Tolerance of Iron-Deficient and -Toxic Soil Conditions in Rice
Abstract
1. Introduction
2. Chemistry of Fe in the Soil
3. Role of Fe in Plants
4. Physiological Basis of Tolerance of FT and FD
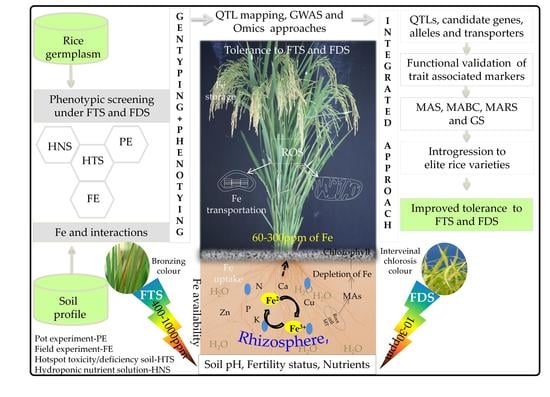
5. Phenotyping for FDS and FTS
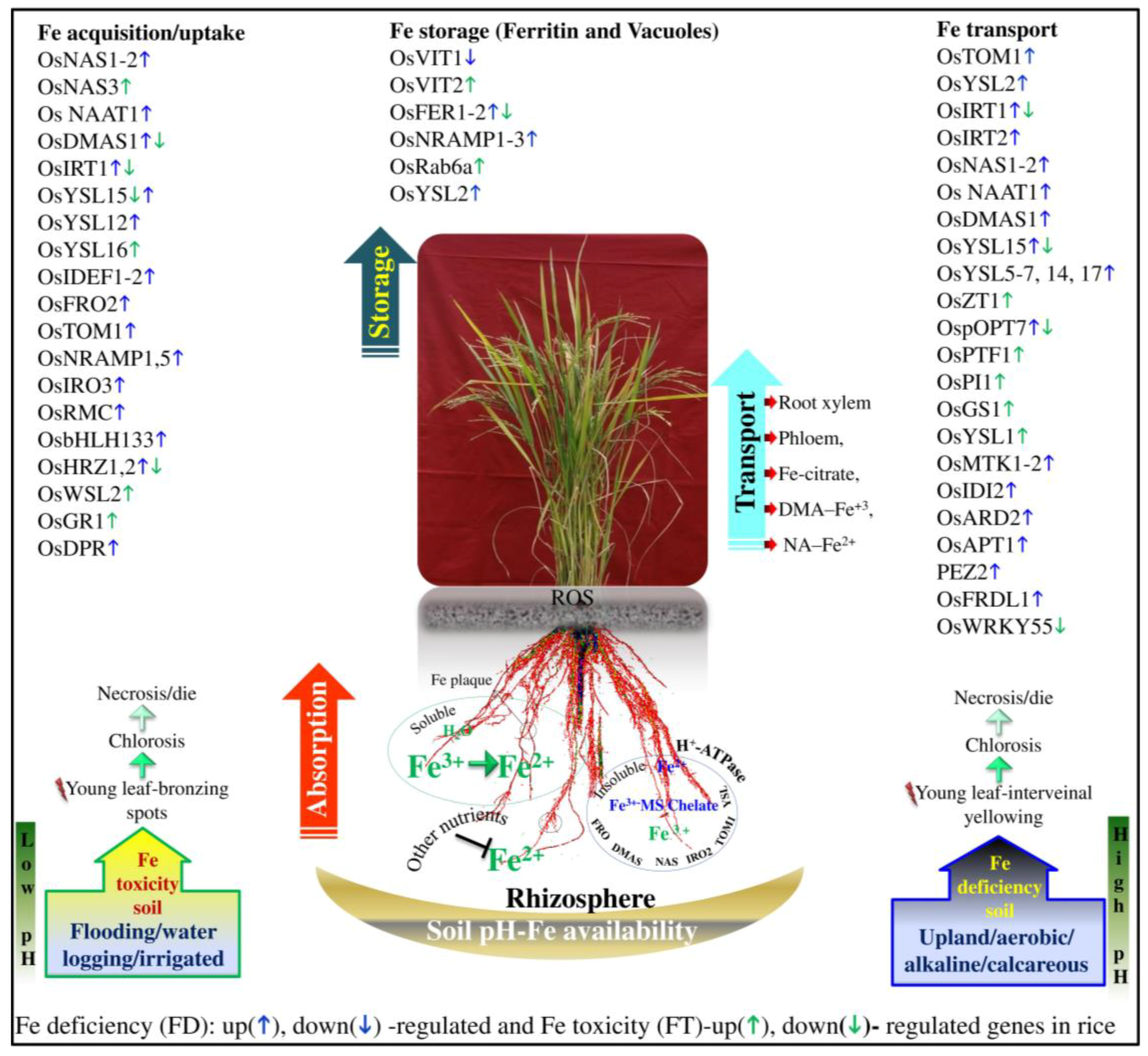
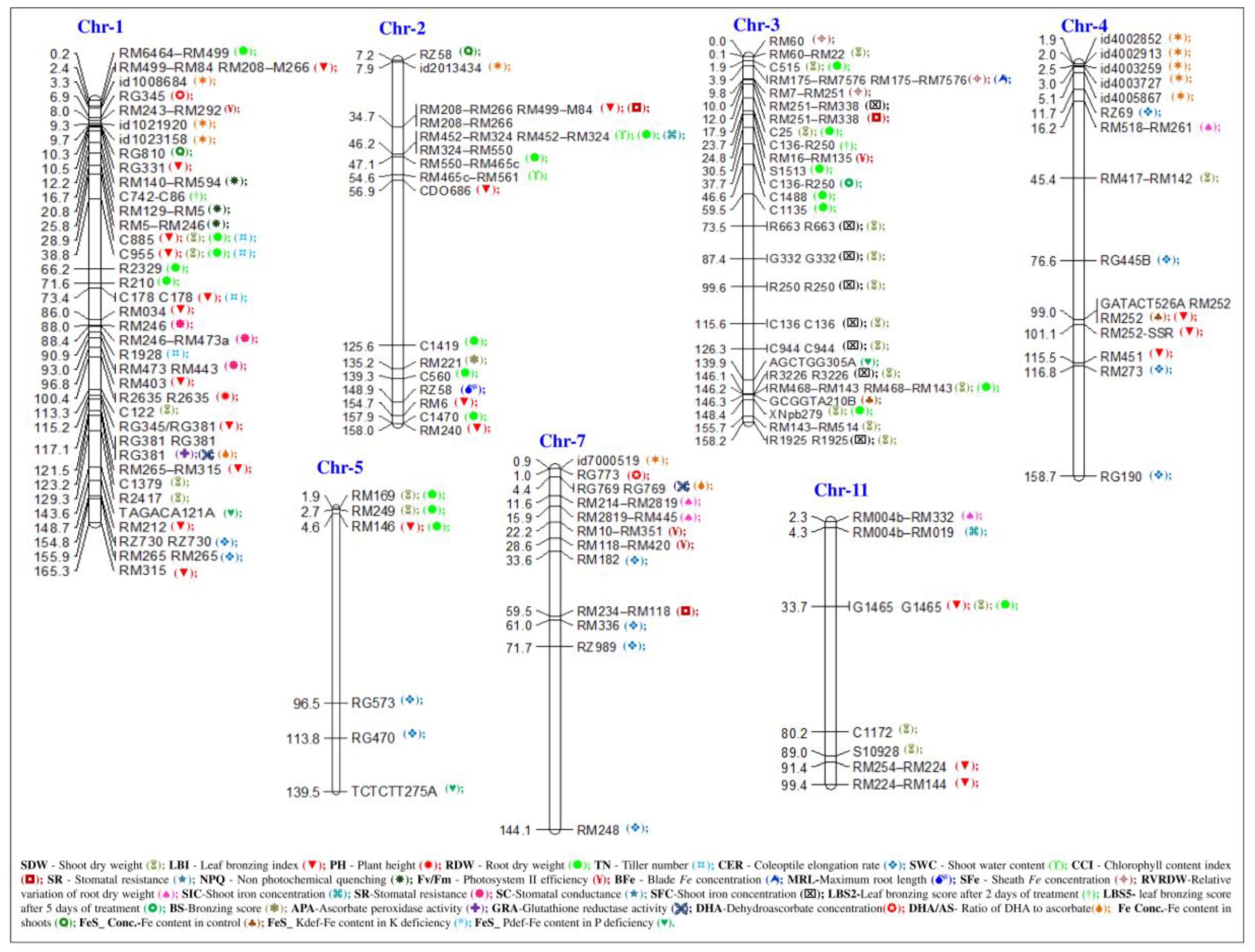
6. The Genetic and Genomic Basis of FD and FT Tolerance
7. Transcriptomics
8. Proteomics
9. Epigenetics
10. Agronomic Practices to Overcome FD
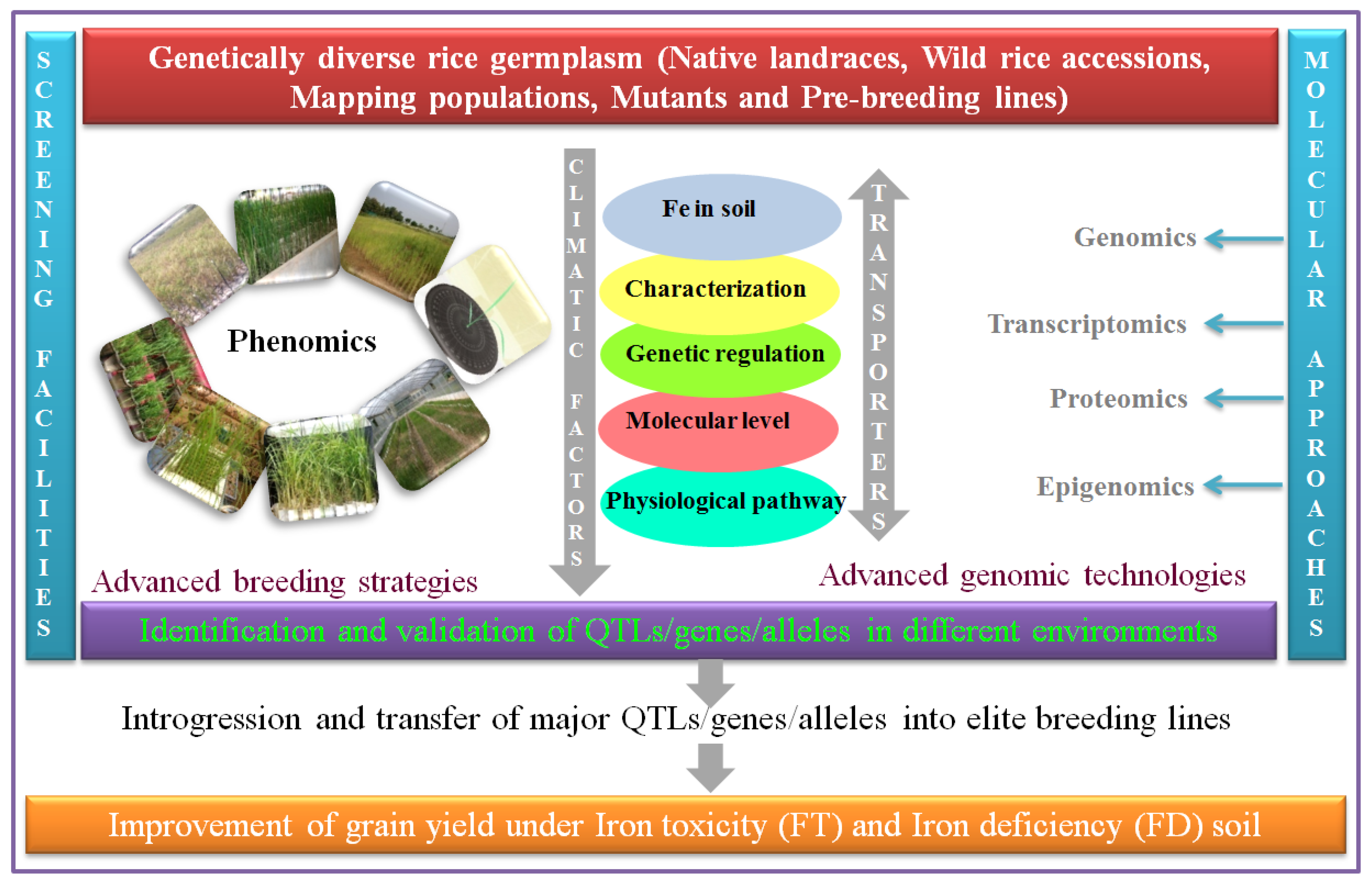
11. Phenotypic Screening and Breeding for FDS and FTS
12. Progress in the Development of FD- and FT-Tolerant Rice Varieties
13. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jaggard, K.W.; Qi, A.; Ober, E.S. Possible changes to arable crop yields by 2050. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 2835–2851. [Google Scholar] [CrossRef] [PubMed]
- Kar, S.; Panda, S.K. Iron homeostasis in rice: Deficit and excess. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2018. [Google Scholar] [CrossRef]
- Sahebi, M.; Hanafi, M.M.; Rafii, M.Y.; Mahmud, T.M.M.; Azizi, P.; Osman, M.; Abiri, R.; Taheri, S.; Kalhori, N.; Shabanimofrad, M. Improvement of drought tolerance in rice (Oryza sativa L.): Genetics, genomic tools, and the WRKY gene family. BioMed Res. Int. 2018, 2018, 315–8474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, Q.; Wang, S.; Hong, Y.; Wang, Z. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 2014, 7, 24. [Google Scholar] [CrossRef]
- van Oort, P.A.J.J. Mapping abiotic stresses for rice in Africa: Drought, cold, iron toxicity, salinity and sodicity. Field Crops Res. 2018, 219, 55–75. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nakanishi Itai, R.; Nishizawa, N.K. Iron deficiency responses in rice roots. Rice 2014, 7, 27. [Google Scholar] [CrossRef]
- Mongon, J.; Chaiwong, N.; Bouain, N.; Prom-U-Thai, C.; Secco, D.; Rouached, H. Phosphorus and iron deficiencies influence rice shoot growth in an oxygen dependent manner: Insight from upland and lowland rice. Int. J. Mol. Sci. 2017, 18, 607. [Google Scholar] [CrossRef]
- Wainaina, C.M.; Makihara, D.; Nakamura, M.; Ikeda, A.; Suzuki, T.; Mizukami, Y.; Nonoyama, T.; Doi, K.; Kikuta, M.; Samejima, H. Identification and validation of QTLs for cold tolerance at the booting stage and other agronomic traits in a rice cross of a Japanese tolerant variety, Hananomai, and a NERICA parent, WAB56-104. Plant Prod. Sci. 2018, 21, 132–143. [Google Scholar] [CrossRef]
- Lamaoui, M.; Jemo, M.; Datla, R.; Bekkaoui, F. Heat and drought stresses in crops and approaches for their mitigation. Front. Chem. 2018, 6, 26. [Google Scholar] [CrossRef]
- Mahender, A.; Anandan, A.; Pradhan, S.K.; Singh, O.N. Traits-related QTLs and genes and their potential applications in rice improvement under low phosphorus condition. Arch. Agron. Soil Sci. 2018, 64, 49–464. [Google Scholar] [CrossRef]
- Ali, J.; Jewel, Z.A.; Mahender, A.; Anandan, A.; Hernandez, J.; Li, Z. Molecular genetics and breeding for nutrient use efficiency in rice. Int. J. Mol. Sci. 2018, 19, 1762. [Google Scholar] [CrossRef]
- Swamy, B.P.M.; Rahman, M.A.; Inabangan-Asilo, M.A.; Amparado, A.; Manito, C.; Chadha-Mohanty, P.; Reinke, R.; Slamet-Loedin, I.H. Advances in breeding for high grain zinc in rice. Rice 2016, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Sikirou, M.; Saito, K.; Achigan-Dako, E.G.; Dramé, K.N.; Ahanchédé, A.; Venuprasad, R.; Adam, A.; Venuprasad, R. Genetic improvement of iron toxicity tolerance in rice: Progress, challenges and prospects in West Africa. Plant Prod. Sci. 2015, 18, 423–434. [Google Scholar] [CrossRef]
- Ma, N.L.; Lah, W.A.C.; Kadir, N.A.; Mustaqim, M.; Rahmat, Z.; Ahmad, A.; Lam, S.D.; Ismail, M.R. Susceptibility and tolerance of rice crop to salt threat: Physiological and metabolic inspections. PLoS ONE 2018, 13, e0192732. [Google Scholar] [CrossRef]
