Early-Life Stress Induced Epigenetic Changes of Corticotropin-Releasing Factor Gene in Anorexic Low Body Weight–Selected Chicks
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design
2.2. Body Composition
2.3. Nuclear Protein Extraction and DNMT Activity
2.4. Genomic DNA Extraction
2.5. Global DNA Methylation Quantification
2.6. Bisulfite Conversion, PCR, and Sub-Cloning
2.7. DNA Sequencing and Sequence Analysis
2.8. Chromatin Immunoprecipitation (ChIP) Assay
2.9. Statistical Analysis
3. Results
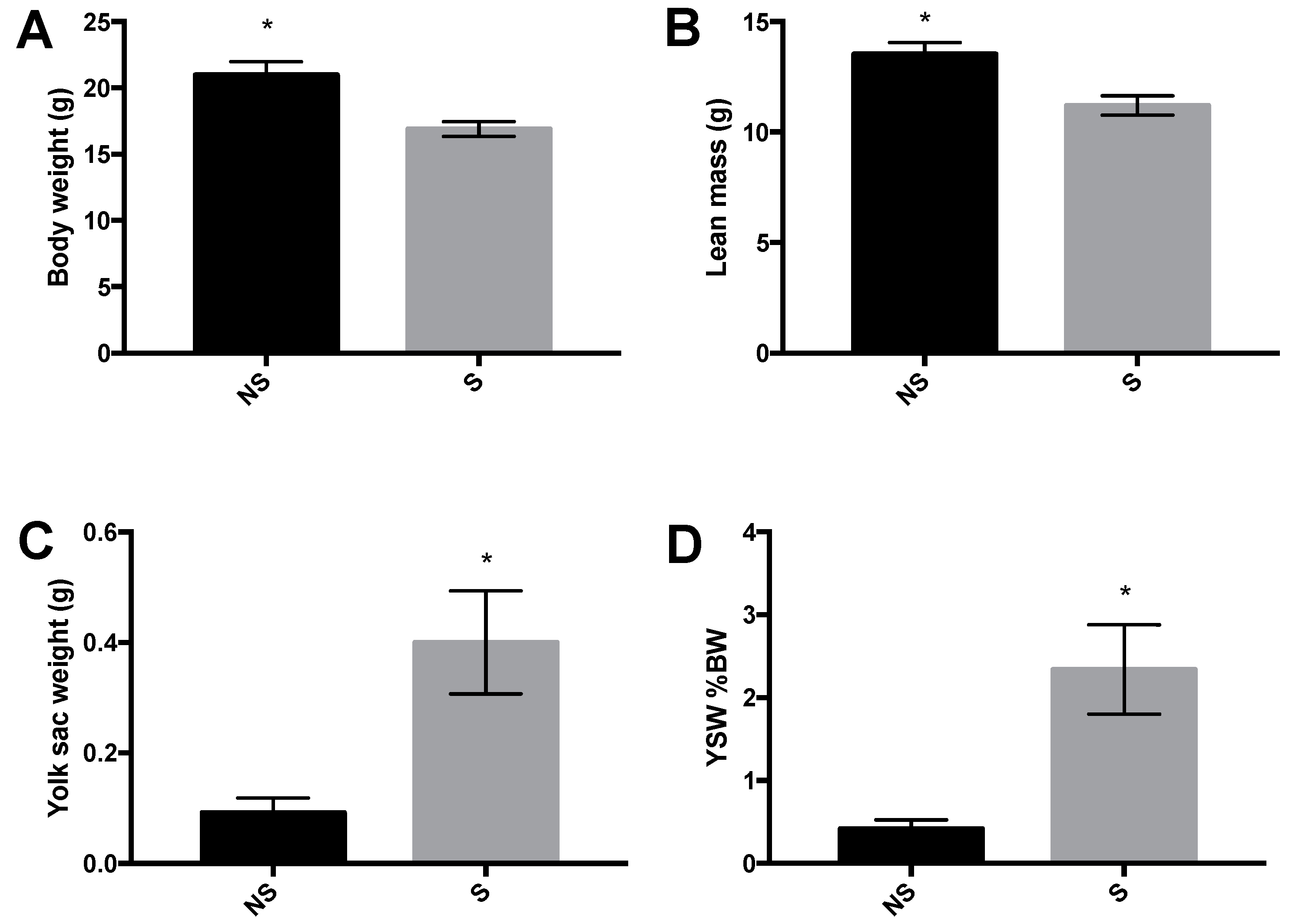
3.1. Body Weight, Body Composition, and Yolk Sac Weight
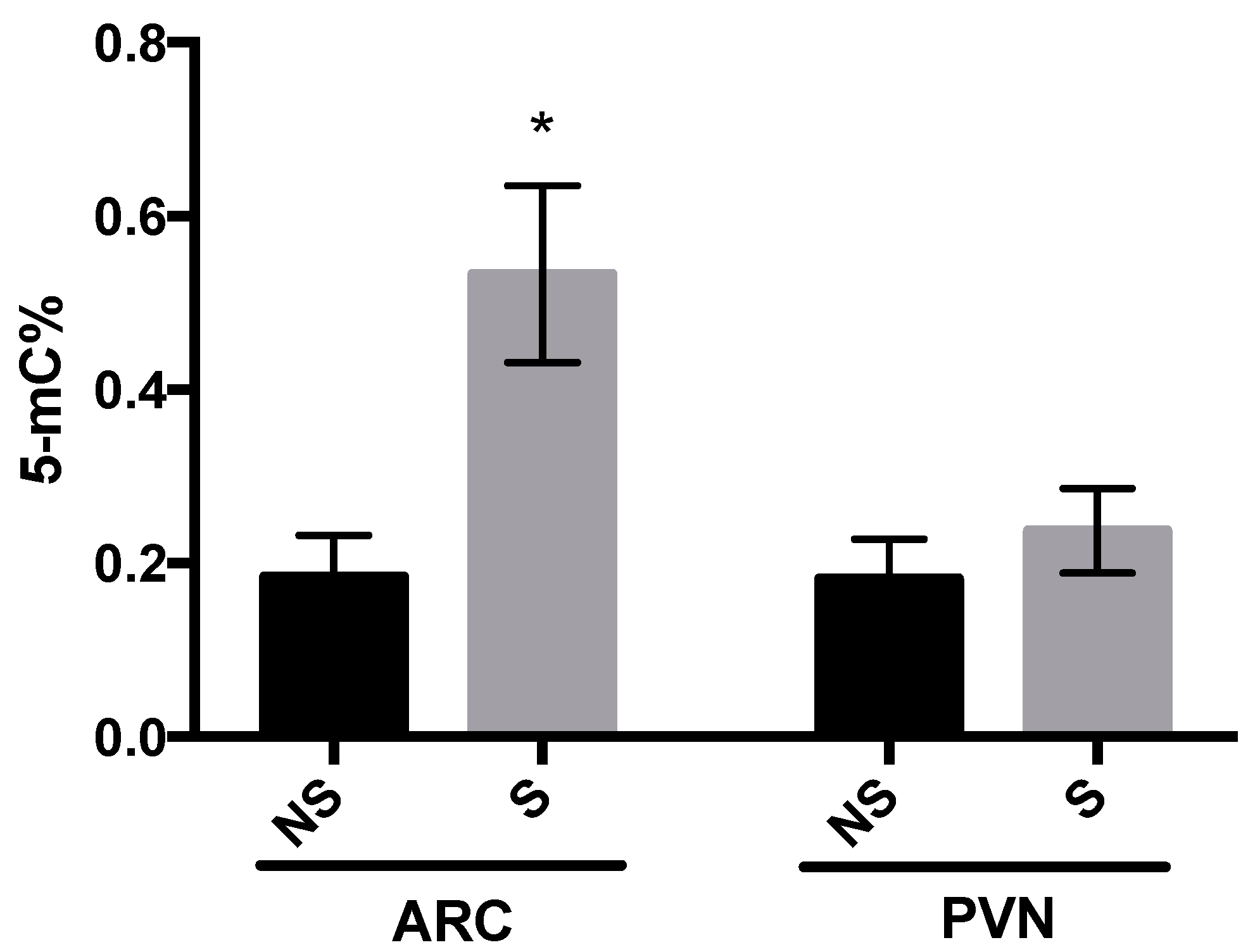
3.2. DNMT Activity and Global DNA Methylation
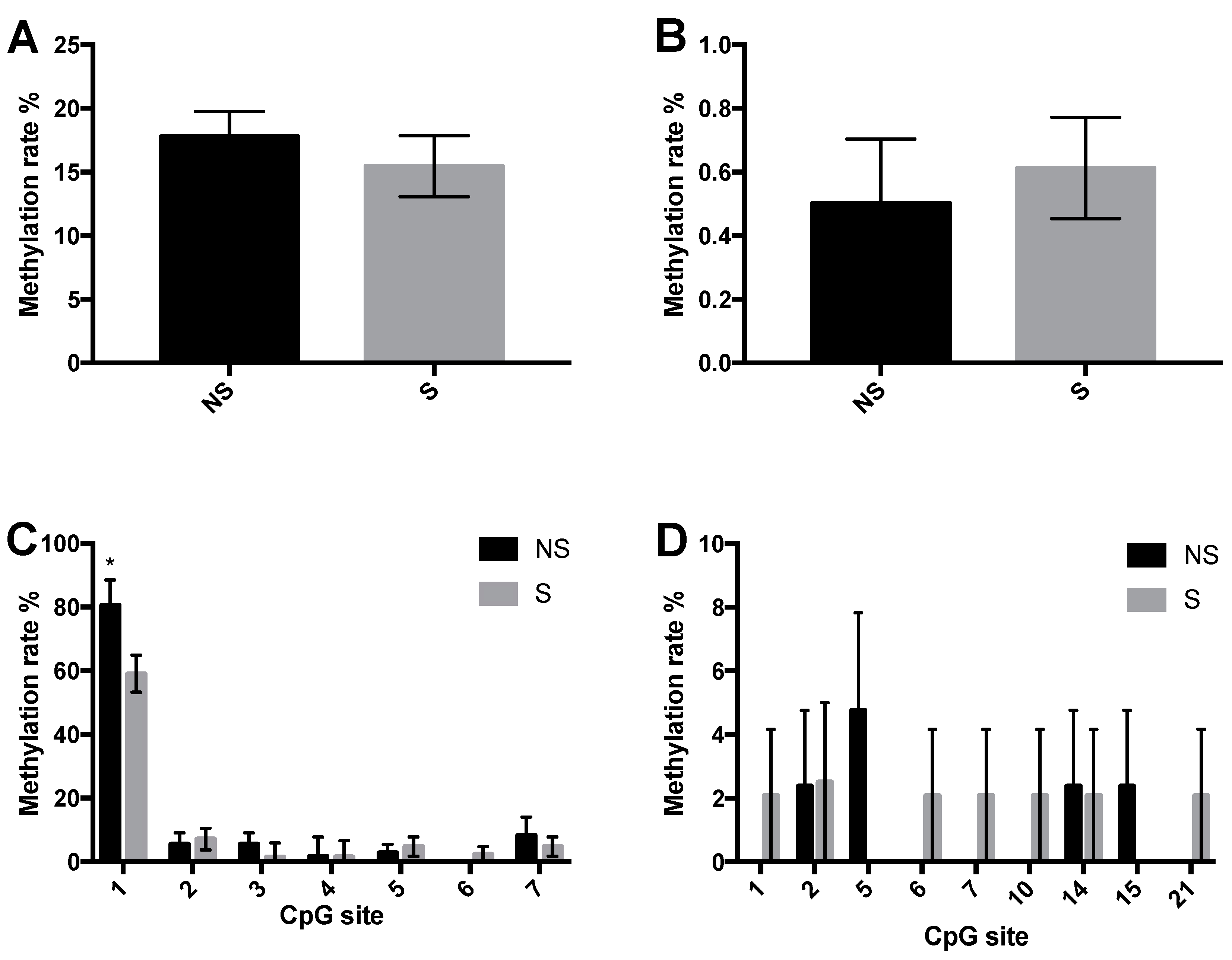
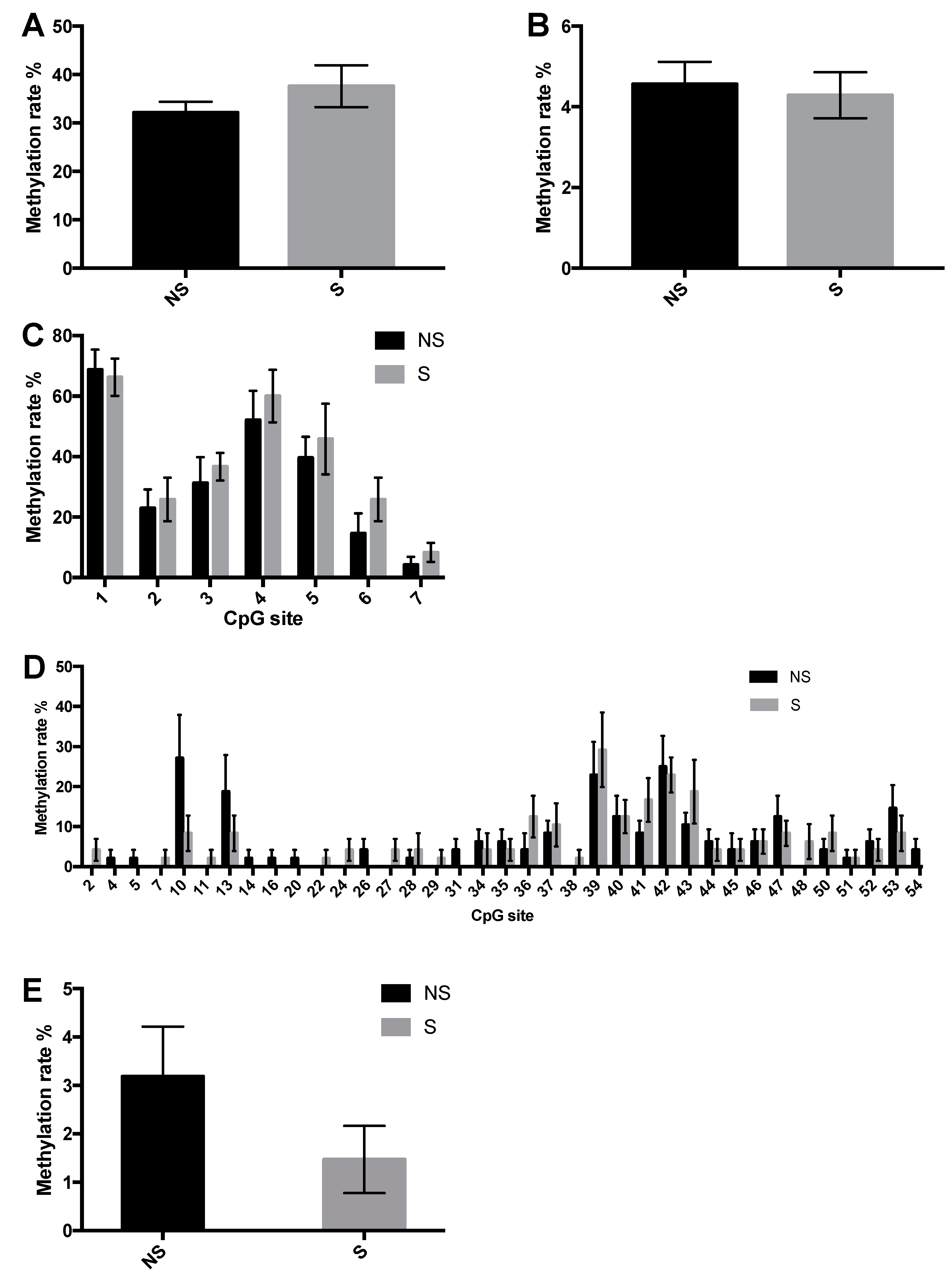
3.3. CpG Site Methylation at the CRF and NPY Gene
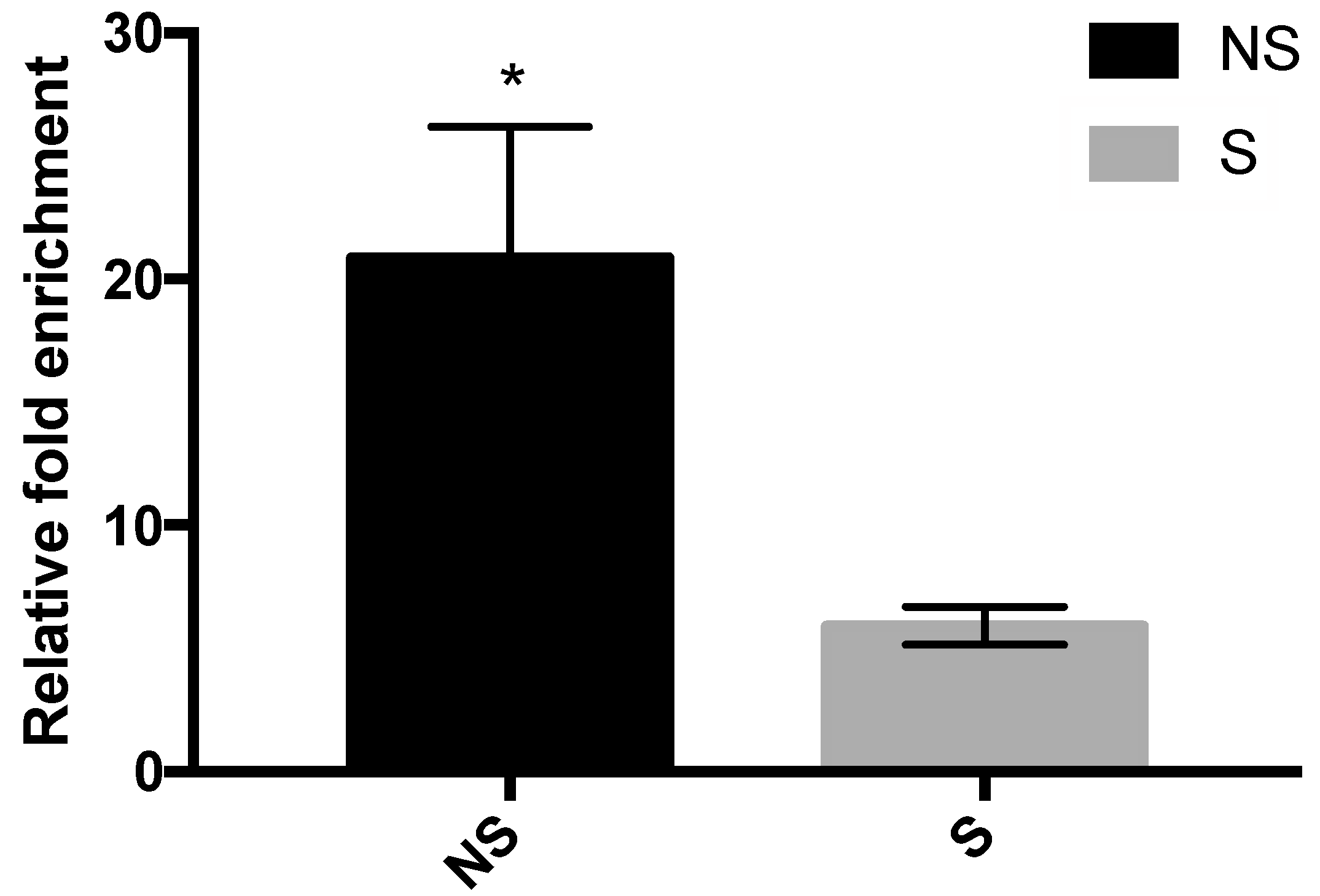
3.4. Binding of MBD2 to the Promoter Region of CRF
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Jambui, M.; Honaker, C.F.; Siegel, P.B. Selection for juvenile body weight in chickens: Standardizing for scaling. Poult. Sci. 2017, 96, 2562–2568. [Google Scholar] [CrossRef] [PubMed]
- Siegel, P.B. Selection for body weight at eight weeks of age1. Short term response and heritabilities. Poult. Sci. 1962, 41, 954–962. [Google Scholar] [CrossRef]
- Newmyer, B.A.; Nandar, W.; Webster, R.I.; Gilbert, E.R.; Siegel, P.B.; Cline, M.A. Neuropeptide Y is associated with changes in appetite-associated hypothalamic nuclei but not food intake in a hypophagic avian model. Behav. Brain Res. 2013, 236, 327–331. [Google Scholar] [CrossRef]
