A Polymorphism in the Catalase Gene Promoter Confers Protection against Severe RSV Bronchiolitis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genotyping for SNPs
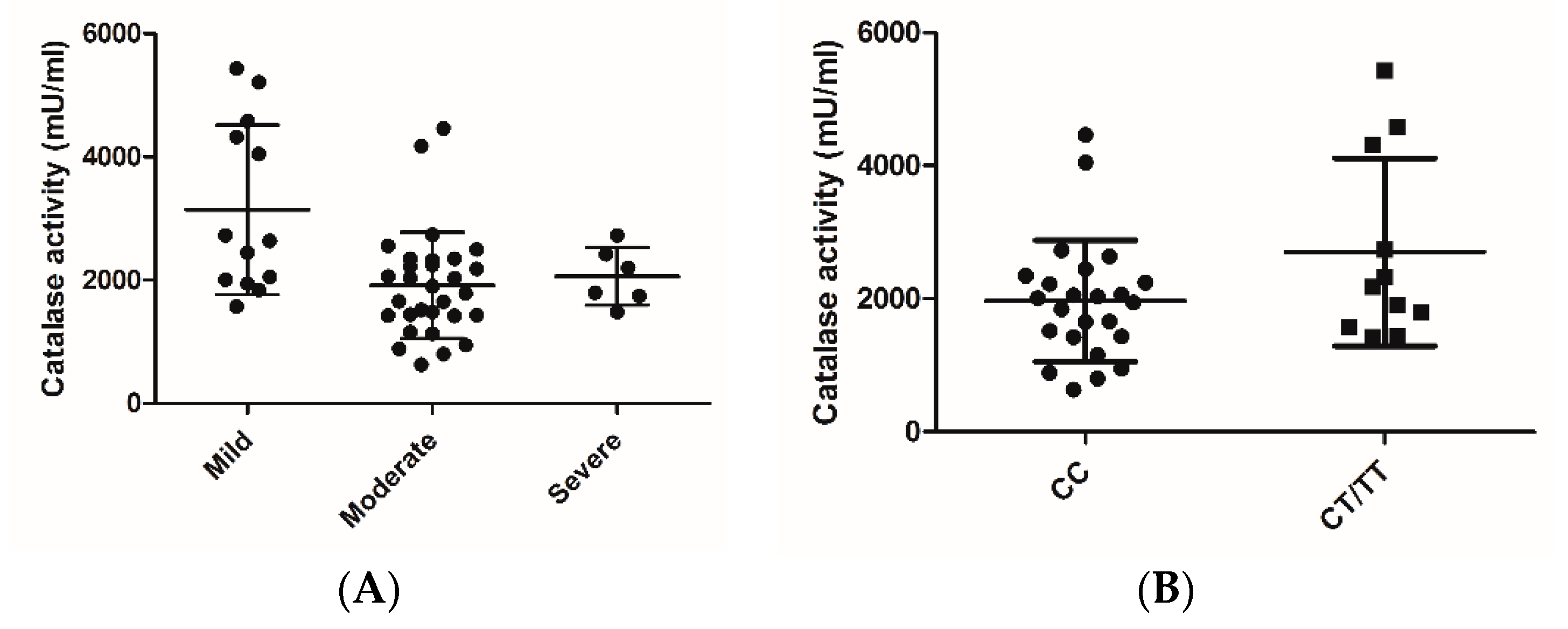
2.3. Catalase Activity
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shi, T.; McAllister, D.A.; O’Brien, K.L.; Simoes, E.A.F.; Madhi, S.A.; Gessner, B.D.; Polack, F.P.; Balsells, E.; Acacio, S.; Aguayo, C.; et al. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: A systematic review and modelling study. Lancet 2017, 390, 946–958. [Google Scholar] [CrossRef]
- Nair, H.; Nokes, D.J.; Gessner, B.D.; Dherani, M.; Madhi, S.A.; Singleton, R.J.; O’Brien, K.L.; Roca, A.; Wright, P.F.; Bruce, N.; et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: A systematic review and meta-analysis. Lancet 2010, 375, 1545–1555. [Google Scholar] [CrossRef]
- The IMpact-RSV Study Group. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. Pediatrics 1998, 102, 531–537. [Google Scholar] [CrossRef]
- Casola, A.; Burger, N.; Liu, T.; Jamaluddin, M.; Brasier, A.R.; Garofalo, R.P. Oxidant tone regulates RANTES gene transcription in airway epithelial cells infected with respiratory syncytial cirus: Role in viral-induced Interferon Regulatory Factor activation. J. Biol. Chem. 2001, 276, 19715–19722. [Google Scholar] [CrossRef]
- Liu, T.; Castro, S.; Brasier, A.R.; Jamaluddin, M.; Garofalo, R.P.; Casola, A. Reactive oxygen species mediate virus-induced STAT activation: Role of tyrosine phosphatases. J. Biol. Chem. 2004, 279, 2461–2469. [Google Scholar] [CrossRef]
- Li, W.; Kong, A.N. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol. Carcinog. 2009, 48, 91–104. [Google Scholar] [CrossRef]
- Hosakote, Y.M.; Liu, T.; Castro, S.M.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus induces oxidative stress by modulating antioxidant enzymes. Am. J. Respir. Cell Mol. Biol. 2009, 41, 348–357. [Google Scholar] [CrossRef]
- Hosakote, Y.M.; Jantzi, P.D.; Esham, D.L.; Spratt, H.; Kurosky, A.; Casola, A.; Garofalo, R.P. Viral-mediated inhibition of antioxidant enzymes contributes to the pathogenesis of severe respiratory syncytial virus bronchiolitis. Am. J. Respir. Crit. Care Med. 2011, 183, 1550–1560. [Google Scholar] [CrossRef]
- Komaravelli, N.; Tian, B.; Ivanciuc, T.; Mautemps, N.; Brasier, A.R.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus infection down-regulates antioxidant enzyme expression by triggering deacetylation-proteasomal degradation of NRF2. Free Radic. Biol. Med. 2015, 88, 391–403. [Google Scholar] [CrossRef]
- Komaravelli, N.; Ansar, M.; Garofalo, R.P.; Casola, A. Respiratory syncytial virus induces NRF2 degradation through a promyelocytic leukemia protein-ring finger protein 4 dependent pathway. Free Radic. Biol. Med. 2017, 113, 494–504. [Google Scholar] [CrossRef]
