1. Introduction
Terpenoids are a large and diverse class of metabolites that include compounds essential for cellular functions and environment interactions [
1]. They are the largest and most diverse class of metabolites, containing over 40,000 substances. The molecules are industrially relevant and are used as flavors, pigments, polymers and drugs [
1,
2].
Terpenoids are produced in all living organisms, but they are most abundant and possess a greater degree of diversity in the plant kingdom [
1]. Their biological functions affect plant membrane structure (sterols), respiration (ubiquinone), photosynthesis (chlorophylls, carotenoids, prenylquinones) and the regulation of plant development (cytokines, brassinosteroids, gibberellins, abscisic acid, strigolactones) [
3].
They are derived from two precursor molecules: isopentenyl diphosphate (IPP) and its isomer, dimethylalyl diphosphate (DMAPP) [
4]. There are two pathways used to produce IPP and DMAPP in higher plants: the mevalonate pathway (MVA) and methylerythritol phosphate (MEP) pathways. The MVA pathway is predominantly in the cytosol but is also distributed between the endoplasmic reticulum and peroxisomes [
5,
6], and the MEP pathway is located in plastids [
7]. The key rate-limiting enzymes of the MEP pathway and the MVA pathway are, respectively, the extensively studied 1-deoxy-D-xylulose-5-phosphate synthase (DXS) and hydroxymethylglutaryl-CoA reductase (HMGR) [
7,
8,
9,
10]. However, there are enzymes in the MVA and MEP pathways that are still poorly addressed.
The
mevalonate kinase (
MVK) gene encodes one important biosynthetic enzyme in the MVA pathway [
3]. It catalyzes the phosphorylation of mevalonate to produce 5-phosphate mevalonate [
3].
MVK activity is involved in plant regeneration and growth, and it is also related to the frequency of shoot and root formation in white pine (reviewed in [
7]).
1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) is also an important enzyme involved in terpenoid biosynthesis via the MEP plastid pathway, and is responsible for catalyzing the second step of terpenoid biosynthesis in the chloroplast [
1]. Inactivation of the
1-deoxy-D-xylulose-5-phosphate reductoisomerase (
DXR) gene results in strong developmental arrest and albino plants in
Arabidopsis [
11].
Only plants are capable of simultaneously synthesizing terpenoids via MEP and MVA pathways in parallel, suggesting that genes involved in terpenoid biosynthesis in plants have adapted evolutionarily in basal to terrestrial plant lineages [
12,
13]. In this sense, it is important to understand the evolutionary relationships and duplication events that have occurred within these gene families [
3]. Gene duplication, mutation and natural selection are essential processes that expand plant genetic diversity and facilitate biological adaptation [
14,
15]. Phylogenetic studies can provide important information needed to enhance our knowledge regarding the mechanism by which
MVK and
DXR genes influence terpenoid biosynthesis in higher plants. These genes are both single copy genes in the model plant
Arabidopsis thaliana. Although previous studies report the phylogenetic analyses of
MVK and
DXR [
16,
17,
18], these studies were focused in species-specific gene characterization and did not seek to further investigate the evolutionary aspects of
MVK and
DXR in plants.
The
Coffea genus (Rubiaceae) is comprised of 124 species [
19].
Coffea arabica and
C. canephora are of the species of greatest economic importance and have garnered worldwide interest [
20]. Few molecular studies have addressed the terpenoid biosynthesis pathways in coffee plants. Previously, researchers investigated the roles of the
3-hydroxy-3-methylglutaryl-CoA reductase (
HMGR) gene, which is responsible for the third step of the MVA pathway, and their respective expression profiles in several organs [
21]. Another study validated the function of three
C. arabica monoterpene synthases (limonene, linalool and β-myrcene synthases), and studied the evolution of the genes, which affect the composition of coffee aroma [
22]. Thus far, there have been no studies addressing the molecular responses of hub genes of these pathways.
Resistance inducers are known to modulate the expression pattern of genes involved in terpenoid synthesis [
23]. Hexanoic acid (Hx) is a short, naturally occurring monocarboxylic acid that induces resistance to pathogens in several plant systems [
24]. In
Citrus plants, Hx has been shown to induce genes involved in the MVA pathway [
25]. This prompted us to hypothesize that terpenoid pathway genes can be modulated by the application of Hx in coffee plants.