- Bashir, K.; Hanada, K.; Shimizu, M.; Seki, M.; Nakanishi, H.; Nishizawa, N.K. Transcriptomic analysis of rice in response to iron deficiency and excess. Rice 2014, 7, 18. [Google Scholar] [CrossRef]
- Dos Santos, R.S.; de Araujo Júnior, A.T.; Pegoraro, C.; de Oliveira, A.C. Dealing with iron metabolism in rice: From breeding for stress tolerance to biofortification. Genet. Mol. Biol. 2017, 40, 312–325. [Google Scholar] [CrossRef]
- Müller, C.; Kuki, K.N.; Pinheiro, D.T.; de Souza, L.R.; Silva, A.I.S.; Loureiro, M.E.; Oliva, M.A.; Almeida, A.M. Differential physiological responses in rice upon exposure to excess distinct iron forms. Plant Soil 2015, 391, 123–138. [Google Scholar] [CrossRef]
- Gross, J.; Stein, R.J.; Fett-Neto, A.G.; Fett, J.P. Iron homeostasis related genes in rice. Genet. Mol. Biol. 2003, 26, 477–497. [Google Scholar] [CrossRef]
- Rout, G.R.; Sahoo, S. Role of iron in plant growth and metabolism. Rev. Agric. Sci. 2015, 3, 1–24. [Google Scholar] [CrossRef]
- Bashir, K.; Nozoye, T.; Nagasaka, S.; Rasheed, S.; Miyauchi, N.; Seki, M.; Nakanishi, H.; Nishizawa, N.K. Paralogs and mutants show that one DMA synthase functions in iron homeostasis in rice. J. Exp. Bot. 2017, 68, 1785–1795. [Google Scholar] [CrossRef]
- Li, W.; Lan, P. The understanding of the plant iron deficiency responses in Strategy I plants and the role of ethylene in this process by omic approaches. Front. Plant Sci. 2017, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Guerinot, M. Lou Mining iron: Iron uptake and transport in plants. FEBS Lett. 2007, 581, 2273–2280. [Google Scholar] [CrossRef]
- Nakanishi, H.; Ogawa, I.; Ishimaru, Y.; Mori, S.; Nishizawa, N.K. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2006, 52, 464–469. [Google Scholar] [CrossRef]
- Pereira, M.P.; Santos, C.; Gomes, A.; Vasconcelos, M.W. Cultivar variability of iron uptake mechanisms in rice (Oryza sativa L.). Plant Physiol. Biochem. 2014, 85, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Roychoudhury, A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2014, 2, 53. [Google Scholar] [CrossRef]
- Rout, G.R.; Sunita, S.; Das, A.B.; Das, S.R. Screening of iron toxicity in rice genotypes on the basis of morphological, physiological and biochmeical analysis. J. Exp. Biol. Sci. 2014, 2, 567–582. [Google Scholar]
- Vose, P.B. Iron nutrition in plants: A world overview. J. Plant Nutr. 1982, 5, 233–249. [Google Scholar] [CrossRef]
- Audebert, A.; Sahrawat, K.L. Mechanisms for iron toxicity tolerance in lowland rice. J. Plant Nutr. 2000, 23, 1877–1885. [Google Scholar] [CrossRef]
- Audebert, A. Iron Toxicity in Rice–Environmental Conditions and Symptoms; Africa Rice Center (WARDA): Cotonou, Benin, 2006; pp. 18–33. [Google Scholar]
- Chérif, M.; Audebert, A.; Fofana, M.; Zouzou, M. Evaluation of iron toxicity on lowland irrigated rice in West Africa. Tropicultura 2009, 27, 88–92. [Google Scholar]
- Dufey, I.; Hakizimana, P.; Draye, X.; Lutts, S.; Bertin, P. QTL mapping for biomass and physiological parameters linked to resistance mechanisms to ferrous iron toxicity in rice. Euphytica 2009, 167, 143–160. [Google Scholar] [CrossRef]
- Saikia, T.; Baruah, K.K. Iron toxicity tolerance in rice (Oryza sativa) and its association with anti-oxidative enzyme activity. J. Crop Sci. 2012, 3, 90. [Google Scholar]
- Mori, A.S.; Nishizawa, N.; Hayashi, H.; Chino, M.; Yoshimura, E. Why are young rice plants highly susceptible to iron deficiency? Plant Soil 2016, 130, 143–156. [Google Scholar] [CrossRef]
- Morrissey, J.; Guerinot, M.L. Iron uptake and transport in plants: The good, the bad, and the ionome. Chem. Rev. 2009, 109, 4553–4567. [Google Scholar] [CrossRef] [PubMed]