- Ciencewicki, J.; Trivedi, S.; Kleeberger, S.R. Oxidants and the pathogenesis of lung diseases. J. Allergy Clin. Immunol. 2008, 122, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Renkonen, J.; Joenvaara, S.; Parviainen, V.; Mattila, P.; Renkonen, R. Network analysis of single nucleotide polymorphisms in asthma. J. Asthma Allergy 2010, 3, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Yuan, B.; Lopez, E.; Bai, C.; Wang, X. Gene polymorphisms and chronic obstructive pulmonary disease. J. Cell Mol. Med. 2014, 18, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Marzec, J.; Kleeberger, S.R. Functional polymorphisms in Nrf2: Implications for human disease. Free Radic. Biol. Med. 2015, 88, 362–372. [Google Scholar] [CrossRef]
- Canova, C.; Dunster, C.; Kelly, F.J.; Minelli, C.; Shah, P.L.; Caneja, C.; Tumilty, M.K.; Burney, P. PM10-induced hospital admissions for asthma and chronic obstructive pulmonary disease: The modifying effect of individual characteristics. Epidemiology 2012, 23, 607–615. [Google Scholar] [CrossRef]
- Malling, T.H.; Sigsgaard, T.; Brasch-Andersen, C.; Frischknecht, L.; Andersen, H.R.; Kruse, T.A.; Sherson, D.; Skadhauge, L.R.; Thomsen, G.; Baelum, J.; et al. Genetic polymorphisms in antioxidative enzymes are associated to forced expiratory volume in 1 s (FEV1) in smokers independently of asthma. Clin. Respir. J. 2012, 6, 46–55. [Google Scholar] [CrossRef]
- Chambliss, J.; Kelley, J.P.; Palkowetz, K.; Spratt, H.; Garofalo, R.P.; Casola, A. A Polymorphism in the Catalase Gene Promoter Confers Protection Against Severe RSV Bronchiolitis. J. Allergy Clin. Immunol. 2018, 141, AB10. [Google Scholar] [CrossRef]
- Garofalo, R.P.; Patti, J.; Hintz, K.A.; Hill, V.; Ogra, P.L.; Welliver, R.C. Macrophage inflammatory protein 1-alpha, and not T-helper type 2 cytokines, is associated with severe forms of bronchiolitis. J. Infect. Dis. 2001, 184, 393–399. [Google Scholar] [CrossRef]
- Laham, F.R.; Trott, A.A.; Bennett, B.L.; Kozinetz, C.A.; Jewell, A.M.; Garofalo, R.P.; Piedra, P.A. LDH concentration in nasal-wash fluid as a biochemical predictor of bronchiolitis severity. Pediatrics 2010, 125, e225–e233. [Google Scholar] [CrossRef]
- Mehta, R.; Scheffler, M.; Tapia, L.; Aideyan, L.; Patel, K.D.; Jewell, A.M.; Avadhanula, V.; Mei, M.; Garofalo, R.P.; Piedra, P.A. Lactate dehydrogenase and caspase activity in nasopharyngeal secretions are predictors of bronchiolitis severity. Influenza Other Respir. Viruses 2014, 8, 617–625. [Google Scholar] [CrossRef]
- Kodydkova, J.; Vavrova, L.; Kocik, M.; Zak, A. Human catalase, its polymorphisms, regulation and changes of its activity in different diseases. Folia Biol. (Praha) 2014, 60, 153–167. [Google Scholar] [PubMed]
- Fridovich, I. Biological effects of the superoxide radical. Arch. Biochem. Biophys. 1986, 247, 1–11. [Google Scholar] [CrossRef]
