Comprehensive Profiling of Surface Gangliosides Extracted from Various Cell Lines by LC-MS/MS
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Membrane Extraction
2.3. Purification and Enrichment of Gangliosides from Cell Membrane
2.4. Negative Nano-LC Chip Q-TOF MS Analysis of Gangliosides
2.5. Ganglioside Identification
3. Results
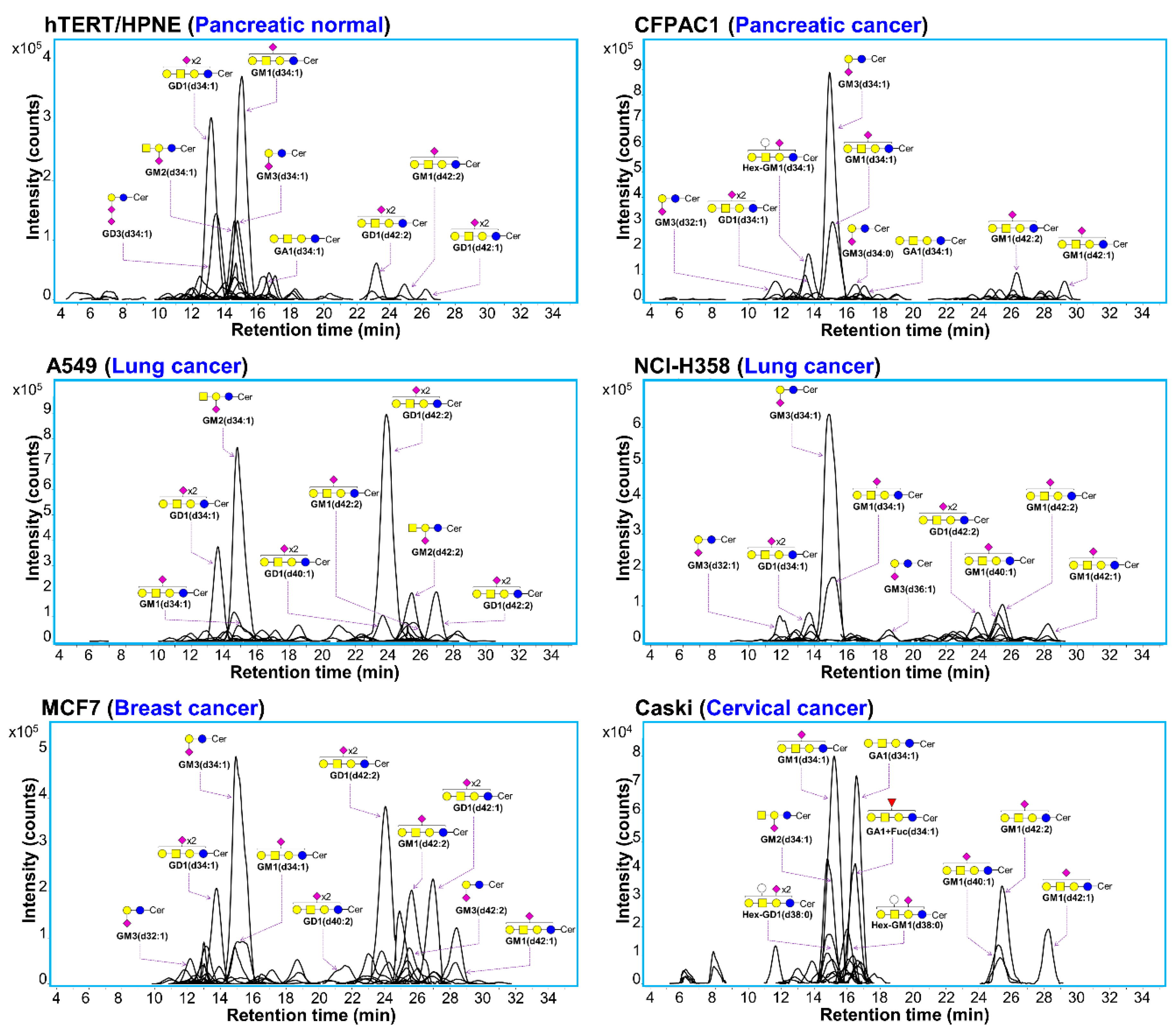
3.1. Profiling of Gangliosides by Negative Ion Detection Mode LC/MS
3.2. Identification of Cell Surface Gangliosides
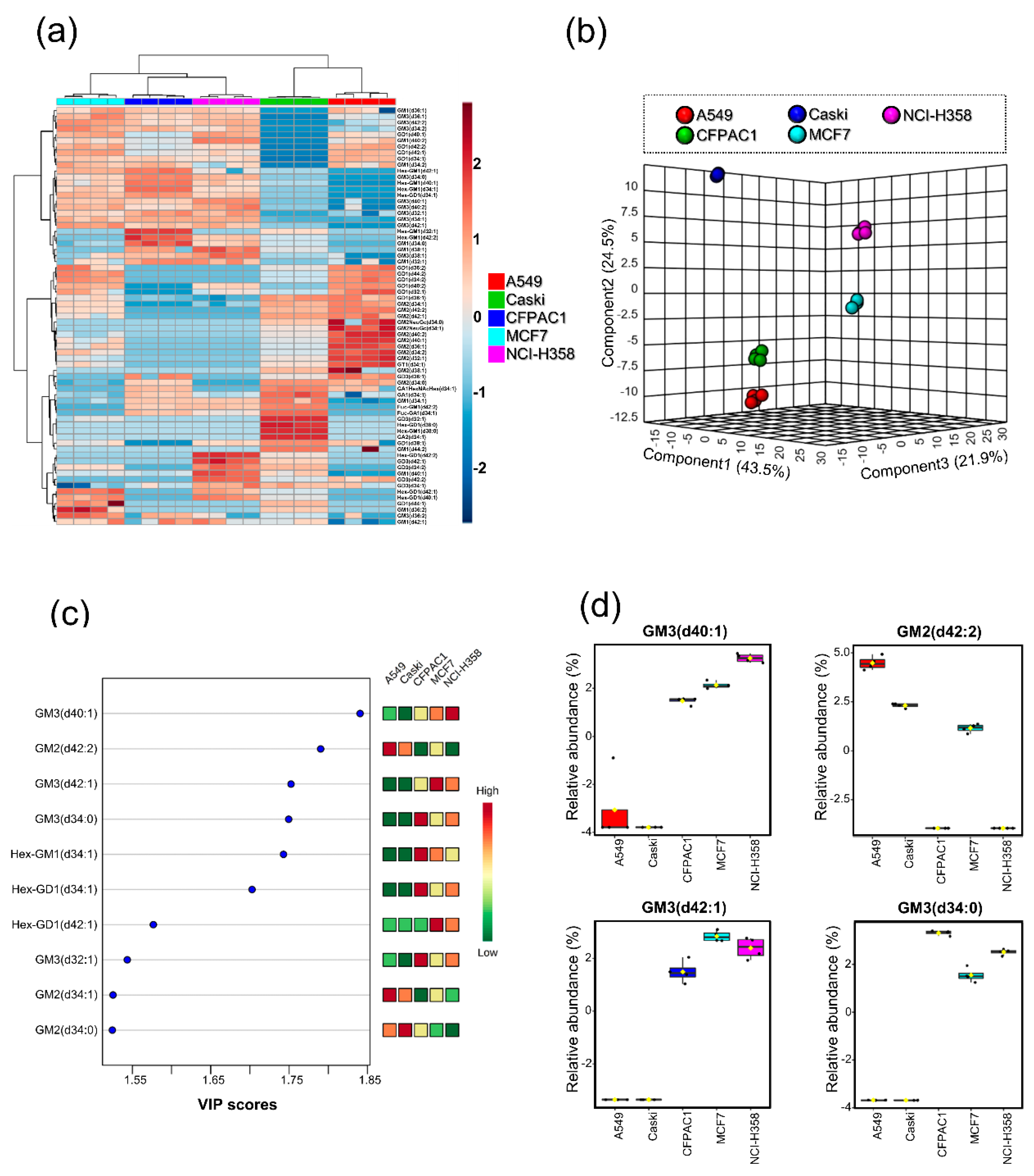
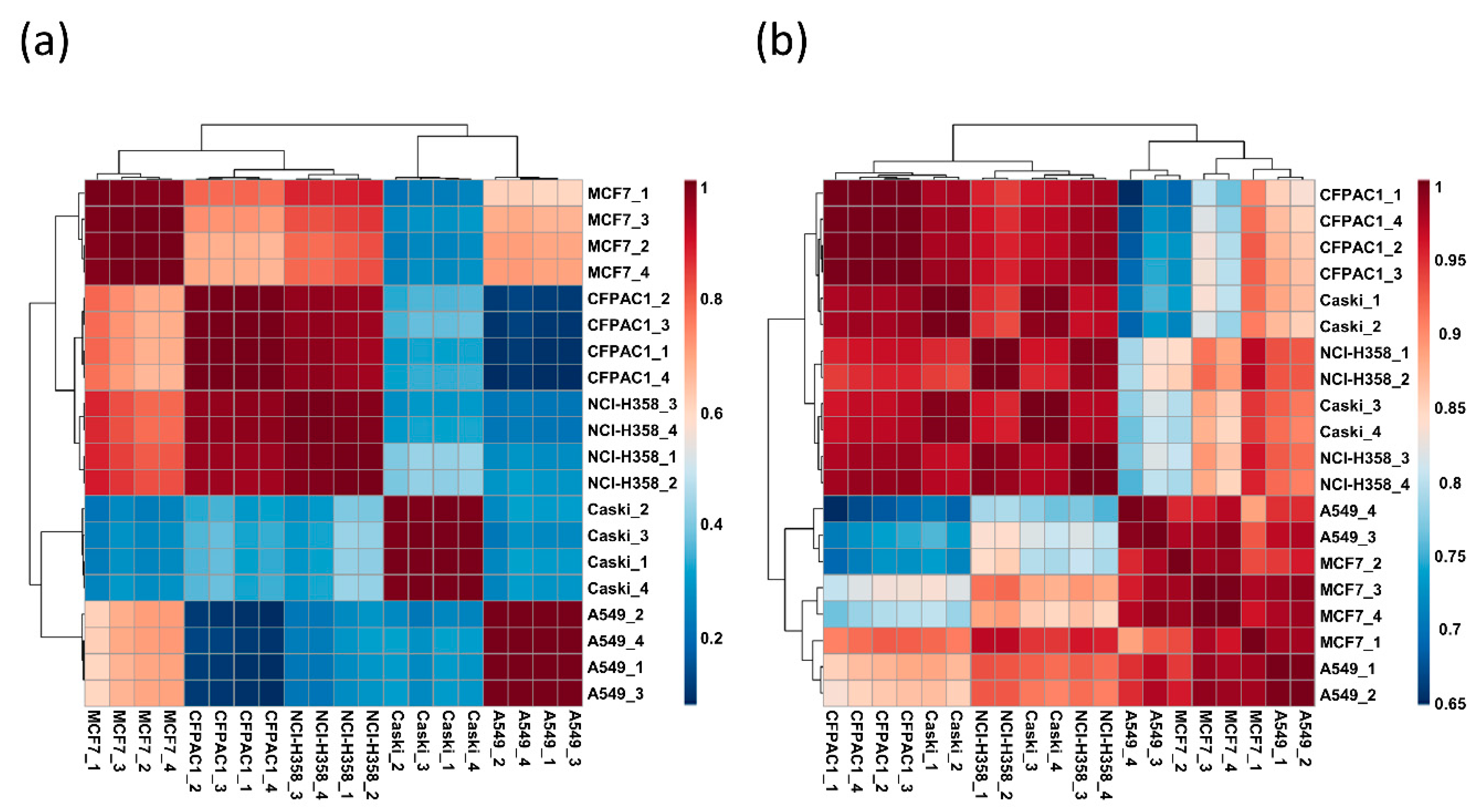
3.3. Differences in Ganglioside Profiles between Cell Lines
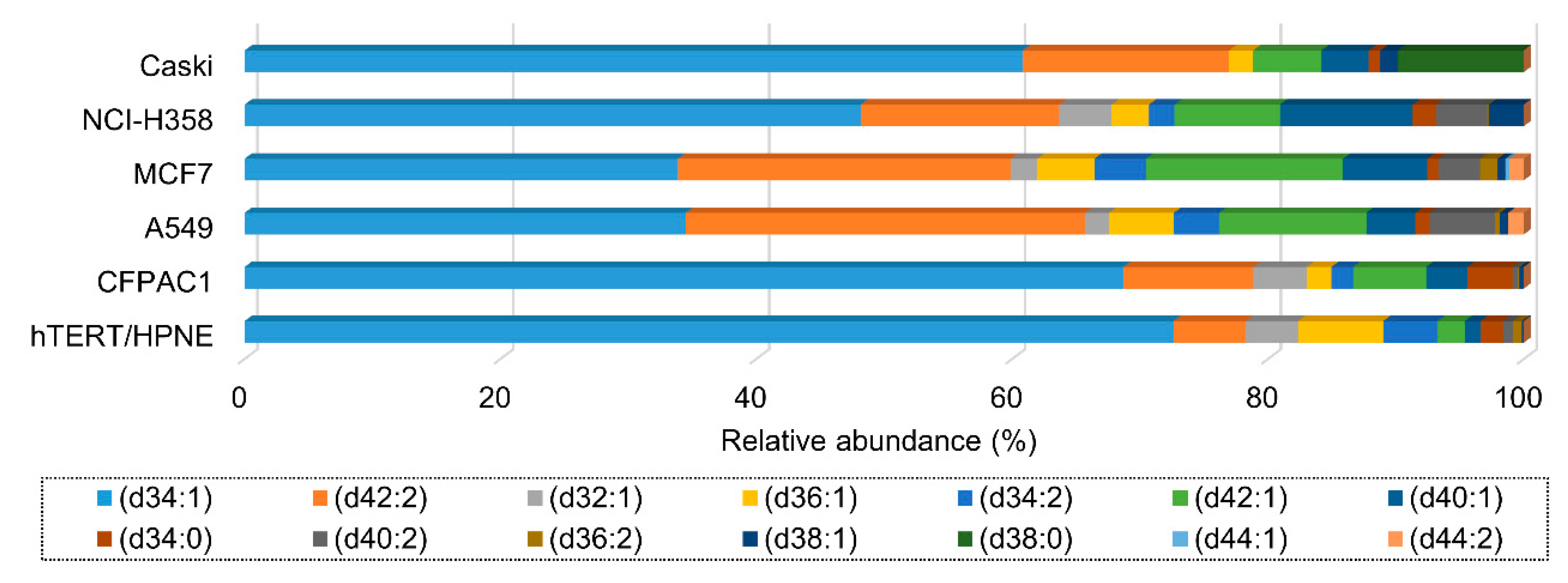
3.4. Ganglioside Differences between Cell Lines Comes from the Heterogeneity of Glycan Moiety
3.5. Ganglioside Differences between Cancerous and Noncancerous Cell Line
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bretscher, M.S.; Raff, M.C. Mammalian plasma membranes. Nature 1975, 258, 43. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Kannagi, R.; Toole, B.P. Glycosylation changes in cancer. In Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press, Cold Spring Harbor: New York City, NY, USA, 2009. [Google Scholar]