The main goal of this study was to enhance our understanding of the evolution of both DXR and MVK genes in angiosperm species, and to characterize the transcriptional profiles of these genes in C. arabica plants. Our results provide important knowledge regarding the evolutionary dynamics of the DXR and MVK gene families, and evaluate differences in expression via assessment of gene transcription. In addition, these genes can be further analyzed to verify whether they are involved in plant defense mechanisms aimed at enhancing the biosynthesis of terpenoids.
3. Discussion
MVK and
DXR are key genes involved in MVA and MEP terpenoid biosynthesis pathways. They produce several terpenoid precursors, however, little is known about their molecular evolution [
3]. These genes are present in several plant kingdom lineages, such as algae, mosses, monocotyledons and eudicotyledons [
9,
10,
29]. This study revealed that
MVK and
DXR are low copy number genes in all 24 species analyzed. In contrast to
Arabidopsis, which only has one copy of each gene, several species have more than one copy of
MVK and
DXR.
We observed that the outgroup species,
C. reinhardtii, does not have the
MVK gene. A previous study has shown that another MVA pathway gene,
HMGR, was also absent in the species [
10]. These data suggest that throughout evolution the MVA terpene pathway may have been lost in Chlorophyta. Unusual patterns of MVA pathway has already been reported in a previous study in bacteria [
29].
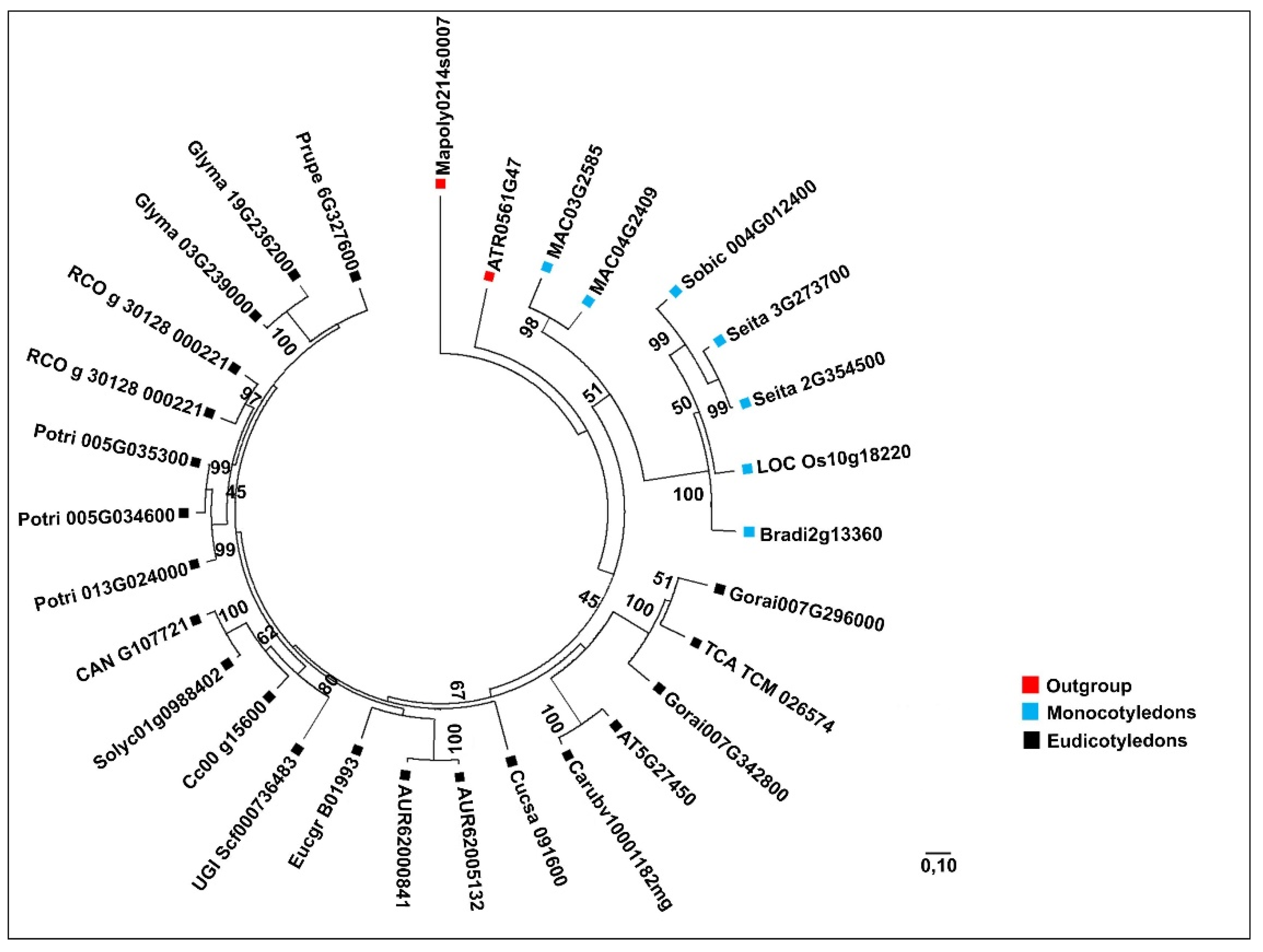
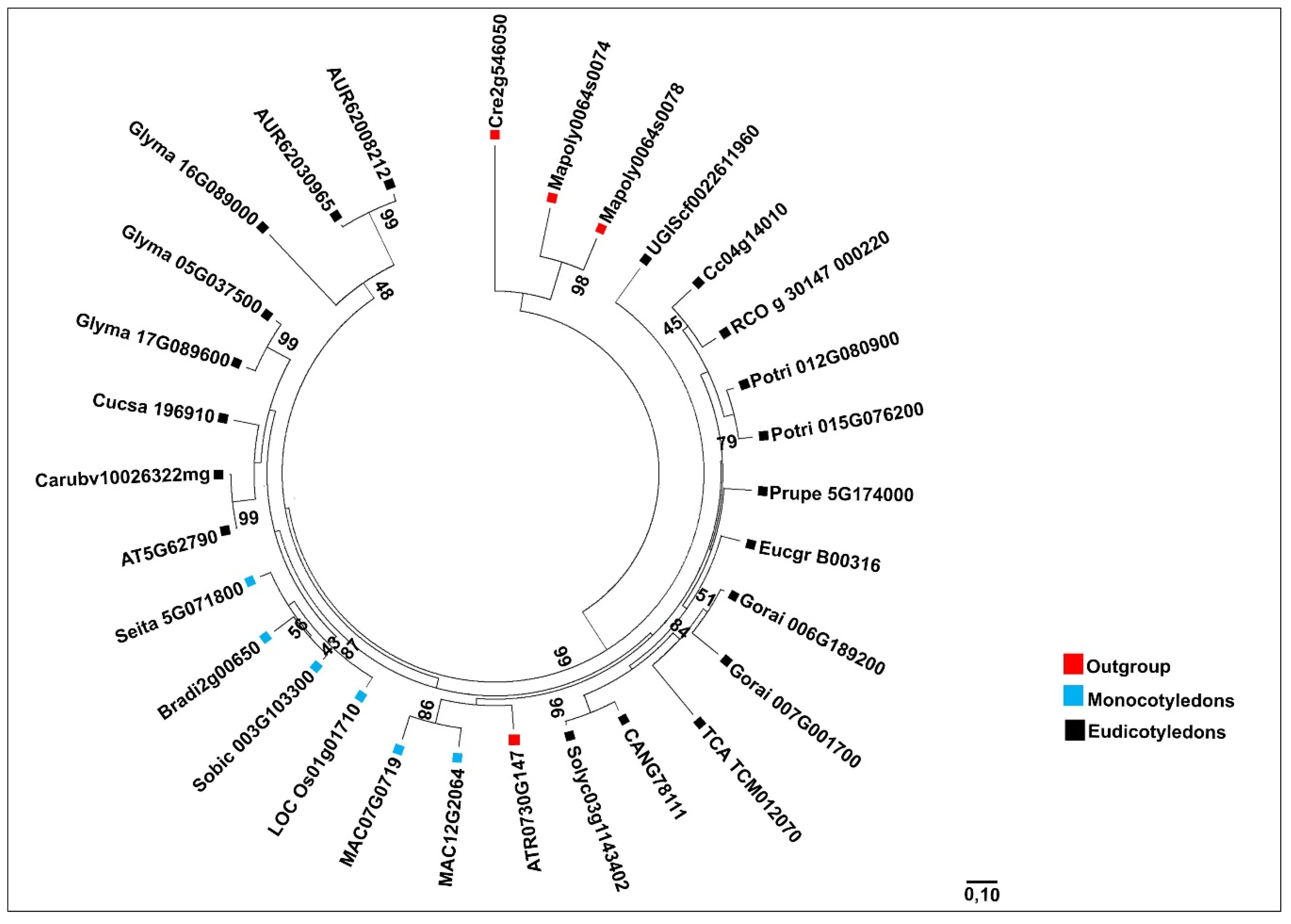
When analyzing the evolutionary relationships of MVK and DXR genes among angiosperm species, we observed that the phylogenetic trees displayed distinct clades, which divided outgroup species, monocotyledon and eudicotyledon lineages.
