Abstract
Several new products with innovative formulations are being proposed to facilitate oral care. Here, we evaluated the effects of a commercially available product, a toothpaste and chewing gum named Biorepair Peribioma, on oral microorganisms of healthy subjects. Saliva from six volunteers was collected during 20 min of mastication of a traditional gum (gum A) and the Biorepair Peribioma gum (gum P). Orthodontic elastics (OE) were in vitro contaminated with salivary samples, both A and P, and subsequently exposed or not to a Biorepair Peribioma toothpaste-conditioned supernatant (Tp-SUP). The salivary samples were tested for initial microbial load; hence, the contaminated OE were assessed for microbial growth, adhesion, biofilm formation and persistence; moreover, species identification was assessed. We found that the salivary samples A and P had similar microbial load; upon contamination, microbial adhesion onto the OE was detected to a lower extent when using saliva P with respect to saliva A. Microbial growth and biofilm formation, assessed at 24 h, remained at lower levels in OE exposed to saliva P, compared to saliva A. This difference between salivary samples A and P was confirmed when measuring biofilm persistence (48 h), while it was lost in terms of microbial re-growth (48 h). The Tp-SUP treatment drastically affected microbial load at 24 h and strongly impaired biofilm formation/persistence, in OE exposed to both salivary samples A and P. Finally, such treatment resulted in consistent overgrowth of Lactobacilli, bacterial species originally present both in the Biorepair Peribioma toothpaste and gum. In conclusion, by an in vitro pilot study, we show that the Biorepair Peribioma toothpaste and gum deeply affect oral microorganisms’ behavior, drastically impairing their ability to contaminate and produce plaque onto orthodontic devices.
1. Introduction
The oral cavity is a highly complex ecological niche, comprising resident microbial communities that crucially participate at the maintenance of local homeostasis. The latter can be affected by several factors, such as poor oral hygiene, poor diet, and use of drugs, as well as occurrence of dental appliances. In addition, immunodeficiency or a variety of systemic diseases greatly impact on oral health [1,2]. When the homeostasis of the oral microbiota is perturbed by external factors, high numbers of opportunistic pathogens can prevail locally and organize themselves in microbial biofilms. The latter predispose to the onset of oral diseases, by promoting a persistent and deleterious inflammatory process [3].
Saliva is a fundamental element in oral health. It is responsible for continuous clearing of substances and acids neutralization as well; in addition, because of its high content in inorganic ions, it also helps the remineralization process, constantly occurring on teeth surfaces [4]. The protection of oral tissues provided by saliva occurs also by moisturizing and buffering them with high calcium and phosphate concentrations [5]. Saliva contains antimicrobial compounds, such as lysozyme, lactoferrin and lactoperoxidase, which control both resident microbiota and potential oral pathogens, by interfering with their multiplication rate or by a direct killing activity. Lysozyme acts on peptidoglycan, the fundamental component of bacterial cell wall. Lactoferrin acts as an iron sequestering agent, therefore depriving bacteria of an important element for their metabolic processes. Peroxidase intervenes in the redox balance of the oral environment, by neutralizing the hydrogen peroxide produced by oral bacteria [6]. Saliva also contains essential biological components, such as salivary mucins and glycoproteins, which promote epithelial cell health as well as early dental plaque development [7]. Mucins are very resistant to a variety of proteolytic enzymes occurring in the bacterial plaque around the teeth and in the crevicular area, especially in patients with periodontitis. In addition, mucin is protective and prevents ulceration in soft tissues membranes. In inflammatory diseases, proteases are also generated by polymorphonuclear (PMN) leukocytes. Bacterial and PMN proteases (such as elastase, collagenase and cathepsin) affect the integrity of the mucous membranes [8]. Upon secretion, saliva is sterile, but soon it becomes an important vehicle for microorganisms in the oral cavity. Depending on their ability to closely adhere to biotic and abiotic surfaces and to aggregate with each other, some oral microorganisms can easily produce biofilm both on teeth and on orthodontic appliances and dental prostheses [9]. As a result, they can exert their pathogenic potential locally, negatively affecting oral homeostasis and/or directly damaging oral tissues, including mucous membrane, enamel and cement [10,11,12,13].
The most frequent oral diseases are caries and periodontitis, both associated with the presence of disease-promoting bacterial biofilms on tooth surfaces. Dental caries prevalence is steadily increasing [14] as well as periodontitis, which is commonly observed as a chronic disease, generally developing from gingivitis [15,16]. The microbial species most commonly implicated in caries lesions are Streptococcus mutans (S. mutans) and Streptococcus sobrinus (S. sobrinus) [17]. Differently, Streptococcus salivarius (S. salivarius) is considered for its role in prevention of dental caries [18], periodontal disease [19], and halitosis [20]. In this scenario, the possible synergistic or antagonistic relationships among different microbial species and their impact on tooth structure and/or on periodontium inflammation are being investigated. Interestingly, probiotic bacteria have been proposed as a means to counteract the onset of periodontal disease, thanks to their capacity to inhibit the proliferation of pathogenic germs within the periodontium. In particular, initial studies describe the use of probiotics, such as Lactobacillus and Bifidobacterium spp, to contain the level of S. mutans, thus exerting a beneficial role for the oral environment homeostasis [21,22].
Therefore, the “bacteriotherapy” represents a novel approach with enormous potential in the management of oral health and in the containment of local dysmicrobism. In addition, recent studies highlight the effectiveness of various natural substances on supporting oral homeostasis; besides exerting direct antimicrobial activity, such substances have the ability to counteract dental plaque, caries formation, tooth staining, gingival inflammation; also, promotion of enamel recalcification has been documented [23,24].
Several dental malocclusions need to be treated through removable orthodontic clear aligners (Frankel, Bionator, etc.), or fixed orthodontic appliances, such as brackets, tubes or bands, several types of archwires, ligating materials and others. These orthodontic materials, that per se offer a reliable support for microbial adhesion, significantly reduce the efficacy of patient’s oral hygiene, making way for debris retention areas; the latter will ultimately facilitate microbial persistence and growth with subsequent development of dental caries, periodontal diseases or other oral infections [25].
Clinical experience reveals that, because of the complexity of brackets design and/or ligation methods, is uncommon to encounter patients with cleaned fixed appliances and microbial plaque carefully removed [26,27]. According to the literature, among different types of orthodontic materials, elastomeric chains or single elastics are highly involved in favoring microbial adhesion/colonization and in turn cross-infections [28,29]. When using these polyurethane devices, it should also be considered that they may undergo alterations to different degree upon contact with physical/chemical agents [30].