- Rodriguez-Emmenegger, C.; Xiao, Q.; Kostina, N.Y.; Sherman, S.E.; Rahimi, K.; Partridge, B.E.; Li, S.; Sahoo, D.; Perez, A.M.R.; Buzzacchera, I. Encoding biological recognition in a bicomponent cell-membrane mimic. Proc. Natl. Acad. Sci. USA 2019, 116, 5376–5382. [Google Scholar] [CrossRef] [PubMed]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, A.; Relvas-Santos, M.; Azevedo, R.; Santos, L.L.; Ferreira, J.A. Protein glycosylation and tumour microenvironment alterations driving cancer hallmarks. Front. Oncol. 2019, 9, 380. [Google Scholar] [CrossRef]
- Hua, S.; Saunders, M.; Dimapasoc, L.M.; Jeong, S.H.; Kim, B.J.; Kim, S.; So, M.; Lee, K.-S.; Kim, J.H.; Lam, K.S. Differentiation of cancer cell origin and molecular subtype by plasma membrane N-glycan profiling. J. Proteome Res. 2014, 13, 961–968. [Google Scholar] [CrossRef]
- Council, N.R. Transforming Glycoscience: A Roadmap for the Future; National Academies Press: Washington, DC, USA, 2012. [Google Scholar]
- Yoshida, S.; Fukumoto, S.; Kawaguchi, H.; Sato, S.; Ueda, R.; Furukawa, K. Ganglioside GD2 in small cell lung cancer cell lines: Enhancement of cell proliferation and mediation of apoptosis. Cancer Res. 2001, 61, 4244–4252. [Google Scholar]
- Fuentes, R.; Allman, R.; Mason, M. Ganglioside expression in lung cancer cell lines. Lung Cancer 1997, 18, 21–33. [Google Scholar] [CrossRef]