- Lynch, S.R. Why nutritional iron deficiency persists as a worldwide problem, 2. J. Nutr. 2011, 141, 763S–768S. [Google Scholar] [CrossRef] [PubMed]
- Pennock, D.; McKenzie, N.; Montanarella, L. Status of the World’s Soil Resources; FAO: Rome, Italy, 2015. [Google Scholar]
- Mwangi, M.N.; Phiri, K.S.; Abkari, A.; Gbané, M.; Bourdet-Sicard, R.; Braesco, V.A.; Zimmermann, M.B.; Prentice, A.M. Iron for Africa—Report of an Expert Workshop. Nutrients 2017, 9, 576. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.; Asch, F. Iron toxicity in rice—Conditions and management concepts. J. Plant Nutr. Soil Sci. 2005, 168, 558–573. [Google Scholar] [CrossRef]
- Nugraha, Y.; Ardie, S.W.; Ghulamahdi, M.; Aswidinnoor, H.; Suwarno; Aswidinnoor, H. Generation mean analysis of leaf bronzing associated with iron toxicity in rice seedlings using digital imaging methods. SABRAO J. Breed. Genet. 2016, 48, 453–464. [Google Scholar]
- Gridley, H.E.; Efisue, A.; Tolou, B.; Bakayako, T. Breeding for Tolerance to Iron Toxicity at WARDAl; Africa Rice Center (WARDA): Cotonou, Benin, 2006; pp. 96–111. [Google Scholar]
- Fageria, N.K. Yield physiology of rice. J. Plant Nutr. 2007, 30, 843–879. [Google Scholar] [CrossRef]
- Asch, F.; Becker, M.; Kpongor, D.S. A quick and efficient screen for resistance to iron toxicity in lowland rice. J. Plant Nutr. Soil Sci. 2005, 168, 764–773. [Google Scholar] [CrossRef]
- Chandel, G.; Banerjee, S.; Chandel, G. Understanding the role of metal homeostasis related candidate genes in Fe/Zn uptake, transport and redistribution in rice using semi-quantitative RT-PCR. J. Plant Mol. Biol. Biotechnol. 2011, 2, 33–46. [Google Scholar]
- Connorton, J.M.; Balk, J.; Rodríguez-Celma, J. Iron homeostasis in plants: A brief overview. Metallomics 2017, 9, 813–823. [Google Scholar] [CrossRef]
- Bashir, K.; Ishimaru, Y.; Nishizawa, N.K. Iron uptake and loading into rice grains. Rice 2010, 3, 122–130. [Google Scholar] [CrossRef]
- Quinet, M.; Vromman, D.; Clippe, A.; Bertin, P.; Lequeux, H.; Dufey, I.; Lutts, S.; Lefevre, I. Combined transcriptomic and physiological approaches reveal strong differences between short-and long-term response of rice (Oryza sativa) to iron toxicity. Plant. Cell Environ. 2012, 35, 1837–1859. [Google Scholar] [CrossRef] [PubMed]
- Bodek, I. Environmental Inorganic Chemistry: Properties, Processes, and Estimation Methods; Pergamon Press: Elmsford, NY, USA, 1988; ISBN 0080368336. [Google Scholar]
- Audebert, A.; Narteh, L.T.; Kiepe, P.; Millar, D.; Beks, B. Iron Toxicity in Rice-Based Systems in West Africa; Africa Rice Centre (WARDA): Cotonou, Benin, 2006. [Google Scholar]
- Singh, M.V. Micronutrient nutritional problems in soils of India and improvement for human and animal health. Indian J. Fertil. 2009, 5, 11–56. [Google Scholar]
- Barton, L.L.; Abadía, J. Iron Nutrition in Plants and Rhizospheric Microorganisms; Springer Science & Business Media: Dordrecht, The Netherlands, 2007; ISBN 1402066236. [Google Scholar]
- Fageria, N.K.; Baligar, V.C.; Wright, R.J. Iron nutrition of plants: An overview on the chemistry and physiology of its deficiency and toxicity. Pesqui. Agropecu. Bras. 1990, 25, 553–570. [Google Scholar]
- Dotaniya, M.; Meena, H.M.; Lata, M.; Kumar, K. Role of phytosiderophores in iron uptake by plants. Agric. Sci. Dig. 2013, 33, 73–76. [Google Scholar]
- Yi, Y.; Saleeba, J.A.; Guerinot, M.L. Iron uptake in Arabidopsis thaliana. In The Biochemistry of Metal Micronutrients in the Rhizosphere; Manthey, J., Crowley, D.E., Luster, D.G., Eds.; CRC Press: Lewis, Boca Raton, FL, USA, 1994; pp. 295–307. [Google Scholar]
- Radzki, W.; Mañero, F.J.G.; Algar, E.; García, J.A.L.; García-Villaraco, A.; Solano, B.R. Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Van Leeuwenhoek 2013, 104, 321–330. [Google Scholar] [CrossRef]