- Kinnula, V.L.; Crapo, J.D. Superoxide dismutases in the lung and human lung diseases. Am. J. Respir. Crit. Care Med. 2003, 167, 1600–1619. [Google Scholar] [CrossRef] [PubMed]
- Rahman, I. Redox signaling in the lungs. Antioxid. Redox Signal. 2005, 7, 1–5. [Google Scholar] [CrossRef]
- Betsuyaku, T.; Fuke, S.; Inomata, T.; Kaga, K.; Morikawa, T.; Odajima, N.; Adair-Kirk, T.; Nishimura, M. Bronchiolar epithelial catalase is diminished in smokers with mild COPD. Eur. Respir. J. 2013, 42, 42–53. [Google Scholar] [CrossRef]
- Hebert-Schuster, M.; Fabre, E.E.; Nivet-Antoine, V. Catalase polymorphisms and metabolic diseases. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 397–402. [Google Scholar] [CrossRef]
- Forsberg, L.; Lyrenas, L.; de Faire, U.; Morgenstern, R. A common functional C-T substitution polymorphism in the promoter region of the human catalase gene influences transcription factor binding, reporter gene transcription and is correlated to blood catalase levels. Free Radic. Biol. Med. 2001, 30, 500–505. [Google Scholar] [CrossRef]
- Mak, J.C.; Leung, H.C.; Ho, S.P.; Ko, F.W.; Cheung, A.H.; Ip, M.S.; Chan-Yeung, M.M. Polymorphisms in manganese superoxide dismutase and catalase genes: Functional study in Hong Kong Chinese asthma patients. Clin. Exp. Allergy 2006, 36, 440–447. [Google Scholar] [CrossRef]
- Islam, T.; McConnell, R.; Gauderman, W.J.; Avol, E.; Peters, J.M.; Gilliland, F.D. Ozone, oxidant defense genes, and risk of asthma during adolescence. Am. J. Respir. Crit. Care Med. 2008, 177, 388–395. [Google Scholar] [CrossRef]
| All | Mild | Moderate and Severe | Moderate Only | Severe Only | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n = 136 | n = 64 | n = 72 | n = 57 | n = 15 | ||||||
| Median Age, Months | 6 | 9 | 4 | 5 | 3 | |||||
| Gender | ||||||||||
| Males: n (%) | 86 | 63.2% | 40 | 62.5% | 46 | 63.9% | 37 | 64.9% | 9 | 60.0% |
| Females: n (%) | 50 | 36.7% | 24 | 37.5% | 26 | 36.1% | 20 | 35.1% | 6 | 40.0% |
| Race | ||||||||||
| Caucasian: n (%) | 67 | 49.3% | 33 | 51.6% | 34 | 47.2% | 27 | 47.4% | 7 | 46.7% |
| Hispanic: n (%) | 37 | 27.2% | 14 | 21.9% | 23 | 31.9% | 20 | 37.1% | 3 | 20.0% |
| African American: n (%) | 32 | 23.5% | 17 | 26.7% | 15 | 20.8% | 10 | 17.5% | 5 | 33.3% |
| Median Length of Hospital Stay (Days) | 2 | 1 | 6 | 5 | 8 | |||||
| Percent Hospitalized | 64.1% | 100% | ||||||||
| Preterm (<38 Weeks Gestation): n (%) | 21 | 15.5% | 2 | 3% | 19 | 26.4% | 15 | 26.3% | 4 | 26.7% |
| Smoke Exposure: n (%) | ||||||||||
| Yes: n (%) | 28 | 20.6% | 11 | 17.2% | 17 | 23.6% | 11 | 19.3% | 6 | 40.0% |
| No: n (%) | 89 | 65.5% | 40 | 62.5% | 49 | 68.1% | 40 | 70.2% | 9 | 60.0% |
| Not documented: n (%) | 19 | 14.0% | 13 | 20.3% | 6 | 8.3% | 6 | 10.5% | 0 | 0.0% |
| Personal Atopic History | ||||||||||