- Hamamura, K.; Furukawa, K.; Hayashi, T.; Hattori, T.; Nakano, J.; Nakashima, H.; Okuda, T.; Mizutani, H.; Hattori, H.; Ueda, M. Ganglioside GD3 promotes cell growth and invasion through p130Cas and paxillin in malignant melanoma cells. Proc. Natl. Acad. Sci. USA 2005, 102, 11041–11046. [Google Scholar] [CrossRef]
- Péguet-Navarro, J.; Sportouch, M.; Popa, I.; Berthier, O.; Schmitt, D.; Portoukalian, J. Gangliosides from human melanoma tumors impair dendritic cell differentiation from monocytes and induce their apoptosis. J. Immunol. 2003, 170, 3488–3494. [Google Scholar] [CrossRef]
- Dong, L.; Liu, Y.; Colberg-Poley, A.M.; Kaucic, K.; Ladisch, S. Induction of GM1a/GD1b synthase triggers complex ganglioside expression and alters neuroblastoma cell behavior; a new tumor cell model of ganglioside function. Glycoconj. J. 2011, 28, 137. [Google Scholar] [CrossRef]
- Berois, N.; Osinaga, E. Glycobiology of neuroblastoma: Impact on tumor behavior, prognosis, and therapeutic strategies. Front. Oncol. 2014, 4, 114. [Google Scholar] [PubMed]
- Fleurence, J.; Cochonneau, D.; Fougeray, S.; Oliver, L.; Geraldo, F.; Terme, M.; Dorvillius, M.; Loussouarn, D.; Vallette, F.; Paris, F. Targeting and killing glioblastoma with monoclonal antibody to O-acetyl GD2 ganglioside. Oncotarget 2016, 7, 41172. [Google Scholar] [CrossRef] [PubMed]