- Wang, J.; Yi, J.; Siegel, P.B.; Cline, M.A.; Gilbert, E.R. Stress-induced suppression of neuropeptide Y-induced hunger in anorexic chicks involves corticotrophin-releasing factor signalling and the paraventricular nucleus of the hypothalamus. J. Neuroendocrinol. 2017, 29, e12555. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Delp, M.S.; Gilbert, E.R.; Siegel, P.B.; Cline, M.A. Anorexia is associated with stress-dependent orexigenic responses to exogenous neuropeptide Y. J. Neuroendocrinol. 2016, 28, e12378. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M. Central regulation of food intake in the neonatal chick. J. Anim. Sci. 2002, 73, 83–94. [Google Scholar] [CrossRef]
- Suzuki, Y.; Nakahara, K.; Maruyama, K.; Okame, R.; Ensho, T.; Inoue, Y.; Murakami, N. Changes in mRNA expression of arcuate nucleus appetite-regulating peptides during lactation in rats. J. Mol. Endocrinol. 2014, 52, 97–109. [Google Scholar] [CrossRef]
- Stanley, S.; Wynne, K.; McGowan, B.; Bloom, S. Hormonal regulation of food intake. Physiol. Rev. 2005, 85, 1131–1158. [Google Scholar] [CrossRef]
- Andermann, M.L.; Lowell, B.B. Toward a Wiring Diagram Understanding of Appetite Control. Neuron 2017, 95, 757–778. [Google Scholar] [CrossRef]
- Maniam, J.; Antoniadis, C.P.; Wang, K.W.; Morris, M.J. Early life stress induced by limited nesting material produces metabolic resilience in response to a high-fat and high-sugar diet in male rats. Front. Endocrinol. 2015, 6, 6. [Google Scholar] [CrossRef]
