxidative stress (GO:0034599) | 4 | 34.09 | 9.14 × 10−6 | |
| 4 | protein ubiquitination (GO:0016567) | 5 | 7.14 | 7.89 × 10−4 | |
| 4 | DNA repair (GO:0006281) | 3 | 3.97 | 4.15 × 10−2 | |
| 4 | cellular response to stimulus (GO:0051716) | 8 | 2.88 | 7.25 × 10−3 | |
| 5 | glycolytic process (GO:0006096) | 4 | 83.87 | 2.62 × 10−7 | |
| 5 | cellular amino acid biosynthetic process (GO:0008652) | 6 | 37.47 | 1.96 × 10−8 | |
| 5 | protein transmembrane import into intracellular organelle (GO:0044743) | 3 | 37.47 | 8.65 × 10−5 |
| Meme Motif Discovery | GOMo Motif Ontology Enrichment | |||||
|---|---|---|---|---|---|---|
| R. | Motif | E-Val | Sites | GO Term | q-Val | GO Name |
| Increased (+WFVv. +WF) | 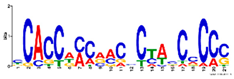 | GO:0009507 | 1 × 10−3 | Chloroplast 1 | ||
| GO:0009941 | 1 × 10−2 | Chloroplast envelope 1 | ||||
| 3 × 10−7 | 13 | GO:0003700 | 1 × 10−2 | Transcription factor activity 2 | ||
| GO:0006355 | 2 × 10−2 | Transcription regulation, DNA-dependent 3 | ||||
| GO:0003677 | 3 × 10−2 | DNA binding 2 | ||||
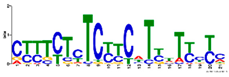 | GO:0003700 | 9 × 10−5 | Transcription factor activity 2 | |||
| GO:0048366 | 4 × 10−4 | Leaf development 3 | ||||
| 4 × 10−4 | 20 | GO:0007623 | 5 × 10−3 | Circadian rhythm 3 | ||
| GO:0004842 | 5 × 10−3 | Ubiquitin–protein ligase activity 2 | ||||
| GO:0010599 | 4 × 10−2 | Production of lsiRNA involved in RNAi 3 | ||||
| Decreased (+WFV v. +WF) | 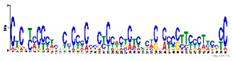 | GO:0009507 | 2 × 10−4 | Chloroplast 1 | ||
| GO:0003700 | 2 × 10−4 | Transcription factor activity 2 | ||||
| 6 × 10−24 | 10 | GO:0003677 | 2 × 10−4 | DNA binding 2 | ||
| GO:0048366 | 4 × 10−2 | Leaf development 3 | ||||
| GO:0009941 | 4 × 10−2 | Chloroplast envelope 1 | ||||
 | ||||||
| GO:0009507 | 1 × 10−3 | Chloroplast 1 | ||||
| 6 × 10−10 | 14 | GO:0003700 | 2 × 10−3 | Transcription factor activity 2 | ||
 | GO:0003700 | 2 × 10−4 | Transcription factor activity 2 | |||
| GO:0006355 | 2 × 10−3 | Transcription regulation, DNA-dependent 3 | ||||
| 2 × 10−2 | 20 | GO:0009414 | 5 × 10−3 | Response to water deprivation 3 | ||
| GO:0009956 | 2 × 10−2 | Radial pattern formation 3 | ||||
| GO:0009753 | 4 × 10−2 | Response to jasmonic acid stimulus 3 | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogden, A.J.; Boukari, W.; Nava, A.; Lucinda, N.; Sunter, G.; Curtis, W.R.; Adkins, J.N.; Polston, J.E. Characterization of Local and Systemic Impact of Whitefly (Bemisia tabaci) Feeding and Whitefly-Transmitted Tomato Mottle Virus Infection on Tomato Leaves by Comprehensive Proteomics. Int. J. Mol. Sci. 2020, 21, 7241. https://doi.org/10.3390/ijms21197241
Ogden AJ, Boukari W, Nava A, Lucinda N, Sunter G, Curtis WR, Adkins JN, Polston JE. Characterization of Local and Systemic Impact of Whitefly (Bemisia tabaci) Feeding and Whitefly-Transmitted Tomato Mottle Virus Infection on Tomato Leaves by Comprehensive Proteomics. International Journal of Molecular Sciences. 2020; 21(19):7241. https://doi.org/10.3390/ijms21197241
Chicago/Turabian StyleOgden, Aaron J., Wardatou Boukari, Alba Nava, Natalia Lucinda, Garry Sunter, Wayne R. Curtis, Joshua N. Adkins, and Jane E. Polston. 2020. "Characterization of Local and Systemic Impact of Whitefly (Bemisia tabaci) Feeding and Whitefly-Transmitted Tomato Mottle Virus Infection on Tomato Leaves by Comprehensive Proteomics" International Journal of Molecular Sciences 21, no. 19: 7241. https://doi.org/10.3390/ijms21197241
APA StyleOgden, A. J., Boukari, W., Nava, A., Lucinda, N., Sunter, G., Curtis, W. R., Adkins, J. N., & Polston, J. E. (2020). Characterization of Local and Systemic Impact of Whitefly (Bemisia tabaci) Feeding and Whitefly-Transmitted Tomato Mottle Virus Infection on Tomato Leaves by Comprehensive Proteomics. International Journal of Molecular Sciences, 21(19), 7241. https://doi.org/10.3390/ijms21197241