- Chahlavi, A.; Rayman, P.; Richmond, A.L.; Biswas, K.; Zhang, R.; Vogelbaum, M.; Tannenbaum, C.; Barnett, G.; Finke, J.H. Glioblastomas induce T-lymphocyte death by two distinct pathways involving gangliosides and CD70. Cancer Res. 2005, 65, 5428–5438. [Google Scholar] [CrossRef] [PubMed]
- Cazet, A.; Julien, S.; Bobowski, M.; Burchell, J.; Delannoy, P. Tumour-associated carbohydrate antigens in breast cancer. Breast Cancer Res. 2010, 12, 204. [Google Scholar] [CrossRef]
- Ruckhäberle, E.; Karn, T.; Rody, A.; Hanker, L.; Gätje, R.; Metzler, D.; Holtrich, U.; Kaufmann, M. Gene expression of ceramide kinase, galactosyl ceramide synthase and ganglioside GD3 synthase is associated with prognosis in breast cancer. J. Cancer Res. Clin. Oncol. 2009, 135, 1005–1013. [Google Scholar] [CrossRef]
- Birkle, S.; Zeng, G.; Gao, L.; Yu, R.; Aubry, J. Role of tumor-associated gangliosides in cancer progression. Biochimie 2003, 85, 455–463. [Google Scholar] [CrossRef]
- Zhang, Q.; Furukawa, K.; Chen, H.-H.; Sakakibara, T.; Urano, T.; Furukawa, K. Metastatic potential of mouse Lewis lung cancer cells is regulated via ganglioside GM1 by modulating the matrix metalloprotease-9 localization in lipid rafts. J. Biol. Chem. 2006, 281, 18145–18155. [Google Scholar] [CrossRef]