- Moghadam, H.K.; Johnsen, H.; Robinson, N.; Andersen, O.; Jørgensen, E.H.; Johnsen, H.K.; Baehr, V.J.; Tveiten, H. Impacts of early life stress on the methylome and transcriptome of Atlantic salmon. Sci. Rep. 2017, 7, 5023. [Google Scholar] [CrossRef] [PubMed]
- Schiavone, S.; Colaianna, M.; Curtis, L. Impact of early life stress on the pathogenesis of mental disorders: Relation to brain oxidative stress. Curr. Pharm. Des. 2015, 21, 1404–1412. [Google Scholar] [CrossRef] [PubMed]
- Vaiserman, A.M. Epigenetic programming by early-life stress: Evidence from human populations. Dev. Dyn. 2015, 244, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, C.; Larriva, M.; Boogert, N.J.; Spencer, K.A. Transgenerational transmission of a stress-coping phenotype programmed by early-life stress in the Japanese quail. Sci. Rep. 2017, 7, 46125. [Google Scholar] [CrossRef]
- Murgatroyd, C. Epigenetic changes in the immune systems following early-life stress. In Inflammation and Immunity in Depression; Baune, B.T., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 325–338. [Google Scholar]
- Cramer, T.; Rosenberg, T.; Kisliouk, T.; Meiri, N. Early-life epigenetic changes along the corticotropin-releasing hormone (CRH) gene influence resilience or vulnerability to heat stress later in life. Mol. Psychiatry 2018, 24, 1013–1026. [Google Scholar] [CrossRef]