The
MVK gene tree, clearly divided between monocots and eudicots, is highly similar to
HMGR [
10], suggesting a similar pattern of gene evolution among the MVA pathway. However,
DXR in eudicotyledons have a reticulate pattern, which was not observed for
MVK, suggesting differential evolutionary forces acting on the two genes. A reticulated pattern of gene evolution was also observed in
DXS, the rate-limiting enzyme of the MEP pathway [
9], suggesting that this pattern is common for genes related to the MEP pathway.
Previous studies have shown that
DXR has a greater impact on carotenoid and steroid synthesis than
MVK [
30] and the drastic effect of
DXR loss on plant development is well known [
11]. In this sense, we can also speculate that these observations might be associated with a greater impact of
DXR on terpenoid metabolism.
Most duplications of these genes were associated with recent WGD (whole-genome duplication) events detected in plant species [
15], or to polyploid characteristics of species analyzed. In
C. quinoa, duplication could be explained by the polyploid character of its genome [
31]. In
G. max,
G. raimondii,
P. trichocarpa and
M. acuminata the identification of more than one copy of
MVK can be also associated to recent genome duplication events [
15]. Interestingly,
R. communis (
Table 4) and
S. italica (
Table 5) possessed more than one copy of
MVK and did not experience any recent genome duplication events, but increased the number of genes nonetheless. They underwent a possible local gene species-specific duplication of