- Hamasaki, H.; Aoyagi, M.; Kasama, T.; Handa, S.; Hirakawa, K.; Taki, T. GT1b in human metastatic brain tumors: GT1b as a brain metastasis-associated ganglioside. Biochim. Biophys. Acta -Mol. Cell Biol. Lipids 1999, 1437, 93–99. [Google Scholar] [CrossRef]
- Krengel, U.; Bousquet, P.A. Molecular recognition of gangliosides and their potential for cancer immunotherapies. Front. Immunol. 2014, 5, 325. [Google Scholar] [CrossRef]
- Sait, S.; Modak, S. Anti-GD2 immunotherapy for neuroblastoma. Expert Rev. Anticancer Ther. 2017, 17, 889–904. [Google Scholar] [CrossRef]
- Gholamin, S.; Mirzaei, H.; Razavi, S.M.; Hassanian, S.M.; Saadatpour, L.; Masoudifar, A.; ShahidSales, S.; Avan, A. GD2-targeted immunotherapy and potential value of circulating microRNAs in neuroblastoma. J. Cell. Physiol. 2018, 233, 866–879. [Google Scholar] [CrossRef] [PubMed]
- Castel, V.; Segura, V.; Cañete, A. Treatment of high-risk neuroblastoma with anti-GD2 antibodies. Clin. Transl. Oncol. 2010, 12, 788–793. [Google Scholar] [CrossRef] [PubMed]
- Bethke, U.; Müthing, J.; Schauder, B.; Conradt, P.; Mühlradt, P. An improved semi-quantitative enzyme immunostaining procedure for glycosphingolipid antigens on high performance thin layer chromatograms. J. Immunol. Methods 1986, 89, 111–116. [Google Scholar] [CrossRef]