- Rengel, Z. Availability of Mn, Zn and Fe in the rhizosphere. J. Soil Sci. Plant Nutr. 2015, 15, 397–409. [Google Scholar] [CrossRef]
- Mengel, K.; Kirkby, E.A.; Kosegarten, H.; Appel, T. Boron. In Principles of Plant Nutrition, 5th Ed.; Mengel, K., Kirkby, E., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2001; pp. 621–638. [Google Scholar]
- Mori, S.; Nishizawa, N.; Hayashi, H.; Chino, M.; Yoshimura, E.; Ishihara, J. Why are young rice plants highly susceptible to iron deficiency? In Iron Nutrition and Interactions in Plants; Springer: Dordrecht, The Netherlands, 1991; pp. 175–188. [Google Scholar]
- Fageria, N.K.; Baligar, V.C.; Li, Y.C. The role of nutrient efficient plants in improving crop yields in the twenty first century. J. Plant Nutr. 2008, 31, 1121–1157. [Google Scholar] [CrossRef]
- Da Silveira, V.C.; de Oliveira, A.P.; Sperotto, R.A.; Espindola, L.S.; Amaral, L.; Dias, J.F.; da Cunha, J.B.; Fett, J.P. Influence of iron on mineral status of two rice (Oryza sativa L.) cultivars. Braz. J. Plant Physiol. 2007, 19, 127–139. [Google Scholar] [CrossRef]
- Curie, C.; Briat, J.-F. Iron transport and signaling in plants. Annu. Rev. Plant Biol. 2003, 54, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Koh, S.; Lee, S.-C.; Kim, M.-K.; Koh, J.H.; Lee, S.; An, G.; Choe, S.; Kim, S.-R. T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 2007, 65, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Onaga, G.; Edema, R.; Asea, G. Tolerance of rice germplasm to iron toxicity stress and the relationship between tolerance, Fe2+, P and K content in the leaves and roots. Arch. Agron. Soil Sci. 2013, 59, 213–229. [Google Scholar] [CrossRef]
- White, P.J.; Brown, P.H. Plant nutrition for sustainable development and global health. Ann. Bot. 2010, 105, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.R. Bioavailability of iron, zinc, and other trace minerals from vegetarian diets. Am. J. Clin. Nutr. 2003, 78, 633S–639S. [Google Scholar] [CrossRef] [PubMed]
- Römheld, V. Different strategies for iron acquisition in higher plants. Physiol. Plant. 1987, 70, 231–234. [Google Scholar] [CrossRef]
- Vejchasarn, P.; Lynch, J.P.; Brown, K.M. Genetic variability in phosphorus responses of rice root phenotypes. Rice 2016, 9, 29. [Google Scholar] [CrossRef]
- Frei, M.; Tetteh, R.N.; Razafindrazaka, A.L.; Fuh, M.A.; Wu, L.-B.; Becker, M. Responses of rice to chronic and acute iron toxicity: Genotypic differences and biofortification aspects. Plant Soil 2016, 408, 149–161. [Google Scholar] [CrossRef]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef]
- Bruins, M.R.; Kapil, S.; Oehme, F.W. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 2000, 45, 198–207. [Google Scholar] [CrossRef]
- Nworie, O.E.; Qin, J.; Lin, C. Differential effects of low-molecular-weight organic acids on the mobilization of soil-borne arsenic and trace metals. Toxics 2017, 5, 18. [Google Scholar] [CrossRef]
- Schmidt, H.; Eickhorst, T. Detection and quantification of native microbial populations on soil-grown rice roots by catalyzed reporter deposition-fluorescence in situ hybridization. FEMS Microbiol. Ecol. 2014, 87, 390–402. [Google Scholar] [CrossRef]
- Ponnamperuma, F.N. The chemistry of submerged soils. In Advances in Agronomy; Elsevier: New York, NY, USA, 1972; Volume 24, pp. 29–96. ISBN 0065-2113. [Google Scholar]
- Brumbarova, T.; Bauer, P.; Ivanov, R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015, 20, 124–133. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef]
- Balk, J.; Schaedler, T.A. Iron cofactor assembly in plants. Annu. Rev. Plant Biol. 2014, 65, 125–153. [Google Scholar] [CrossRef]
- Wang, Y.; Frei, M.; Wissuwa, M. An agar nutrient solution technique as a screening tool for tolerance to zinc deficiency and iron toxicity in rice. Soil Sci. Plant Nutr. 2008, 54, 744–750. [Google Scholar] [CrossRef]