MVK, which occurred less than 10 and 5 million years ago, respectively—an unusual occurrence in angiosperms.
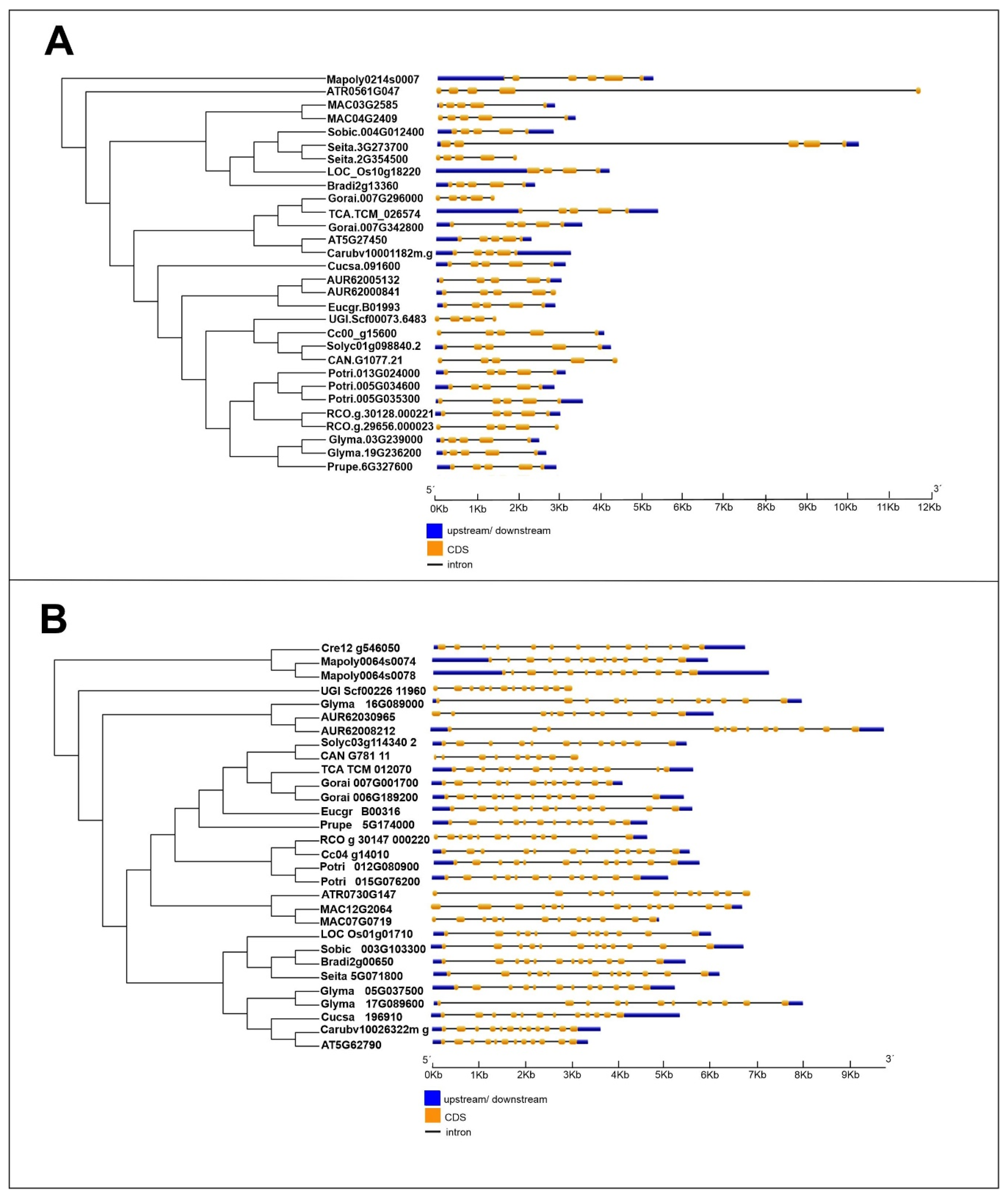
Gene structure and organization analyses revealed that the number of introns and exons within these genes are similar among species. However, the
DXR gene has a more complex exon/intron organization than
MVK, which may also explain its distinct phylogenetic pattern. This observation is in accordance with our findings, indicating that a greater number of recombination events occurred within the
DXR gene than the
MVK gene (
Table 6 and
Table 7). Gene structure and organization analyses revealed that the number of introns and exons within these genes are similar among species.
We observed that the
MVK and
DXR gene families in angiosperms experienced a strong purifying pressure (dN/dS < 1). This fact highlights the functional importance of these genes during their evolutionary process [
32]. Some positive sites indicate a possible diversification between strains of both genes in plants [
32]. Thus, in sites under positive selection, duplication accelerated, which facilitated functional divergence and resulted in the formation of gene subgroups.
Throughout evolution, gene duplication events may be exposed to divergent selection pressure, which leads to non-functionalization (loss of original functions), neofunctionalization (acquisition of new functions) or sub-functionalization (partition of original functions) [
9,
10,
14]. In this study, dN/dS ratios for duplicated
MVK and
DXR genes were less than 1, suggesting that genes were predominantly under strong purifying selection pressure.