- Müthing, J. TLC in structure and recognition studies of glycosphingolipids. In Glycoanalysis Protocols; Springer: New York City, NY, USA, 1998; pp. 183–195. [Google Scholar]
- Kailemia, M.J.; Xu, G.; Wong, M.; Li, Q.; Goonatilleke, E.; Leon, F.; Lebrilla, C.B. Recent advances in the mass spectrometry methods for glycomics and cancer. Anal. Chem. 2017, 90, 208–224. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.; Xu, G.; Park, D.; Barboza, M.; Lebrilla, C.B. Intact glycosphingolipidomic analysis of the cell membrane during differentiation yields extensive glycan and lipid changes. Sci. Rep. 2018, 8, 10993. [Google Scholar] [CrossRef] [PubMed]
- Kirsch, S.; Bindila, L. Nano-LC and HPLC-chip–ESI–MS: An emerging technique for glycobioanalysis. Bioanalysis 2009, 1, 1307–1327. [Google Scholar] [CrossRef]
- Merrill, A.H.; Stokes, T.H.; Momin, A.; Park, H.; Portz, B.J.; Kelly, S.; Wang, E.; Sullards, M.C.; Wang, M.D. Sphingolipidomics: A valuable tool for understanding the roles of sphingolipids in biology and disease. J. Lipid Res. 2009, 50, S97–S102. [Google Scholar] [CrossRef]
- ATCC Home Page. Available online: https://www.atcc.org (accessed on 10 October 2018).