- Banakar, R.; Alvarez Fernández, Á.; Abadía, J.; Capell, T.; Christou, P. The expression of heterologous Fe (III) phytosiderophore transporter Hv YS 1 in rice increases Fe uptake, translocation and seed loading and excludes heavy metals by selective Fe transport. Plant Biotechnol. J. 2017, 15, 423–432. [Google Scholar] [CrossRef]
- Audebert, A.; Fofana, M. Rice yield gap due to iron toxicity in West Africa. J. Agron. Crop Sci. 2009, 195, 66–76. [Google Scholar] [CrossRef]
- Onaga, G.; Dramé, K.N.; Ismail, A.M. Understanding the regulation of iron nutrition: can it contribute to improving iron toxicity tolerance in rice? Funct. Plant Biol. 2016, 43, 709–726. [Google Scholar] [CrossRef]
- Pereira, E.G.; Oliva, M.A.; Rosado-Souza, L.; Mendes, G.C.; Colares, D.S.; Stopato, C.H.; Almeida, A.M. Iron excess affects rice photosynthesis through stomatal and non-stomatal limitations. Plant Sci. 2013, 201, 81–92. [Google Scholar] [CrossRef]
- Hell, R.; Stephan, U.W. Iron uptake, trafficking and homeostasis in plants. Planta 2003, 216, 541–551. [Google Scholar]
- Li, G.; Kronzucker, H.J.; Shi, W. Root developmental adaptation to Fe toxicity: Mechanisms and management. Plant Signal. Behav. 2016, 11, e1117722. [Google Scholar] [CrossRef]
- Wu, L.-B.; Shhadi, M.Y.; Gregorio, G.; Matthus, E.; Becker, M.; Frei, M. Genetic and physiological analysis of tolerance to acute iron toxicity in rice. Rice 2014, 7, 8. [Google Scholar] [CrossRef]
- Sikirou, M.; Saito, K.; Dramé, K.N.; Saidou, A.; Dieng, I.; Ahanchédé, A.; Venuprasad, R. Soil-based screening for iron toxicity tolerance in rice using pots. Plant Prod. Sci. 2016, 19, 489–496. [Google Scholar] [CrossRef]
- Nugraha, Y.; Ardie, S.W.; Ghulamahdi, M.; Suwarno, S.; Aswidinnoor, H. Implication of gene action and heritability under stress and control conditions for selection iron toxicity tolerant in rice. AGRIVITA, J. Agric. Sci. 2016, 38, 282–295. [Google Scholar] [CrossRef]
- Nugraha, Y.; Ardie, S.W.; Ghulammahdi, M.; Aswidinnoor, H. Nutrient culture media with agar is effective for early and rapid screening of iron toxicity tolerance in rice. J. Crop Sci. Biotechnol. 2016, 19, 61–70. [Google Scholar] [CrossRef]
- Dobermann, A. Rice: Nutrient Disorders & Nutrient Management; Potash & Phosphate Institute of Canada, Singapore, and International Rice Research Institute: Los Baños, Philippines, 2000; Volume 1, ISBN 9810427425. [Google Scholar]
- Dramé, K.N.; Saito, K.; Koné, B.; Chabi, A.; Dakouo, D.; Annan-Afful, E.; Monh, S.; Abo, E.; Sié, M. Coping with iron toxicity in the lowlands of sub-Saharan Africa: Experience from Africa Rice Center. In Proceedings of the 2nd Africa Rice Congress, Innovation and Partnerships to Realize Africa’s Rice Potential, Bamako, Mali, 22–26 March 2010; pp. 191–198. [Google Scholar]
- Trejo-Téllez, L.I.; Gómez-Merino, F.C. Nutrient solutions for hydroponic systems. In Hydroponics: A Standard Methodology for Plant Biological Researches; Asao, T., Ed.; InTech: Rijeka, Croatia, 2012; pp. 1–20. [Google Scholar]
- Mengel, K. Iron availability in plant tissues-iron chlorosis on calcareous soils. Plant Soil 1994, 165, 275–283. [Google Scholar] [CrossRef]
- Motesharezadeh, B.; Hesam-Arefi, A.; Savaghebi, G.R. The effect of bicarbonate on iron (Fe) and zinc (Zn) uptakes by soybean varieties. Desert 2017, 22, 145–155. [Google Scholar]
- Jain, A.; Connolly, E.L. Mitochondrial iron transport and homeostasis in plants. Front. Plant Sci. 2013, 4, 348. [Google Scholar] [CrossRef]
- López-Millán, A.F.; Duy, D.; Philippar, K. Chloroplast iron transport proteins–function and impact on plant physiology. Front. Plant Sci. 2016, 7, 178. [Google Scholar] [CrossRef]