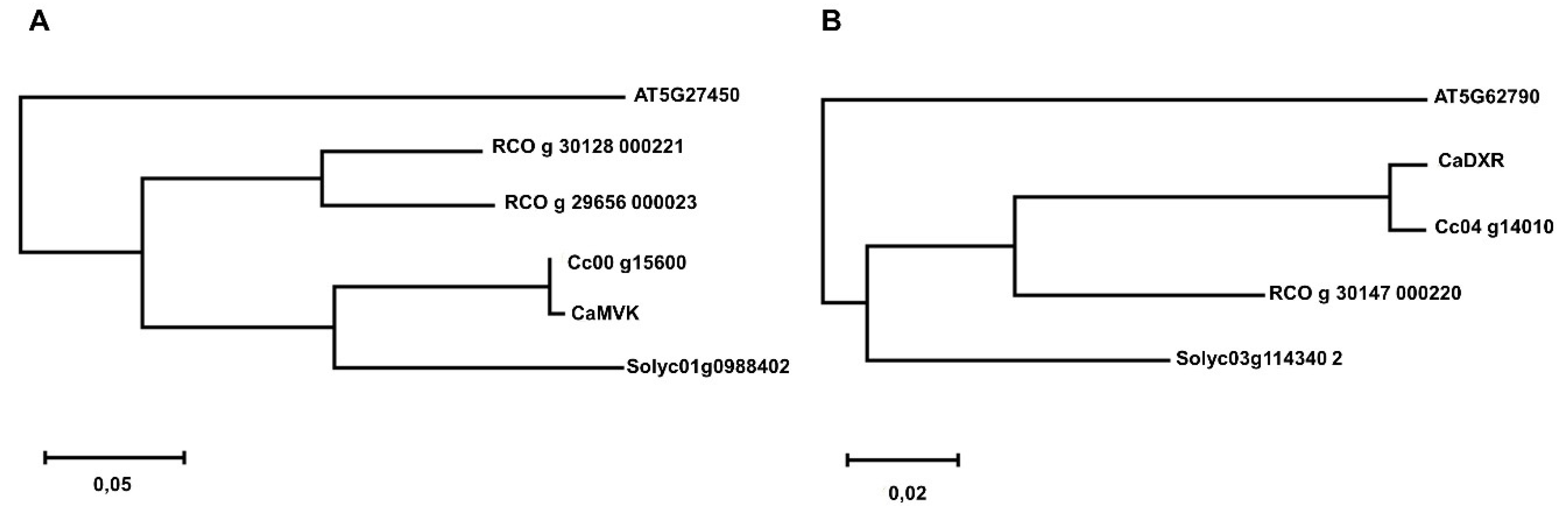
To better understand selective pressure, we estimated the age of MVK and DXR gene duplications using dN/dS data. This confirmed the recent tandem duplication of MVK in P. trichocarpa and WGD duplications in G. max.
We also observed a direct relation between copy number among
MVK and
DXR with the respective rate-limiting genes from these pathways—
HMGR and
DXS. As examples of gene duplication in
MVK, we can highlight
G. raimondii,
G. max and
P. trichocarpa. These species were also the ones with higher number of copies of
HMGR [
10]. In
DXR,
Arabidopsis thaliana and
Eucalyptus grandis are species with a single copy of
DXR; both species have three copies of
DXS [
9]. Conversely,
G. max, the species with the highest number of DXR copies, also harbored the highest number of
DXS copies in a similar analysis [
9].
Using MEME analysis, we identified amino acid sites that experienced episodic positive selection. We observed an opposite pattern of positively selected sites in
MVK and
DXR genes: while more positively selected sites were identified in monocot
MVK genes, for
DXR most positively selected sites were in dicot genes (
Table 3). The amount of positively selected sites helped us to highlight natural selection effects on individual sites that are usually hindered by the low averaged dN/dS values.The analysis of dN/dS among individual amino acid residues in