The importance of plaque control in maintaining oral health leads to the continuous search for innovative products. In this context, it has recently produced a new fluorine-free toothpaste and a chewing gum, both named Biorepair® PERIBIOMATM (Coswell S.p.A., Bologna, Italy), have been recently put on the market. These two innovative products contain microRepair®BIOMA (Coswell S.p.A., Bologna, Italy), consisting of biomimetic hydroxyapatite microcrystals combined with selected probiotics aimed at promoting balance of oral microbiota. Such products are expected to repair tooth enamel, protect from microbial plaque formation and help to contain/prevent inflammation and gingival bleeding; in particular, hydroxyapatite crystals are able to bind enamel and dentine, reducing sensitivity and favoring tooth enamel remineralization. The hydroxyapatite crystals also mediate antimicrobial functions by releasing locally calcium, phosphate and zinc ions, especially when the tooth is affected by dental plaque or under acid pH conditions [31,32]. In accordance with the manufacturer’s guidelines, they can be used for clinical application at all ages, even in kids, with no risks related to ingestion. Actually, children are possibly tempted to the idea of chewing a gum and therefore are easily encouraged towards oral hygiene.
Nowadays, there is a lack of microbiological research in this field, which makes the present work a novelty in the field of oral hygiene maintenance, providing an original input for the introduction/implementation of novel easy-to-use tools in clinical practice.
The aim of the present study was to evaluate in vitro the effects of the Biorepair Peribioma toothpaste and gum on oral cavity microorganisms. Accordingly, salivary samples, collected from healthy volunteers during gum mastication, were used to contaminate in vitro orthodontic elastics (OE); then, microbial growth, biofilm formation and persistence were analyzed at different times in the presence or absence of the toothpaste. Consistent antimicrobial effects were observed.
The clinical implications of these findings are discussed.
2. Materials and Methods
2.1. Chewing Gums and Toothpaste-Conditioned Supernatant Preparation
Two different types of chewing gums, a traditional gum (Vigorsol) and the Biorepair® PERIBIOMATM gum (Coswell S.p.A., Bologna, Italy), both sugar-free, hereafter indicated as gum A and gum P, were provided to the volunteers for saliva collection, as detailed below (see Section 2.2).
The Biorepair® PERIBIOMATM toothpaste was used to prepare a toothpaste-conditioned supernatant. In particular, 50 gr of toothpaste were suspended in 100 mL of saline solution and incubated for 18 h at 37 °C, under gentle shaking. After incubation, the opalescent solution was centrifuged at 1200 rpm for 10 min and the supernatant collected, aliquoted and frozen at −20 °C (hereafter indicated as Tp-SUP).
2.2. Volunteer Selection and Saliva Collection
Six healthy volunteers were selected according to several inclusions and exclusions criteria, in line with other studies on the efficacy of toothpastes [33,34]. In particular, the inclusion criteria were: both genders, age between 18–64 years, self-declaration of no basic pathologies and no pregnancy; the exclusion criteria were: non-compliance of the subjects regarding to oral hygiene instructions, use of mouthwashes and antibiotics during the last month. For all the volunteers enrolled, mouthwashes use was prohibited for the entire duration of the study; food and drink were allowed up to 1 h before saliva collection. The enrolled subjects were asked to provide their saliva at least in 3 sessions (every 2 weeks). Here below, are presented the peculiarities of the volunteers: sex: 3 males and 3 females, age range: 25–51; body mass index range: 23, 9–28, 7; no diseases; no pregnancy.
Saliva collection was performed as detailed in the flow chart (Supplementary Material, Figure S1). Firstly, three volunteers (1, 2 and 3) chewed the gum A and the other three volunteers (4, 5 and 6) chewed the gum P, for 20 min, during which saliva samples were collected and named A1, A2, A3 and P4 P5, P6. Secondly, the volunteers were asked to rinse their mouths with fresh water and then the volunteers 1, 2 and 3 chewed the gum P while the volunteers 4, 5 and 6 chewed the gum A, for further 20 min. During that time, a second series of saliva samples (Saliva A4, A5 and A6, and saliva P1, P2 and P3) were collected. Subsequently, all the saliva samples were delivered to the microbiology laboratory, where saliva A1–A6 were pooled as well as the saliva P1–6 (equal volumes from each volunteer were mixed) and immediately used as detailed below.
The present study had been approved by the local Ethics Committee (Prot. AOU: 14075/20; Prot. EC: 0014230/20, dated 21 May 2020).
2.3. Assessment of Microbial Load and Identification of the Main Culturable Species
Initially, the saliva A and P pools were analyzed to establish the microbial load, by Colony Forming Units (CFU) assay on selective growth media (Tryptic Soy Agar, Sabouraud Dextrose Agar, Mitis Salivarius Agar and De Mann-Rogosa-Sharpe Agar; OXOID; Milan, Italy) under aerobic conditions. The colonies grown after 24 h were phenotypically clustered by color, morphology, size and counted. Subsequently, representative colonies of each type were sub-cultured in Columbia agar plates (OXOID; Milan, Italy) and then identified by MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) and by API® IDENTIFICATION KIT (Biomerieux, Marcy l’Etoile, France).
2.4. Contamination of the Orthodontic Elastics by Saliva
For the present study, the orthodontic elastics (OE) were provided by Leone S.p.A. Florence, Italy. The OE were sterilized by autoclave at 121 °C, for a cycle of 15 min, as indicated elsewhere [35,36]. Then, the OE were exposed to the saliva A and P pools for in vitro contamination, in line with previously described protocols [37]. Briefly, the OE were placed in a 1.5 mL tube containing saliva A or P (100 µL/elastic) and incubated for 1 h at 37 °C with 5% CO2, under gentle shaking.
2.5. Evaluation of Microbial Adhesion onto OE, Growth and Biofilm Formation
During the 1 h contamination, the samples were concomitantly exposed to the Carboxy-fluorescein Diacetate, Acetoxymethyl Ester (CFDA), that measures both enzymatic activity and cell-membrane integrity (Thermo Fisher Scientific, Waltham, MA, USA), according to manufacturer’s instructions. After the 1 h incubation, the elastics were washed with warm saline and transferred to 96 well black microtiter plate, where each well was filled with 100 µL of fresh Tryptic Soy Broth (TSB), supplemented with 2% sucrose (OXOID; Milan, Italy); then, the plate was read at the Fuoroskan microplate reader (Thermo Fisher Scientific, Waltham, MA, USA). The fluorescence signal (excitation/emission: 485/538 nm) was expressed as relative fluorescence units (RFU) and considered as a measure of microbial adhesion to the OE (RFU/OE).
The plate was further incubated for 23 h at 37 °C with 5% CO2. Then, samples were exposed to saline or Tp-SUP (100 μL/well) and the CFDA reagent was added again for additional 1 h. At the end of such incubation time (24 h), total microbial load/well was assessed by CFU assay and species identification was performed by MALDI-TOF MS and API system, as detailed above. In parallel samples, the OE were washed twice with warm saline, transferred to new wells with fresh TSB plus 2% sucrose and the RFU was measured by Fluoroskan microplate reader. The fluorescence signal (excitation/emission: 485/538 nm) was expressed as RFU and taken as a measure of biofilm formation onto the OE (RFU/OE).