- Matosin, N.; Cruceanu, C.; Binder, E.B. Preclinical and clinical evidence of DNA methylation changes in response to trauma and chronic stress. Chronic Stress 2017, 1. [Google Scholar] [CrossRef]
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef]
- Painter, R.C.; Roseboom, T.J.; Bleker, O.P. Prenatal exposure to the Dutch famine and disease in later life: An overview. Reprod. Toxicol. 2005, 20, 345–352. [Google Scholar] [CrossRef]
- Khulan, B.; Manning, J.R.; Dunbar, D.R.; Seckl, J.R.; Raikkonen, K.; Eriksson, J.G.; Drake, A.J. Epigenomic profiling of men exposed to early-life stress reveals DNA methylation differences in association with current mental state. Transl. Psychiatry 2014, 4, e448. [Google Scholar] [CrossRef]
- Murgatroyd, C.; Patchev, A.V.; Wu, Y.; Micale, V.; Bockmühl, Y.; Fischer, D.; Holsboer, F.; Wotjak, C.T.; Almeida, O.F.X.; Spengler, D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat. Neurosci. 2009, 12, 1559. [Google Scholar] [CrossRef]
- Van der Doelen, R.H.; Arnoldussen, I.A.; Ghareh, H.; van Och, L.; Homberg, J.R.; Kozicz, T. Early life adversity and serotonin transporter gene variation interact to affect DNA methylation of the corticotropin-releasing factor gene promoter region in the adult rat brain. Dev. Psychopathol. 2015, 27, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Elliott, E.; Ezra-Nevo, G.; Regev, L.; Neufeld-Cohen, A.; Chen, A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat. Neurosci. 2010, 13, 1351. [Google Scholar] [CrossRef]