| Yes: n (%) | 34 | 25.0% | 15 | 23.4% | 19 | 26.4% | 17 | 29.8% | 2 | 13.3% |
| No: n (%) | 102 | 75.0% | 49 | 76.6% | 53 | 73.6% | 40 | 70.2% | 13 | 86.7% |
| Atopic Family History | ||||||||||
| Yes: n (%) | 28 | 20.6% | 13 | 20.3% | 15 | 20.8% | 11 | 19.3% | 4 | 26.7% |
| No: n (%) | 73 | 53.7% | 28 | 43.8% | 45 | 62.5% | 36 | 63.2% | 9 | 60.0% |
| Not documented: n (%) | 35 | 25.7% | 23 | 35.9% | 12 | 16.7% | 10 | 17.6% | 2 | 13.3% |
| SNPs Analyzed | p-Value | Odd Ratio | BH Q-Value | |
|---|---|---|---|---|
| Catalase | rs1001179 | 0.0122 | 0.38 (0.14, 0.93) | 0.0854 |
| Glutathione peroxidase 1 | rs1050450 | 1 | 0.99 (0.5, 1.96) | 1 |
| Glutathione S-transferase P | rs1695 | 0.617 | 1.28 (0.63, 2.61) | 1 |
| rs1138272 | 1 | 1.18 (0.3, 5) | 1 | |
| NRF2 promoter | rs6721961 | 0.385 | 0.65 (0.29, 1.43) | 1 |
| NRF2 intron | rs1806649 | 0.561 | 1.37 (0.62, 3.14) | 1 |
| Superoxide dismutase 2 | rs4880 | 1 | 0.97 (0.49, 1.92) | 1 |
| African American Population | Study Sample | p-Value | Caucasian Population | Study Sample | p-Value | Hispanic Population | Study Sample | p-Value | |
|---|---|---|---|---|---|---|---|---|---|
| CC | 90.2% | 90.6% | NS | 54.5% | 74.6% | <0.001 | 71.9% | 89.2% | 0.017 |
| CT/TT | 9.8% | 9.4% | 45.5% | 25.4% | 28.1% | 10.8% |
| African American, n = 32 | CC | p-Value | CT/TT | ||
| Mild | 16 | 94% | NS | 1 | 6% |
| Moderate and Severe | 13 | 87% | NS | 2 | 13% |
| Severe only | 5 | 100% | 0 | 0% | |
| Caucasian, n = 67 | |||||
| Mild | 20 | 61% | NS | 13 | 39% |
| Moderate and Severe | 30 | 88% | 4.0 × 10−5 | 4 | 12% |
| Severe only | 7 | 100% | 0 | 0% | |
| Hispanic, n = 37 | |||||
| Mild | 12 | 86% | NS | 2 | 14% |
| Moderate and Severe | 21 | 91% | 0.03 | 2 | 9% |
| Severe only | 3 | 100% | 0 | 0% | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chambliss, J.M.; Ansar, M.; Kelley, J.P.; Spratt, H.; Garofalo, R.P.; Casola, A. A Polymorphism in the Catalase Gene Promoter Confers Protection against Severe RSV Bronchiolitis. Viruses 2020, 12, 57. https://doi.org/10.3390/v12010057
Chambliss JM, Ansar M, Kelley JP, Spratt H, Garofalo RP, Casola A. A Polymorphism in the Catalase Gene Promoter Confers Protection against Severe RSV Bronchiolitis. Viruses. 2020; 12(1):57. https://doi.org/10.3390/v12010057
Chicago/Turabian StyleChambliss, Jeffrey M., Maria Ansar, John P. Kelley, Heidi Spratt, Roberto P. Garofalo, and Antonella Casola. 2020. "A Polymorphism in the Catalase Gene Promoter Confers Protection against Severe RSV Bronchiolitis" Viruses 12, no. 1: 57. https://doi.org/10.3390/v12010057
APA StyleChambliss, J. M., Ansar, M., Kelley, J. P., Spratt, H., Garofalo, R. P., & Casola, A. (2020). A Polymorphism in the Catalase Gene Promoter Confers Protection against Severe RSV Bronchiolitis. Viruses, 12(1), 57. https://doi.org/10.3390/v12010057