- Folch, J.; Lees, M.; Stanley, G.S. A simple method for the isolation and purification of total lipides from animal tissues. J. Boil. Chem. 1957, 226, 497–509. [Google Scholar]
- Hájek, R.; Jirásko, R.; Lísa, M.; Cífková, E.; Holčapek, M. Hydrophilic interaction liquid chromatography–mass spectrometry characterization of gangliosides in biological samples. Anal. Chem. 2017, 89, 12425–12432. [Google Scholar] [CrossRef]
- Hu, T.; Jia, Z.; Zhang, J.-L. Strategy for Comprehensive Profiling and Identification of Acidic Glycosphingolipids Using Ultra-High-Performance Liquid Chromatography Coupled with Quadrupole Time-of-Flight Mass Spectrometry. Anal. Chem. 2017, 89, 7808–7816. [Google Scholar] [CrossRef]
- Kirsch, S.; Souady, J.; Mormann, M.; Bindila, L.; Peter-Katalinić, J. Ceramide Profiles of Human Serum Gangliosides GM2 and GD1a exhibit Cancer-associated Alterations. J. Glycomics Lipidomics 2012, 1. [Google Scholar] [CrossRef]
- Daniotti, J.L.; Vilcaes, A.A.; Torres Demichelis, V.; Ruggiero, F.M.; Rodriguez-Walker, M. Glycosylation of glycolipids in cancer: Basis for development of novel therapeutic approaches. Front. Oncol. 2013, 3, 306. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Kainuma, M.; Jiang, H.; Velasco, H.; Vogt, P.K.; Hakomori, S. Reversion of the Jun-induced oncogenic phenotype by enhanced synthesis of sialosyllactosylceramide (GM3 ganglioside). Proc. Natl. Acad. Sci. USA 2004, 101, 16204–16209. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, T.; Takahashi, K.; Hata, K.; Shiozaki, K.; Yamaguchi, K. Sialidase significance for cancer progression. Glycoconj. J. 2012, 29, 567–577. [Google Scholar] [CrossRef]
- Yin, J.; Miyazaki, K.; Shaner, R.L.; Merrill, A.H.; Kannagi, R. Altered sphingolipid metabolism induced by tumor hypoxia–new vistas in glycolipid tumor markers. FEBS Lett. 2010, 584, 1872–1878. [Google Scholar] [CrossRef]
- Kawamura, S.; Sato, I.; Wada, T.; Yamaguchi, K.; Li, Y.; Li, D.; Zhao, X.; Ueno, S.; Aoki, H.; Tochigi, T. Plasma membrane-associated sialidase (NEU3) regulates progression of prostate cancer to androgen-independent growth through modulation of androgen receptor signaling. Cell Death Differ. 2012, 19, 170. [Google Scholar] [CrossRef]
- Hutchinson, E. Screening: Fingerprinting cancer cells. Nat. Rev. Cancer 2002, 2, 557. [Google Scholar] [CrossRef]
- Karagoz, K.; Sinha, R.; Arga, K.Y. Triple negative breast cancer: A multi-omics network discovery strategy for candidate targets and driving pathways. Omics J. Integr. Biol. 2015, 19, 115–130. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55. [Google Scholar] [CrossRef]