- Inoue, H.; Kobayashi, T.; Nozoye, T.; Takahashi, M.; Kakei, Y.; Suzuki, K.; Nakazono, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Rice OsYSL15 is an iron-regulated iron (III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 2009, 284, 3470–3479. [Google Scholar] [CrossRef]
- Zhao, A.Q.; Bao, Q.L.; Tian, X.H.; Lu, X.C.; William, J.G. Combined effect of iron and zinc on micronutrient levels in wheat (Triticum aestivum L.). J. Environ. Biol. 2011, 32, 235–239. [Google Scholar]
- Nozoye, T.; Nagasaka, S.; Kobayashi, T.; Takahashi, M.; Sato, Y.; Sato, Y.; Uozumi, N.; Nakanishi, H.; Nishizawa, N.K. Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 2011, 286, 5446–5454. [Google Scholar] [CrossRef]
- Pal, S.; Datta, S.P.; Rattan, R.K.; Singh, A.K. Diagnosis and amelioration of iron deficiency under aerobic rice. J. Plant Nutr. 2008, 31, 919–940. [Google Scholar] [CrossRef]
- Nogiya, M.; Pandey, R.N.; Singh, B. Physiological basis of iron chlorosis tolerance in rice (Oryza sativa) in relation to the root exudation capacity. J. Plant Nutr. 2016, 39, 1536–1546. [Google Scholar] [CrossRef]
- Xue, Y.; Xia, H.; Christie, P.; Zhang, Z.; Li, L.; Tang, C. Crop acquisition of phosphorus, iron and zinc from soil in cereal/legume intercropping systems: A critical review. Ann. Bot. 2016, 117, 363–377. [Google Scholar] [CrossRef]
- Lindsay, W.L.; Schwab, A.P. The chemistry of iron in soils and its availability to plants. J. Plant Nutr. 1982, 5, 821–840. [Google Scholar] [CrossRef]
- Meda, A.R.; Scheuermann, E.B.; Prechsl, U.E.; Erenoglu, B.; Schaaf, G.; Hayen, H.; Weber, G.; von Wirén, N. Iron acquisition by phytosiderophores contributes to cadmium tolerance. Plant Physiol. 2007, 143, 1761–1773. [Google Scholar] [CrossRef]
- Krohling, C.A.; Eutrópio, F.J.; Bertolazi, A.A.; Dobbss, L.B.; Campostrini, E.; Dias, T.; Ramos, A.C. Ecophysiology of iron homeostasis in plants. Soil Sci. Plant Nutr. 2016, 62, 39–47. [Google Scholar] [CrossRef]
- Inoue, H.; Takahashi, M.; Kobayashi, T.; Suzuki, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Identification and localisation of the rice nicotianamine aminotransferase gene OsNAAT1 expression suggests the site of phytosiderophore synthesis in rice. Plant Mol. Biol. 2008, 66, 193–203. [Google Scholar] [CrossRef]
- Kobayashi, T.; Suzuki, M.; Inoue, H.; Itai, R.N.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J. Exp. Bot. 2005, 56, 1305–1316. [Google Scholar] [CrossRef]
- Brumbarova, T.; Bauer, P. Iron uptake and transport in plants. Chem. Rev. 2009, 109, 4553–4567. [Google Scholar] [CrossRef]
- Kim, S.A.; Punshon, T.; Lanzirotti, A.; Li, L.; Alonso, J.M.; Ecker, J.R.; Kaplan, J.; Guerinot, M.L. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 2006, 314, 1295–1298. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Yi, H.; Gong, J. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012, 72, 400–410. [Google Scholar] [CrossRef]
- Koike, S.; Inoue, H.; Mizuno, D.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004, 39, 415–424. [Google Scholar] [CrossRef]
- Itai, R.N.; Ogo, Y.; Kobayashi, T.; Nakanishi, H.; Nishizawa, N.K. Rice genes involved in phytosiderophore biosynthesis are synchronously regulated during the early stages of iron deficiency in roots. Rice 2013, 6, 16. [Google Scholar] [CrossRef]
- Nozoye, T.; Inoue, H.; Takahashi, M.; Ishimaru, Y.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. The expression of iron homeostasis-related genes during rice germination. Plant Mol. Biol. 2007, 64, 35–47. [Google Scholar] [CrossRef]
- Walker, E.L.; Connolly, E.L. Time to pump iron: Iron-deficiency-signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 2008, 11, 530–535. [Google Scholar] [CrossRef]
- Vigani, G.; Bashir, K.; Ishimaru, Y.; Lehmann, M.; Casiraghi, F.M.; Nakanishi, H.; Seki, M.; Geigenberger, P.; Zocchi, G.; Nishizawa, N.K. Knocking down mitochondrial iron transporter (MIT) reprograms primary and secondary metabolism in rice plants. J. Exp. Bot. 2015, 67, 1357–1368. [Google Scholar] [CrossRef]