- Kuenzel, W.J.; Masson, M. A Stereotaxic Atlas of the Brain of the Chick (Gallus Domesticus); Johns Hopkins University Press: Baltimore, MD, USA, 1988. [Google Scholar]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef] [PubMed]
- Scarsdale, J.N.; Webb, H.D.; Ginder, G.D.; Williams, D.C., Jr. Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence. Nucleic Acids Res. 2011, 39, 6741–6752. [Google Scholar] [CrossRef] [PubMed]
- Noy, Y.; Sklan, D. Energy utilization in newly hatched chicks. Poult. Sci. 1999, 78, 1750–1756. [Google Scholar] [CrossRef]
- Bhanja, S.K.; Devi, C.A.; Panda, A.K.; Sunder, G.S. Effect of post hatch feed deprivation on yolk-sac utilization and performance of young broiler chickens. Asian-Australas. J. Anim. Sci. 2009, 22, 1174–1179. [Google Scholar] [CrossRef]
- Mikec, M.; Biđin, Z.; Valentić, A.; Savić, V.; Zelenika, T.A.; Raguž-Đurić, R.; Novak, I.L.; Baleńovic, M. Influence of environmental and nutritional stressors on yolk sac utilization, development of chicken gastrointestinal system and its immune status. Worlds Poult. Sci. J. 2006, 62, 31–40. [Google Scholar] [CrossRef]
- Ipek, A.; Sahan, U. Effects of cold stress on broiler performance and ascites susceptibility. Asian-Australas. J. Anim. Sci. 2006, 19, 734–738. [Google Scholar] [CrossRef]
- Finger, B.C.; Dinan, T.G.; Cryan, J.F. High-fat diet selectively protects against the effects of chronic social stress in the mouse. Neuroscience 2011, 192, 351–360. [Google Scholar] [CrossRef]