DXR (
Figures S4 and S5) also indicated that higher values were in the N-terminal, where the protein has its targeting signal to organelles [
33].
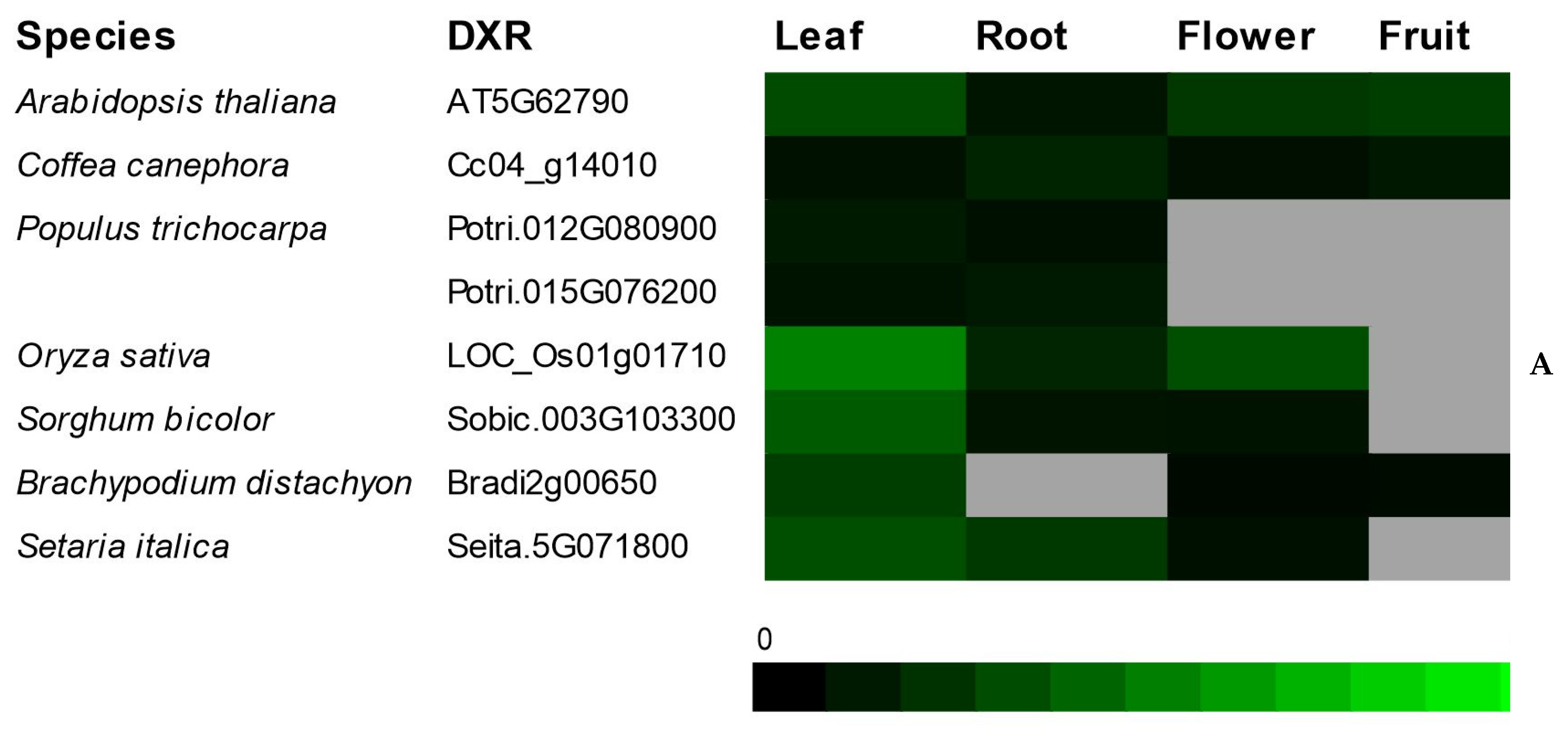
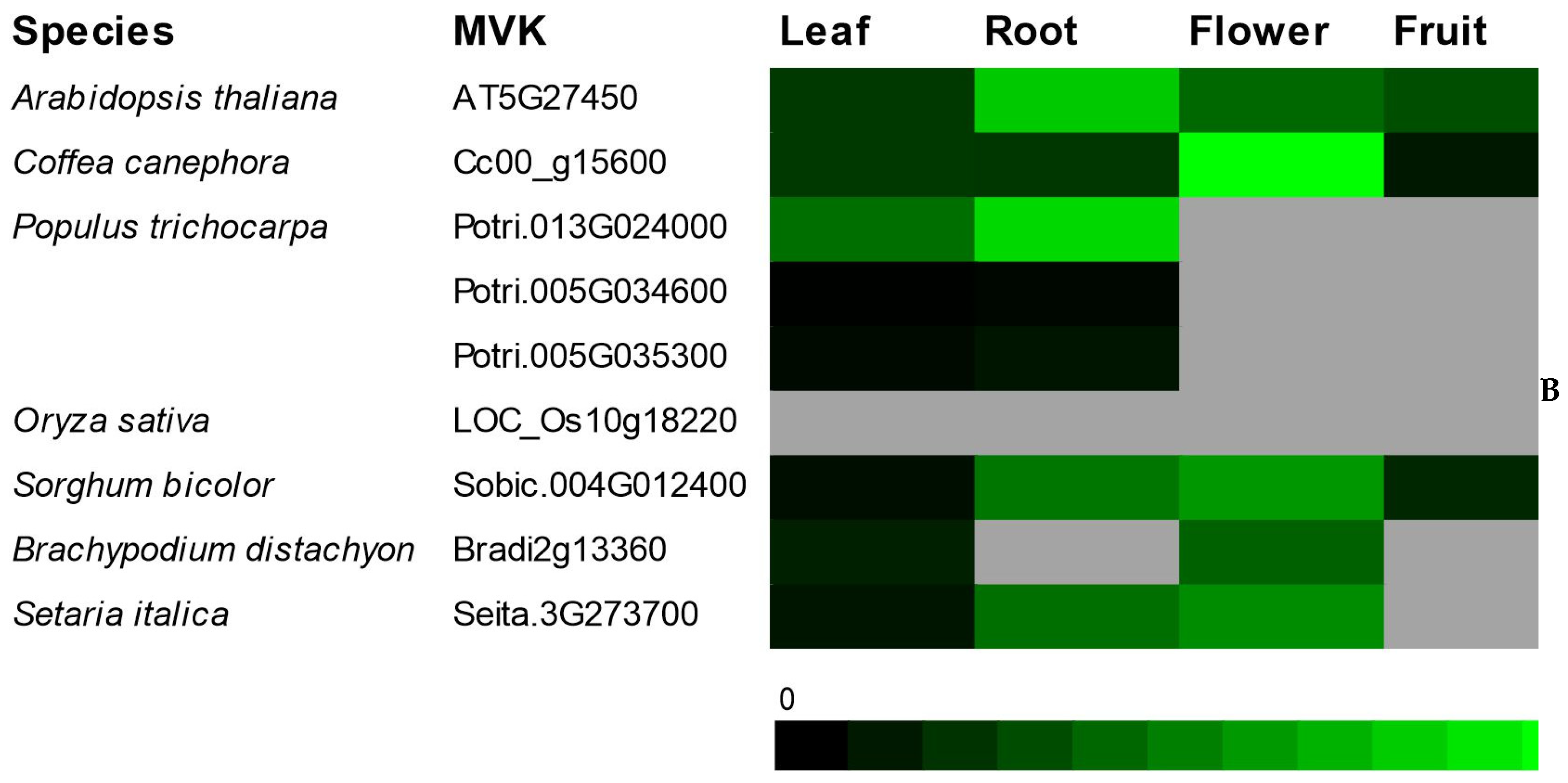
The transcriptional profile of genes involved in the initial steps of terpenoid biosynthesis in the
Coffea genus has, thus far, been poorly investigated. However, the
MVK and
DXR transcriptional profiles in model plants were already extensively studied [
8,
34].
MVK was highly expressed in roots and flowers, while the transcript levels of
DXR was more abundant in photosynthetic tissues [
7,
8,
34].
We verified the expression levels of
CaMVK and
CaDXR genes in
C. arabica organs using RNA-seq data and RT-qPCR. We observed higher transcript levels of these genes in flowers for both analyses. The high levels of expression observed in flowers can be explained by the fact that several terpenes are produced in this organ [
34,
35], most of which are plant volatile compounds, such as limonene, one of the main monoterpene compounds responsible for the aroma of coffee [
22]. In addition, we observed in the RNA-seq data that
CaMVK and
CaDXR are upregulated in coffee fruits under MJ treatment compared to mock-control plants, including most of the genes involved in the MVA and MEP pathways. Several studies already described methyl jasmonate function as a potent elicitor of secondary metabolite production [
36,
37] and hexanoic acid as a plant resistance inductor by producing natural products [
24,
25].
Our RT-qPCR results showed that hexanoic acid is able to upregulate expression of
CaMVK in
C. arabica cv. Obatã roots, a result that is in agreement with previous reports investigating gene expression in
Citrus plants [
25]. In the same Hx experiment, the
DXR gene was downregulated in the leaves of
C. arabica cv. Catuaí Vermelho, suggesting that modulation of expression might be genotype-specific. The upregulation of transcription of the
MVK gene in Hx-treated roots was detected only in the Obatã cultivar. Upregulation of the terpenoid pathway can provide protection against stresses, as previously observed in studies investigating tobacco [
38], tomato [
39] and maize [
40]. In addition, it is important to highlight that Obatã and Catuaí Vermelho cultivars have distinct breeding stories, and they are classified in distinct genetic groups [
41]. In this study, we provide insights to show that distinct genetic groups generated by coffee breeding might result in dissimilar transcriptional responses.