2.6. Evaluation of Microbial Re-Growth and Biofilm Persistence
The OE were further incubated up to 48 h from contamination. Then, microbial re-growth was assessed by CFU analysis and the isolates were identified as detailed above. In parallel groups, the CFDA reagent was added; 1 h later, the OE were washed with warm saline and then residual biofilm was measured by fluorescence analysis, as previously described. The fluorescence emission by live cells (excitation/emission: 485/538 nm) was expressed as RFU and taken as a measure of residual biofilm onto the OE (RFU/OE).
2.7. Statistical Analysis
Data depicted in the graphs are the mean ± SEM from replicate samples of at least 3 independent experiments. Quantitative variables were tested for normal distribution by Shapiro–Wilk test. Statistical differences between groups were analyzed according to Mann–Whitney U test (Figure 1) or Kruskal–Wallis followed by Dunn’s multiple comparisons tests (Figure 2 and Figure 3) by using GraphPad Prism 8. Data depicted in the tables are the CFU mean values of 3 independent experiments.
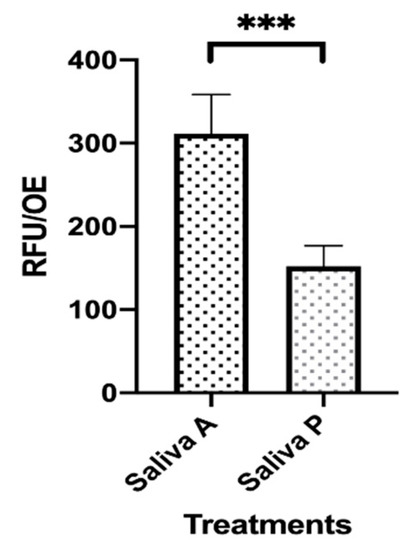
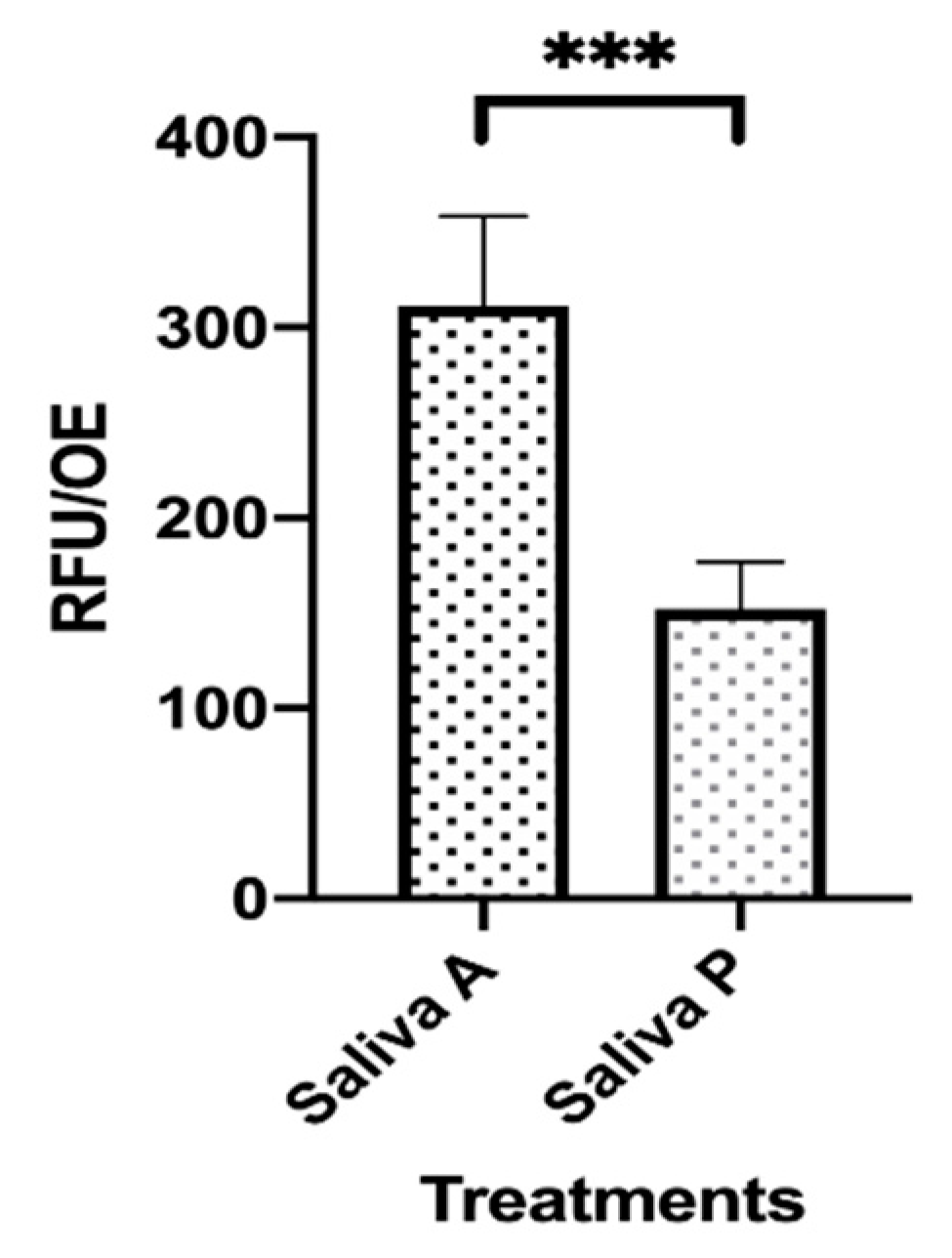
Figure 1.
Microbial adhesion onto saliva-contaminated orthodontic elastics (OE). Two sets of OE were exposed to saliva A and P for 1 h at 37 °C. During that time, fluorescence staining of viable cells was allowed by Carboxy-fluorescein Diacetate, Acetoxymethyl Ester (CFDA) addition. Then, the OE were washed with warm saline and transferred to new wells. The plate was read by the Fluoroskan reader and the fluorescence signal was recorded. The depicted values represent the mean (RFU/OE) ± SEM of 48 replicates obtained by 3 independent experiments. *** p < 0.001.