- Kaufman, D.; Banerji, M.A.; Shorman, I.; Smith, E.L.; Coplan, J.D.; Rosenblum, L.A.; Kral, J.G. Early-life stress and the development of obesity and insulin resistance in juvenile bonnet macaques. Diabetes 2007, 56, 1382–1386. [Google Scholar] [CrossRef]
- Yam, K.Y.; Naninck, E.F.G.; Abbink, M.R.; la Fleur, S.E.; Schipper, L.; van den Beukel, J.C.; Grefhorst, A.; Oosting, A.; van der Beek, E.M.; Lucassen, P.J.; et al. Exposure to chronic early-life stress lastingly alters the adipose tissue, the leptin system and changes the vulnerability to western-style diet later in life in mice. Psychoneuroendocrinology 2017, 77, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, N.; Hu, X.; Li, J.; Du, Z.; Chen, L.; Yin, G.; Duan, J.; Zhang, H.; Zhao, Y.; et al. Genome-wide mapping of DNA methylation in chicken. PLoS ONE 2011, 6, e19428. [Google Scholar] [CrossRef] [PubMed]
- Binder, E.B.; Nemeroff, C.B. The CRF system, stress, depression and anxiety-insights from human genetic studies. Mol. Psychiatry 2010, 15, 574–588. [Google Scholar] [CrossRef] [PubMed]
- Mueller, B.R.; Bale, T.L. Sex-Specific Programming of Offspring Emotionality after Stress Early in Pregnancy. J. Neurosci. 2008, 28, 9055. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Luu, P.L.; Stirzaker, C.; Clark, S.J. Methyl-CpG-binding domain proteins: Readers of the epigenome. Epigenomics 2015, 7, 1051–1073. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Ballestar, E.; Montoya, G.; Taysavang, P.; Wade, P.A.; Esteller, M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003, 31, 1765–1774. [Google Scholar] [CrossRef]