- Paul, B.T.; Manz, D.H.; Torti, F.M.; Torti, S. V Mitochondria and iron: Current questions. Expert Rev. Hematol. 2017, 10, 65–79. [Google Scholar] [CrossRef]
- Bashir, K.; Ishimaru, Y.; Shimo, H.; Nagasaka, S.; Fujimoto, M.; Takanashi, H.; Tsutsumi, N.; An, G.; Nakanishi, H.; Nishizawa, N.K. The rice mitochondrial iron transporter is essential for plant growth. Nat. Commun. 2011, 2, 322. [Google Scholar] [CrossRef]
- Li, K.; Wang, H.; Hu, X.; Ma, F.; Wu, Y.; Wang, Q.; Liu, Z.; Huang, C. Genetic and quantitative trait locus analysis of cell wall components and forage digestibility in the Zheng58 × HD568 maize RIL population at anthesis stage. Front. Plant Sci. 2017, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Imam, M.U.; Zhang, S.; Ma, J.; Wang, H.; Wang, F. Antioxidants mediate both iron homeostasis and oxidative stress. Nutrients 2017, 9, 671. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Hu, B.; Liao, C.Y.; Zhu, J.M.; Wu, Y.R.; Senadhira, D.; Paterson, A.H. Characterization of tissue tolerance to iron by molecular markers in different lines of rice. Plant Soil 1998, 203, 217–226. [Google Scholar] [CrossRef]
- Wu, L.B.; Ueda, Y.; Lai, S.K.; Frei, M. Shoot tolerance mechanisms to iron toxicity in rice (Oryza sativa L.). Plant Cell Environ. 2017, 40, 570–584. [Google Scholar] [CrossRef]
- Chen, W.R.; Feng, Y.; Chao, Y.E. Genomic analysis and expression pattern of OsZIP1, OsZIP3, and OsZIP4 in two rice (Oryza sativa L.) genotypes with different zinc efficiency. Russ. J. Plant Physiol. 2008, 55, 400–409. [Google Scholar] [CrossRef]
- Viana, V.E.; Marini, N.; Finatto, T.; Ezquer, I.; Busanello, C.; Dos Santos, R.S.; Pegoraro, C.; Colombo, L.; de Oliveira, A.C. Iron excess in rice: From phenotypic changes to functional genomics of WRKY transcription factors. Genet. Mol. Res. 2017, 16. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Kronzucker, H.J.; Shi, W. The response of the root apex in plant adaptation to iron heterogeneity in soil. Front. Plant Sci. 2016, 7, 344. [Google Scholar] [CrossRef]
- Kronzucker, H.J.; Siddiqi, M.Y.; Glass, A.D.M. Kinetics of NO3-influx in spruce. Plant Physiol. 1995, 109, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Staal, M.; De Cnodder, T.; Simon, D.; Vandenbussche, F.; Van Der Straeten, D.; Verbelen, J.-P.; Elzenga, T.; Vissenberg, K. Apoplastic alkalinisation is instrumental for the inhibition of cell elongation in the Arabidopsis thaliana root by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC). Plant Physiol. 2011, 155, 2049–2055. [Google Scholar] [CrossRef]
- Majerus, V.; Bertin, P.; Lutts, S. Abscisic acid and oxidative stress implications in overall ferritin synthesis by African rice (Oryza glaberrima Steud.) seedlings exposed to short term iron toxicity. Plant Soil 2009, 324, 253. [Google Scholar] [CrossRef]
- Tripathi, R.D.; Tripathi, P.; Dwivedi, S.; Kumar, A.; Mishra, A.; Chauhan, P.S.; Norton, G.J.; Nautiyal, C.S. Roles for root iron plaque in sequestration and uptake of heavy metals and metalloids in aquatic and wetland plants. Metallomics 2014, 6, 1789–1800. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, F.; Mao, D. Effect of iron plaque outside roots on nutrient uptake by rice (Oryza sativa L.). Zinc uptake by Fe-deficient rice. Plant Soil 1998, 202, 33–39. [Google Scholar] [CrossRef]