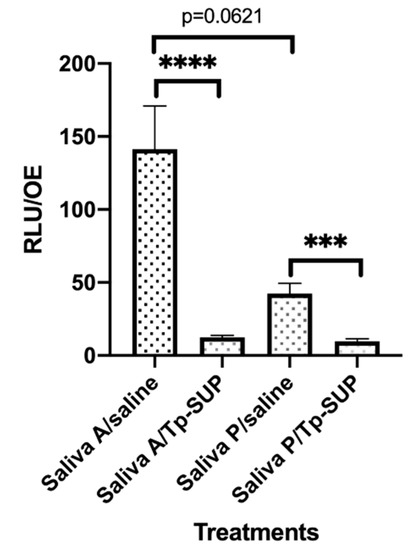
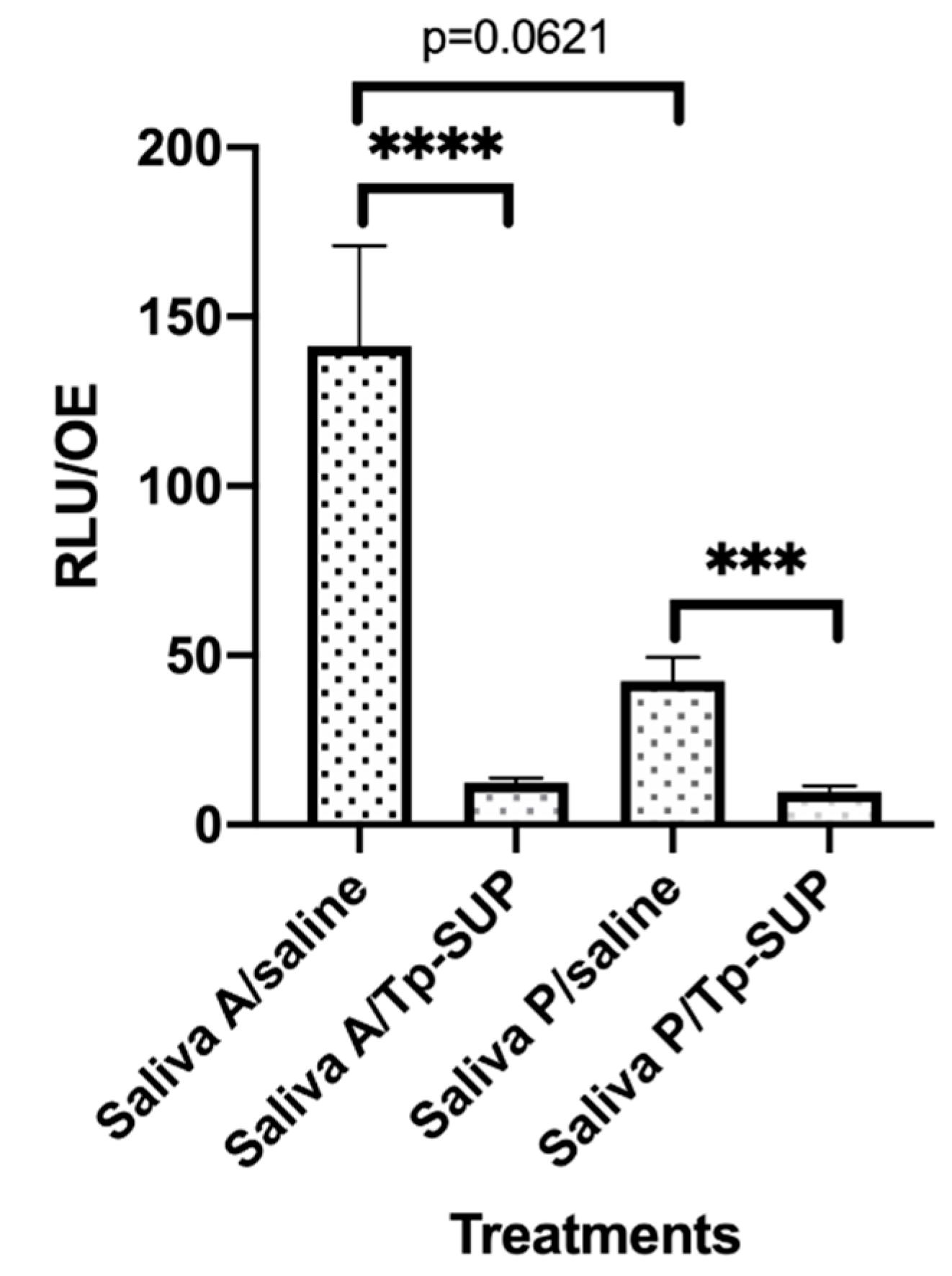
Figure 2.
Early biofilm formation onto saliva-contaminated OE exposed or not to Tp-SUP. The saliva-contaminated OE were incubated for 23 h at 37 °C + 5% CO2 and then exposed to saline or Tp-SUP for additional 1 h, in the presence of CFDA, to allow fluorescence staining. Then, the OE were washed with warm saline and transferred in new wells. The plate was read by the Fluoroskan microplate reader and the fluorescence signal was recorded. The depicted values represent the mean ± SEM (RFU/OE) of 13–16 replicates obtained by 3 independent experiments. **** p < 0.0001; *** p < 0.001.
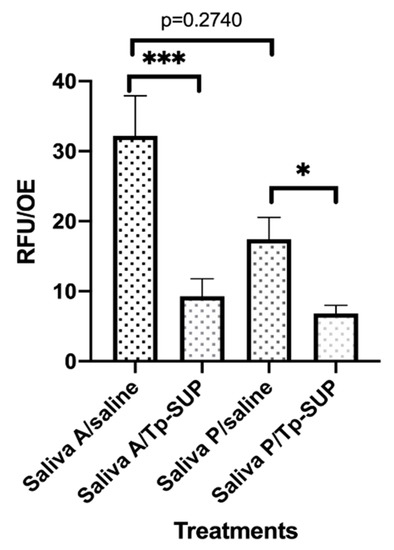
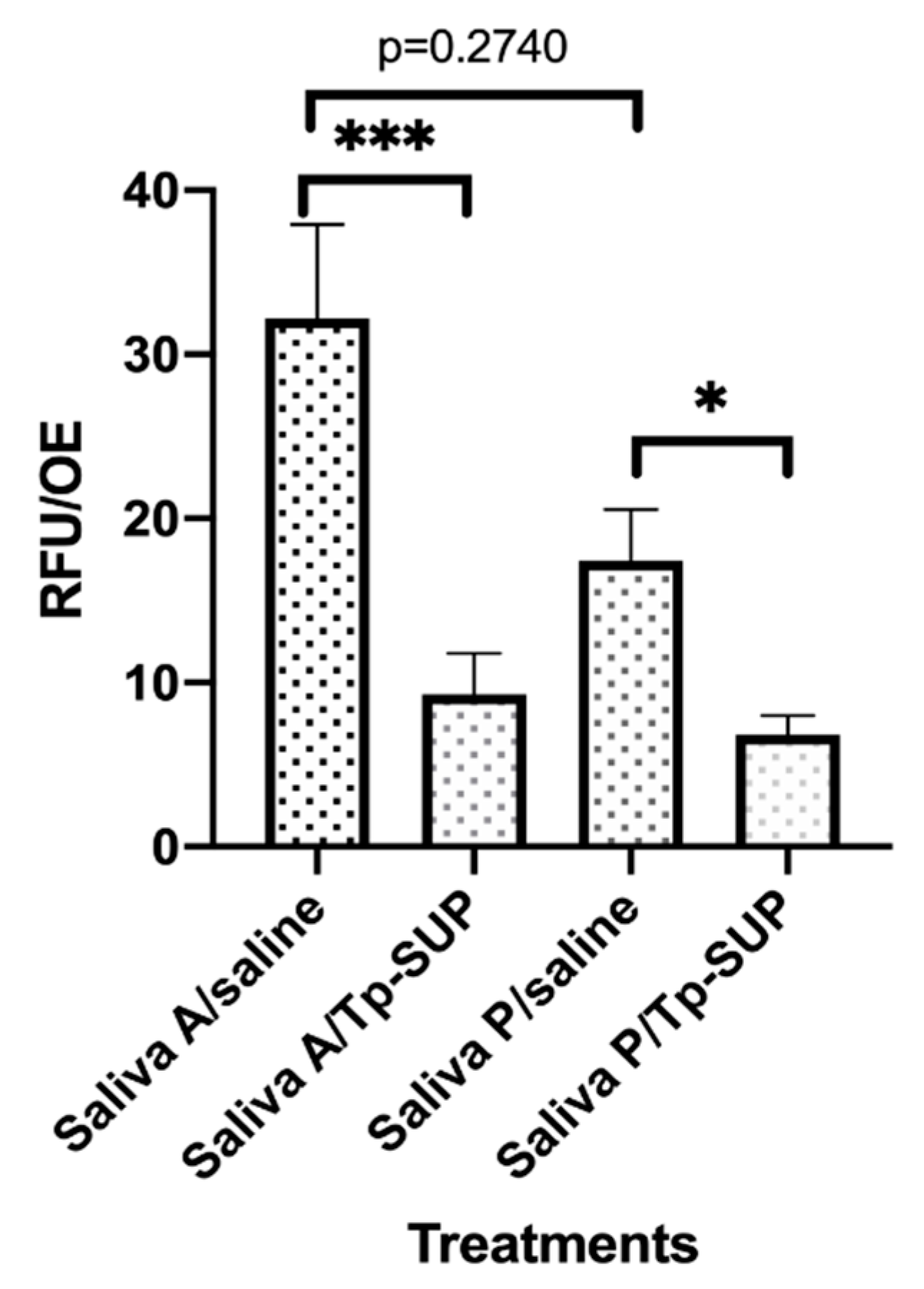
Figure 3.
Biofilm persistence following Tp-SUP treatment. The saliva-contaminated OE, that had been exposed to Tp-Sup or saline (at time 23 h, for 1 h) were further incubated at 37 °C + 5% CO2 up to 48 h in fresh medium. Then, the OE were label with CFDA for 1 h to allow fluorescence staining, washed with warm saline and transferred to new wells containing fresh medium. The plate was read by the Fluoroskan microplate reader and the fluorescence signal was recorded. The depicted values represent the mean RFU/OE ± SEM of 15 replicates obtained by 3 independent experiments. *** p < 0.001; * p ≤ 0.05.
Values of **** p < 0.0001, *** p <0.001 and * p < 0.05 were considered significant.
3. Results
3.1. Initial Microbial Load and Identification of the Main Culturable Species in Saliva A and P
Initially, we evaluated the microbial load and characterized the bacterial species growing in saliva A and P, under aerobic conditions. As shown in Table 1, the microbial load detected at time 0 in saliva A was slightly lower as compared to that obtained from saliva P (379.1 vs. 501 CFU/100 µL × 104, respectively); such difference did not reach statistical significance. Microbial identification revealed that the most representative species were Streptococcus mitis/oralis (S. mitis/oralis), reaching 67.01 and 81%, in saliva A and P, respectively. S. salivarius was the second most abundant species recovered in saliva A (16.8%) but not in saliva P (2.9%); differently, the latter counted 10.9% of Neisseria flava/subflava (N. flava/subflava). The third and fourth most abundant species in saliva A were S. thermophilus (8.9%) and N. flava/subflava (7.1%), while in saliva P, the third and fourth most abundant species were S. thermophilus (4%) followed by S. salivarius (2.9%). Rothia aeria (R. aeria) was identified only occasionally in both saliva A and P, while neither Candida nor Lactobacillus spp. were ever detected in either salivary samples.

Table 1.
Initial microbial load and evaluation of the main microbial species in Saliva A and P.