- Kransdorf, E.P.; Wang, S.Z.; Zhu, S.Z.; Langston, T.B.; Rupon, J.W.; Ginder, G.D. MBD2 is a critical component of a methyl cytosine-binding protein complex isolated from primary erythroid cells. Blood 2006, 108, 2836. [Google Scholar] [CrossRef]
- Desai, M.A.; Webb, H.D.; Sinanan, L.M.; Scarsdale, J.N.; Walavalkar, N.M.; Ginder, G.D.; Williams, D.C., Jr. An intrinsically disordered region of methyl-CpG binding domain protein 2 (MBD2) recruits the histone deacetylase core of the NuRD complex. Nucleic Acids Res. 2015, 43, 3100–3113. [Google Scholar] [CrossRef]
- Blomqvist, A.G.; Soderberg, C.; Lundell, I.; Milner, R.J.; Larhammar, D. Strong evolutionary conservation of neuropeptide Y: Sequences of chicken, goldfish, and torpedo marmorata DNA clones. Proc. Natl. Acad. Sci. USA 1992, 89, 2350–2354. [Google Scholar] [CrossRef]
- Reichmann, F.; Holzer, P. Neuropeptide Y: A stressful review. Neuropeptides 2016, 55, 99–109. [Google Scholar] [CrossRef]
- Hirsch, D.; Zukowska, Z. NPY and stress 30 years later: The peripheral view. Cell Mol. Neurobiol. 2012, 32, 645–659. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer sequence (5′-3′); Forward/Reverse | Genomic Location | Amplicon Length (bp) | Number of CpG Sites | Location (Upstream of TSS) |
|---|---|---|---|---|---|
| CRF | AATCTCATTCAAATATTTTTA/ GAATTTGTGATTAGATTTGG | chr2: 115012110-115012433 | 324 | 24 | −288~35 |