- Yamashita, T.; Wada, R.; Sasaki, T.; Deng, C.; Bierfreund, U.; Sandhoff, K.; Proia, R.L. A vital role for glycosphingolipid synthesis during development and differentiation. Proc. Natl. Acad. Sci. USA 1999, 96, 9142–9147. [Google Scholar] [CrossRef]
- Kwak, D.H.; Seo, B.B.; Chang, K.T.; Choo, Y.K. Roles of gangliosides in mouse embryogenesis and embryonic stem cell differentiation. Exp. Mol. Med. 2011, 43, 379. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kroes, R.A.; He, H.; Emmett, M.R.; Nilsson, C.L.; Leach, F.E.; Amster, I.J.; Marshall, A.G.; Moskal, J.R. Overexpression of ST6GalNAcV, a ganglioside-specific α2, 6-sialyltransferase, inhibits glioma growth in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 12646–12651. [Google Scholar] [CrossRef] [PubMed]
- Cremesti, A.E.; Fischl, A.S. Current methods for the identification and quantitation of ceramides: An overview. Lipids 2000, 35, 937–945. [Google Scholar] [CrossRef] [PubMed]

| hTERT/HPNE | CFPAC1 | A549 | MCF7 | NCI-H358 | Caski | |
|---|---|---|---|---|---|---|
| GM1 | ++++ | +++ | +++ | +++ | +++ | ++++ |
| GD1 | +++ | ++ | ++++ | ++++ | ++ | + |
| GD3 | ++ | + | + | + | + | + |
| GM3 | ++ | ++++ | + | ++++ | ++++ | + |
| GM2 | ++ | + | ++++ | + | + | ++ |
| GA1 | + | + | + | + | + | +++ |
| GT1 | + | ND | + | + | ND | ND |
| GD2 | + | ND | ND | ND | ND | ND |
| Hex-GD1 | + | + | ND | + | + | + |
| GT3 | + | ND | ND | ND | ND | ND |
| GA1+Hexnac+Hex | + | + | + | ND | ND | + |
| Hex-GM1 | + | ++ | ND | + | + | ++ |
| Fuc-GM1 | ND | + | ND | ND | + | + |
| GM2(NeuGc) | ND | ND | + | ND | ND | ND |
| Fuc-GA1 | ND | + | ND | ND | + | ++ |
| GA2 | ND | ND | ND | ND | ND | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Hwang, H.; Kim, S.; Hwang, J.; Yoon, J.; Yin, D.; Choi, S.I.; Kim, Y.-H.; Kim, Y.-S.; An, H.J. Comprehensive Profiling of Surface Gangliosides Extracted from Various Cell Lines by LC-MS/MS. Cells 2019, 8, 1323. https://doi.org/10.3390/cells8111323
Lee J, Hwang H, Kim S, Hwang J, Yoon J, Yin D, Choi SI, Kim Y-H, Kim Y-S, An HJ. Comprehensive Profiling of Surface Gangliosides Extracted from Various Cell Lines by LC-MS/MS. Cells. 2019; 8(11):1323. https://doi.org/10.3390/cells8111323
Chicago/Turabian StyleLee, Jua, Heeyoun Hwang, Sumin Kim, Jaeyun Hwang, Jaekyung Yoon, Dongtan Yin, Sun Il Choi, Yun-Hee Kim, Yong-Sam Kim, and Hyun Joo An. 2019. "Comprehensive Profiling of Surface Gangliosides Extracted from Various Cell Lines by LC-MS/MS" Cells 8, no. 11: 1323. https://doi.org/10.3390/cells8111323
APA StyleLee, J., Hwang, H., Kim, S., Hwang, J., Yoon, J., Yin, D., Choi, S. I., Kim, Y.-H., Kim, Y.-S., & An, H. J. (2019). Comprehensive Profiling of Surface Gangliosides Extracted from Various Cell Lines by LC-MS/MS. Cells, 8(11), 1323. https://doi.org/10.3390/cells8111323