3.2. Microbial Adhesion onto OE, Growth and Biofilm Formation
The ability of microorganisms occurring in the saliva A and P to mediate adhesion onto OE was evaluated. Accordingly, two sets of pre-sterilized OE were exposed to each pool of saliva in the presence of CFDA to allow fluorescence staining of cells. After 1 h at 37 °C, a condition commonly used to allow microbial adhesion onto abiotic surfaces [37], the OE were washed with warm saline, transferred to new wells containing fresh medium and analyzed for fluorescence emission. As shown in Figure 1, the fluorescence signal, indicating the amounts of viable and metabolically active cells, was significantly lower in saliva P, as compared to saliva A contaminated OE (about 50% difference).
Subsequently, the contaminated OE were incubated for further 23 h, under standard culture conditions, to allow microbial growth. Then, each set of OE (saliva A and saliva P contaminated groups) was split in two sub-groups and exposed to saline or Tp-SUP (in both cases, 100 µL/well were added) for additional 1 h at 37 °C. Subsequently, each sample was assessed for microbial growth by CFU assay, followed by MALDI-TOF MS/API colonies identification. As depicted in Table 2, when comparing the total CFU from saliva A- and saliva P-contaminated OE maintained in saline, about 1 log lower values were found in the latter group. Moreover, the treatment with Tp-SUP strongly reduced the CFU, in both saliva A- and saliva P-contaminated OE; particularly, the reduction was approximately of 2 logs when comparing saliva A/saline vs. saliva A/Tp-SUP and of about 1 log considering saliva P/saline vs. saliva P/Tp-SUP.

Table 2.
Microbial load onto saliva-contaminated OE subsequently exposed or not to Tp-SUP.
In parallel, species identification revealed that the most representative one, both in saliva A and P/saline groups, was S. mitis/oralis (97.1% and 99.1%, respectively). Tp-SUP treatment did not substantially change this percentage. Furthermore, when considering the crude numbers, the S. mitis/oralis CFU counted in saliva A/Tp-SUP-treated samples were 2 logs lower than those in saliva A/saline-treated samples; such decrease was of about 1 log when comparing saliva P/Tp-SUP vs. saliva P/saline. The second most representative species in saliva A/saline was S. thermophilus, followed by S. salivarius and traces of N. flava/subflava and R. aeria, while, in all the other groups, these species were consistently below the detection limit of the assay. Subsequently, in parallel groups, the saliva-contaminated OE were incubated for 23 h at 37 °C + 5% CO2 and then exposed to saline or Tp-SUP for additional 1 h, in the presence of CFDA. Then, the OE were washed with warm saline, transferred to new wells, and the fluorescence emission was measured.