| GGATGTGTAATTTGAAGGAGGTAGA/ AACAAATCCCTCTAAAATCCCTTTA | chr2:115012790-115012952 | 161 | 7 | −830~−670 | |
| NPY | AAAACACCATAAAACTATAA/ TTAGGAGAAGGGTAGTTTAG | chr2: 31464313-31464554 | 242 | 16 | 175~416 |
| ATAGTTTTTTAGAAGGTAGTTATGGG/ AATATCAAATCAATACCACAAACTC | chr2: 31463545-31463709 | 165 | 7 | −594~−430 | |
| GTTTAAGGTTTTTTTTGTTTGT/ ACCTCATAATACCCTACATCTAAAC | chr2: 31464042-31464328 | 287 | 38 | −96~190 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Y.; Wang, J.; Siegel, P.B.; Cline, M.A.; Gilbert, E.R. Early-Life Stress Induced Epigenetic Changes of Corticotropin-Releasing Factor Gene in Anorexic Low Body Weight–Selected Chicks. Life 2020, 10, 51. https://doi.org/10.3390/life10050051
Xiao Y, Wang J, Siegel PB, Cline MA, Gilbert ER. Early-Life Stress Induced Epigenetic Changes of Corticotropin-Releasing Factor Gene in Anorexic Low Body Weight–Selected Chicks. Life. 2020; 10(5):51. https://doi.org/10.3390/life10050051
Chicago/Turabian StyleXiao, Yang, Jinxin Wang, Paul B. Siegel, Mark A. Cline, and Elizabeth R. Gilbert. 2020. "Early-Life Stress Induced Epigenetic Changes of Corticotropin-Releasing Factor Gene in Anorexic Low Body Weight–Selected Chicks" Life 10, no. 5: 51. https://doi.org/10.3390/life10050051
APA StyleXiao, Y., Wang, J., Siegel, P. B., Cline, M. A., & Gilbert, E. R. (2020). Early-Life Stress Induced Epigenetic Changes of Corticotropin-Releasing Factor Gene in Anorexic Low Body Weight–Selected Chicks. Life, 10(5), 51. https://doi.org/10.3390/life10050051