As shown in Figure 2, a consistent difference was detected when comparing the RFU onto saliva A/saline vs. saliva P/saline OE although without reaching significance (p = 0.0621). Moreover, Tp-SUP treatment drastically reduced the amounts of biofilm, independently of the groups considered. In particular, when comparing saliva A/saline vs. saliva A/Tp-SUP or saliva P/saline vs. saliva P/Tp-SUP, significant decreases were consistently observed upon Tp-SUP treatment in both cases.
3.3. Microbial Re-Growth and Biofilm Persistence
To assess the microbial regrowth under the different conditions, the 4 groups of OE, were incubated for further 24 h (up to time 48 h) in fresh medium and tested by CFU assay and MALDI-TOF MS/API analysis. Table 3 shows that the total CFU detected in all the groups were similar, ranging between 552 to 953 × 104/100 µL. Interestingly, S. mitis/oralis was the most representative species both in saliva A/saline and saliva P/saline groups (91.01% and 85.3%, respectively), while, these same species were not detected in Tp-SUP treated groups, no matter whether A or P saliva groups were considered. In contrast, in the Tp-SUP-treated groups, most of the microbial population was identified as Lactobacillus spp. (>90%); such bacteria were undetectable (<0.1 CFU × 104/100 µL) in the two groups that had not received the Tp-SUP treatment.

Table 3.
Microbial re-growth onto saliva-contaminated and Tp-SUP treated OE.
To evaluate biofilm persistence, at time 48 h, the OE were washed again, transferred to new wells and labelled with CFDA fluorescent reagent for 1 h.
As shown in Figure 3, the RFU/OE were consistently lower in saliva P/saline than in saliva A/saline groups, although without reaching significance. The exposure to Tp-SUP significantly reduced the RFU/OE in both groups. When comparing saliva A/saline with saliva A/Tp-SUP or saliva P/saline with saliva P/Tp-SUP, statistically significant differences were observed.
4. Discussion
Here, we provide the first in vitro data concerning the effects of the Biorepair-PERIBIOMA toothpaste and chewing gum on the ability of oral microbiota to adhere, grow and produce biofilm onto orthodontic devices.
Orthodontic therapy with fixed appliances is commonly used to treat malocclusions and teeth irregularities; in these cases, orthodontic materials such as brackets, tubes and elastics are widely employed. These materials prevent an accurate oral hygiene and promote an increase in microbial load, deposition of debris and formation of plaque. With the aim of promoting a better oral hygiene, a new toothpaste, fluorine-free, and a chewing gum, Biorepair® PERIBIOMATM, added with probiotics, have recently been developed; little information exists on their mechanisms of action.
Among the most important components in the oral environment, saliva is an integral part of oral health; particularly, it plays a role in caries control and bacterial plaque containment [8]. Although being sterile when secreted [38], saliva immediately becomes a major vehicle of microorganisms, either resident microbiota in healthy subjects as well as pathogens in patients with oral cavity diseases. From here, our choice of using saliva from healthy donors as source of oral microbial population to be employed in an in vitro model that, by mimicking orthodontic materials contamination, would allow to evaluate the efficacy of novel tools for oral hygiene. The Biorepair® PERIBIOMATM gum and a traditional gum, both sugar-free, have been used to facilitate saliva collection.
In line with the literature [39], we show that the microbial species, isolated by a culture-based approach from saliva A and P are mainly S. mitis/oralis and S. salivarius, followed by N. flava/subflava and S. thermophilus. Interestingly, the number of CFU observed in saliva P is slightly higher (397 vs. 501 CFU/100 µL × 104 A vs. P samples) as compared to that of saliva A; unexpectedly, the relative abundance of S. salivarius appears to be enhanced in the latter group (16.88% vs. 2.99%, A vs. P samples). These results suggest that the gum P is slightly more effective in mechanically removing local microorganisms than a traditional gum. The partial fluctuation in relative abundance of some species may be due to their different susceptibly to mechanical gum-mediated detachment and/or to the different ingredients contained in each gum.
Being adhesion a crucial step in microbial biofilm formation, both on oral tissues and abiotic surfaces, here we investigated the ability of microbial communities occurring in saliva A and saliva P to adhere onto OE. As indicated by the fluorescence data, adhesion occurs, irrespectively of the saliva employed; yet, the phenomenon is less pronounced when using saliva P, implying that, although slightly more numerous, this microbial population exhibits a reduced efficacy on binding to an abiotic surface, such as OE. Whether the slightly higher numbers of S. mitis/oralis detected in saliva P may account for the overall less efficient adhesion, remains to be established. Furthermore, we cannot exclude that, in our in vitro model, hydroxyapatite crystals and probiotics, present in both Peribioma products, remain and persist in saliva P samples and, thus, may interfere with microbial adhesion to OE. In line with these data, we show that, after 24 h incubation, the Saliva P/saline group had 1 log lower microbial CFU than the counterpart samples (Saliva A/saline); once again, we may hypothesize that the hydroxyapatite, contained in gum P and likely persisting in Saliva P samples, may have limited microbial growth. To a similar extent, the Saliva P significantly reduces biofilm formation onto the OE (time 24 h). This phenomenon is also evident at a later time, indicating that saliva collected after chewing the gum P consistently affects biofilm persistence more than saliva A (17.42 vs. 33.84 RFU, respectively; at time 48 h).
Here, we have used the Tp-SUP as a simplified and easy-to-use tool, for assessing toothpaste effects against oral microorganisms in vitro. In particular, we show that the Tp-SUP exerts relevant antimicrobial activity, regardless of the saliva used to contaminate the orthodontic devices; specifically, both parameters, namely microbial growth and biofilm formation, are significantly impaired. The Tp-SUP-mediated microbial abatement ranges between 1 and 2 logs, which is already evident at time 24 h, when using the CFU assay. In contrast, at later times (48 h), microbial re-growth occurs comparably in all the groups, irrespective of the gum initially used for saliva collection and independently upon the in vitro Tp-SUP treatment. Furthermore, it should be noted that, at that time, the microbial composition greatly varies upon Tp-SUP exposure; in particular, the streptococci mostly detected in saliva A and saliva P/saline groups (>90%) are drastically displaced by Lactobacilli in saliva A and saliva P/Tp-SUP groups (>90%). This finding may be explained considering that the Biorepair® PERIBIOMATM products also contain probiotics, such as L. reuteri, L. salivarius, L. plantarum and Bifidobacterium. The reason for adding probiotics to such oral care products is that such microbial species may adhere to dental tissues and become part of the oral plaque; in turn, they may compete with cariogenic and periodontal pathogens for nutrients and space, thus preventing caries formation and gingivitis [40,41]. In our hands, the late detection of Lactobacilli is in line with previous results (data not shown) showing that, indeed, Tp-SUP culture allowed growth of colonies subsequently identified as Lactobacilli.
The efficacy of Tp-SUP is further emphasized by the fact that also biofilm persistence is significantly affected after such treatment: the extent of the inhibition is similar both in saliva A and saliva P contaminated OE.
Overall, by an in vitro model, we provide the first evidence on the efficacy of the Biorepair® PERIBIOMATM toothpaste and gum in impairing adhesion, growth and biofilm formation/persistence by oral microorganisms onto orthodontic devices. We favor the idea that, by different pathways, the hydroxyapatite crystals and the probiotics, abundantly present in such oral care products, may act in concert controlling local microbial communities. Moreover, based on the present in vitro data, we may envisage that the combined use of toothpaste and gum can ameliorate oral daily care, thus clinically reducing the risk of developing oral diseases, especially those related to microbial agents.
5. Conclusions
Efficacious strategies helping to maintain a good oral hygiene are necessary to prevent or treat oral diseases, especially when they are associated with microbial biofilms, commonly produced onto oral tissues and dental devices. This in vitro pilot study raises the possibility that the daily use of the Biorepair® PERIBIOMATM gum and toothpaste may, on the one hand, profoundly impact on microbial adhesion, growth and biofilm formation onto abiotic surfaces and, on the other hand, may promote replacement of potential oral pathogens with microorganisms beneficial to oral health. By this in vitro prototype, wide-spectrum studies may be pursued opening to other orthodontic/dentistry materials as well as towards other novel health-care products.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-3417/10/19/6721/s1, Figure S1: Flow-chart of the saliva collection.
Author Contributions
Conceptualization, A.M., A.O., B.C. and E.B.; methodology, B.C., A.M., A.O. and A.L.R.; validation, B.C., A.M., L.B.G., E.P. and E.B.; formal analysis, B.C., E.P. and E.B.; investigation, A.M., B.C. and A.O.; resources, A.O.; data curation, B.C., A.O., A.M., and L.B.G.; writing—original draft preparation, B.C., A.M., A.O., E.P. and L.B.G.; writing—review and editing, B.C., E.P. and E.B.; visualization, A.M., A.O., L.B.G. and B.C.; supervision, E.B.; project administration, B.C. and E.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors thank Andrea Ardizzoni for helpful contribution in English revision.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zarco, M.; Vess, T.; Ginsburg, G.S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Devine, D.; Marsh, P.D.; Meade, J. Modulation of host responses by oral commensal bacteria. J. Oral Microbiol. 2015, 7, 26941. [Google Scholar] [CrossRef]
- Mira, A. Oral Microbiome Studies: Potential Diagnostic and Therapeutic Implications. Adv. Dent. Res. 2018, 29, 71–77. [Google Scholar] [CrossRef]
- Manton, D.J.; Walker, G.D.; Fai, C. Remineralization of enamel subsurface lesions in situ by the use of three commercially available sugar-free gums. Int. J. Paediatr. Dent. 2008, 18, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, L.R.; De Lima, A.A.S.; Machado, M.A.N.; Grégio, A.M.T.; Almeida, P.D.V.D. Saliva Composition and Functions: A Comprehensive Review. J. Contemp. Dent. Pr. 2008, 9, 72–80. [Google Scholar] [CrossRef]
- Morinushi, T.; Murayama, M.; Kinjyo, S. Mutans streptococci, Lactobacilli in saliva and acidity for microorganisms in dental plaque: Changes after restorative treatment. J. Clin. Pediatr. Dent. 2004, 28, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Jakubovics, N.S. Saliva as the sole nutritional source in the development of multispecies communities in dental plaque. Microbiol. Spectr. 2015, 3, 3. [Google Scholar] [CrossRef]
- Kumar, B.; Kashyap, N.; Avinash, A.; Chevvuri, R.; Sagar, M.K.; Shrikant, K. The composition, function and role of saliva in maintaining oral health: A review. Int. J. Contemp. Dent. Med. Rev. 2017, 2017, 1–6. [Google Scholar]
- Zijnge, V.; Van Leeuwen, M.B.M.; John, E.D.; Abbas, F.; Thurnheer, T.; Gmür, R.; Harmsen, H.J.M. Oral biolm architecture on natural teeth. PLoS ONE 2010, 5, e9321. [Google Scholar] [CrossRef] [PubMed]
- Bossù, M.; Matassa, R.; Relucenti, M.; Iaculli, F.; Salucci, A.; Di Giorgio, G.; Familiari, G.; Polimeni, A.; Di Carlo, S. Morpho-Chemical Observations of Human Deciduous Teeth Enamel in Response to Biomimetic Toothpastes Treatment. Materials 2020, 13, 1803. [Google Scholar] [CrossRef] [PubMed]
- Meto, A.; Colombari, B.; Castagnoli, A.; Sarti, M.; Denti, L.; Blasi, E. Efficacy of a Copper–Calcium–Hydroxide Solution in Reducing Microbial Plaque on Orthodontic Clear Aligners: A Case Report. Eur. J. Dent. 2019, 13, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Conserva, E.; Pisciotta, A.; Bertoni, L.; Bertani, G.; Meto, A.; Colombari, B.; Blasi, E.; Bellini, P.; De Pol, A.; Consolo, U.; et al. Evaluation of biological response of stro-1/c-kit enriched human dental pulp stem cells to titanium surfaces treated with two different cleaning systems. Int. J. Mol. Sci. 2019, 20, 1868. [Google Scholar] [CrossRef] [PubMed]
- Meto, A.; Conserva, E.; Liccardi, F.; Colombari, B.; Consolo, U.; Blasi, E. Differential Efficacy of Two Dental Implant Decontamination Techniques in Reducing Microbial Biofilm and Re-Growth onto Titanium Disks In Vitro. Appl. Sci. 2019, 9, 3191. [Google Scholar] [CrossRef]
- Bagramian, R.A.; Garcia-Godoy, F.; Volpe, A.R. The global increase in dental caries. A pending public health crisis. Am. J. Dent. 2009, 22, 3–8. [Google Scholar] [PubMed]
- Kalsbeek, H. Series: Caries prevention in historical perspective. Fluoride. Ned. Tijdschr. voor Tandheelkd. 2018, 125, 257–261. [Google Scholar] [CrossRef]
- Matthews, D.C. Prevention and treatment of periodontal diseases in primary care. Evid. Based Dent. 2014, 15, 68–69. [Google Scholar] [CrossRef][Green Version]
- Costalonga, M.; Herzberg, M.C. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol. Lett. 2014, 162, 22–38. [Google Scholar] [CrossRef]
- Tanzer, J.M.; Kurasz, A.B.; Clive, J. Competitive displacement of mutans streptococci and inhibition of tooth decay by Streptococcus salivarius TOVE-R. Infect. Immun. 1985, 48, 44–50. [Google Scholar] [CrossRef]
- Teughels, W.; Newman, M.; Coucke, W.; Haffajee, A.; Van Der Mei, H.; Haake, S.K.; Schepers, E.; Cassiman, J.-J.; Van Eldere, J.; Van Steenberghe, D.; et al. Guiding Periodontal Pocket Recolonization: A Proof of Concept. J. Dent. Res. 2007, 86, 1078–1082. [Google Scholar] [CrossRef]
- Burton, J.P.; Chilcott, C.; Moore, C.; Speiser, G.; Tagg, J. A preliminary study of the effect of probiotic Streptococcus salivarius K12 on oral malodour parameters. J. Appl. Microbiol. 2006, 100, 754–764. [Google Scholar] [CrossRef]
- Haukioja, A.; Yli-Knuuttila, H.; Loimaranta, V.; Kari, K.; Ouwehand, A.C.; Meurman, J.H.; Tenovuo, J. Oral adhesion and survival of probiotic and other lactobacilli and bifidobacteria in vitro. Oral Microbiol. Immunol. 2006, 21, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Näse, L.; Hatakka, K.; Savilahti, E.; Saxelin, M.; Pönkä, A.; Poussa, T.; Korpela, R.; Meurman, J. Effect of long-term consumption of a probiotic bacterium, Lactobacillus rhamnosus GG, in milk on dental caries and caries risk in children. Caries Res. 2002, 35, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Fiorillo, L.; Cervino, G.; Herford, A.S.; Laino, L.; Cicciù, M. Stannous Fluoride Effects on Enamel: A Systematic Review. Biomimetics 2020, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Biesbrock, A.R.; He, T.; Digennaro, J.; Zou, Y.; Ramsey, D.; Garcia-Godoy, F. The effects of bioavailable gluconate chelated stannous fluoride dentifrice on gingival bleeding: Meta-analysis of eighteen randomized controlled trials. J. Clin. Periodontol. 2019, 46, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Sukontapatipark, W.; El-Agroudi, M.A.; Selliseth, N.J.; Thunold, K.; Selvig, K.A. Bacterial colonization associated with fixed orthodontic appliances-A SEM study. Eur. J. Orthod. 1998, 23, 475–484. [Google Scholar] [CrossRef]
- Kitada, K.; De Toledo, A.; Oho, T. Increase in detectable opportunistic bacteria in the oral cavity of orthodontic patients. Int. J. Dent. Hyg. 2009, 7, 121–125. [Google Scholar] [CrossRef]
- Brusca, M.I.; Chara, O.; Sterin-Borda, L.; Rosa, A.C. Influence of Different Orthodontic Brackets on Adherence of Microorganisms In Vitro. Angle Orthod. 2007, 77, 331–336. [Google Scholar] [CrossRef]
- Dittmer, P.D.M.; Demling, A.; Borchers, L.; Stiesch, M.; Kohorst, P.; Schwestka-Polly, R. The influence of simulated aging on the mechanical properties of orthodontic elastomeric chains without an intermodular link. J. Orofac. Orthop. Fortschr. Kieferorthopädie 2012, 73, 289–297. [Google Scholar] [CrossRef]
- Takla, G.S.; Cunningham, S.J.; Horrocks, E.N.; Wilson, M. The effectiveness of an elastomeric module dispenser in cross-infection control. J. Clin. Orthod. 1998, 32, 721–726. [Google Scholar]
- Josell, S.D.; Leiss, J.B.; Rekow, E.D. Force degradation in elastomeric chains. Semin. Orthod. 1997, 3, 189–197. [Google Scholar] [CrossRef]
- Degli Esposti, L.; Ionescu, A.C.; Brambilla, E.; Tampieri, A.; Iafisco, M. Characterization of a Toothpaste Containing Bioactive Hydroxyapatites and In Vitro Evaluation of Its Efficacy to Remineralize Enamel and to Occlude Dentinal Tubules. Materials 2020, 13, 2928. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Hector, M.; Rampersad, M. Critical pH in resting and stimulated whole saliva in groups of children and adults. Int. J. Paediatr. Dent. 2001, 11, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Sava, S.; Sava, S.; Delean, A.G.; Mihailescu, A.M.; Dumitrescu, L.S.; Moldovan, M.; Festila, D.G. Toothpaste Composition Effect on Enamel Chromatic and Morphological Characteristics: In Vitro Analysis. Materials 2019, 12, 2610. [Google Scholar] [CrossRef]
- Pajor, K.; Pajchel, L.; Kolmas, J. Hydroxyapatite and Fluorapatite in Conservative Dentistry and Oral Implantology-A Review. Materials 2019, 12, 2683. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, H.; Alikhah, H.; Alamdari, N.S.; Behzad, M.N.; Mehrabi, E.; Borzui, L.; Bakhshian, F. Developing the use of quality indicators in sterilization practices Iranian. Iran. J. Public Health 2012, 41, 64–69. [Google Scholar] [PubMed]
- Pithon, M.M.; Ferraz, C.S.; Rosa, F.C.S.; Rosa, L.P. Sterilizing elastomeric chains without losing mechanical properties. Is it possible? Dent. Press J. Orthod. 2015, 20, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Harikrishnan, P.; Subha, T.S.; Kavitha, V.; Gnanamani, A. Microbial Adhesion on Orthodontic Ligating Materials: An in Vitro Assessment. Adv. Microbiol. 2013, 3, 108–114. [Google Scholar] [CrossRef]
- Schrøder, S.; Bardow, A.; Eickhardt-Dalbøge, S.; Johansen, H.K.; Homøe, P. Is parotid saliva sterile on entry to the oral cavity? Acta Oto-Laryngol. 2017, 137, 762–764. [Google Scholar] [CrossRef]
- Dzidic, M.; Collado, M.C.; Abrahamsson, T.; Artacho, A.; Stensson, M.; Jenmalm, M.C.; Mira, A. Oral microbiome development during childhood: An ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. 2018, 12, 2292–2306. [Google Scholar] [CrossRef]
- Haukioja, A.; Loimaranta, V.; Tenovuo, J. Probiotic bacteria affect the composition of salivary pellicle and streptococcal adhesion in vitro. Oral Microbiol. Immunol. 2008, 23, 336–343. [Google Scholar] [CrossRef]
- Nikawa, H.; Makihira, S.; Fukushima, H.; Nishimura, H.; Ozaki, Y.; Ishida, K.; Darmawan, S.; Hamada, T.; Hara, K.; Matsumoto, A.; et al. Lactobacillus reuteri in bovine milk fermented decreases the oral carriage of mutans streptococci. Int. J. Food Microbiol. 2004, 95, 219–223. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).