- Wang, X.; Pang, Y.; Zhang, J.; Wu, Z.; Chen, K.; Ali, J.; Ye, G.; Xu, J.; Li, Z. Genome-wide and gene-based association mapping for rice eating and cooking characteristics and protein content. Sci. Rep. 2017, 7, 17203. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Hao, H.; Fan, X.; Karim, M.R.; Zhang, F.; Zou, C. Responses of aerobic rice (Oryza sativa L.) to iron deficiency. J. Integr. Agric. 2012, 11, 938–945. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Xu, G.; Song, J.; Wu, T.; Mei, X.; Liu, P. Effects of iron toxicity on the morphological and biological characteristics of rice root border cells. J. Plant Nutr. 2017, 40, 332–343. [Google Scholar] [CrossRef]
- Dhakal, R. Screening of Rice Genotypes (Oryza sativa L.) for Yield, Grain Quality Characters and Iron Chlorosis on Vertisols. Ph.D. Thesis, Vasantrao Naik Marathwada Krishi Vidyapeeth, Parbhani, Maharashtra, India, 2014. Available online: http://krishikosh.egranth.ac.in/handle/1/5810056813 (accessed on 22 May 2014).
- Yakop, U.M. Physiological and Genetic Investigations of Iron Deficiency in Field Peas (Pisum sativum L.). Ph.D. Thesis, University of Adelaide, Adelaide, Australia, 2012. [Google Scholar]
- Yu, L.J.; Luo, Y.F.; Liao, B.; Xie, L.J.; Chen, L.; Xiao, S.; Li, J.T.; Hu, S.N.; Shu, W.S. Comparative transcriptome analysis of transporters, phytohormone and lipid metabolism pathways in response to arsenic stress in rice (Oryza sativa). New Phytol. 2012, 195, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, M.W.; Grusak, M.A. Morpho-physiological parameters affecting iron deficiency chlorosis in soybean (Glycine max L.). Plant Soil 2014, 374, 161–172. [Google Scholar] [CrossRef]
- Hoan, N.T.; Rao, U.P.; Siddiq, E.A. Genetics of tolerance to iron chlorosis in rice. Plant Soil 1992, 146, 327–333. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, F.; Chen, Q.; Rengel, Z.; Tang, C.; Song, C. Genotypic difference in seed iron content and early responses to iron deficiency in wheat. J. Plant Nutr. 2002, 25, 1631–1643. [Google Scholar] [CrossRef]
- Mahmoudi, H.; Labidi, N.; Ksouri, R.; Gharsalli, M.; Abdelly, C. Differential tolerance to iron deficiency of chickpea varieties and Fe resupply effects. C. R. Biol. 2007, 330, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Kabir, A.H.; Rahman, M.M.; Haider, S.A.; Paul, N.K. Mechanisms associated with differential tolerance to Fe deficiency in okra (Abelmoschus esculentus Moench.). Environ. Exp. Bot. 2015, 112, 16–26. [Google Scholar] [CrossRef]
- Mamidi, S.; Lee, R.K.; Goos, J.R.; McClean, P.E. Genome-wide association studies identifies seven major regions responsible for iron deficiency chlorosis in soybean (Glycine max). PLoS ONE 2014, 9, e107469. [Google Scholar] [CrossRef] [PubMed]
- Carvalhais, L.C.; Dennis, P.G.; Fedoseyenko, D.; Hajirezaei, M.; Borriss, R.; von Wirén, N. Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. J. Plant Nutr. Soil Sci. 2011, 174, 3–11. [Google Scholar] [CrossRef]
- Sahrawat, K.L. Iron toxicity in wetland rice and the role of other nutrients. J. Plant Nutr. 2004, 27, 1471–1504. [Google Scholar] [CrossRef]
- Takehisa, H.; Sato, T. Stress, Physiological and Genetic Factors of Rice Leaf Bronzing in Paddy Fields. Jpn. J. Plant Sci. 2007, 1, 63–68. [Google Scholar]
- Wang, H.; Inukai, Y.; Yamauchi, A. Root development and nutrient uptake. CRC Crit. Rev. Plant Sci. 2006, 25, 279–301. [Google Scholar] [CrossRef]
- Xiong, J.; Fu, G.; Tao, L.; Zhu, C. Roles of nitric oxide in alleviating heavy metal toxicity in plants. Arch. Biochem. Biophys. 2010, 497, 13–20. [Google Scholar] [CrossRef]
- Pathirana, R.; Wijithawarna, W.A.; Jagoda, K.; Ranawaka, A.L. Selection of rice for iron toxicity tolerance through irradiated caryopsis culture. Plant Cell. Tissue Organ Cult. 2002, 70, 83–90. [Google Scholar] [CrossRef]
- Elec, V.; Quimio, C.A.; Mendoza, R.; Sajise, A.G.C.; Beebout, S.E.J.; Gregorio, G.B.; Singh, R.K. Maintaining elevated Fe2+ concentration in solution culture for the development of a rapid and repeatable screening technique for iron toxicity tolerance in rice (Oryza sativa L.). Plant Soil 2013, 372, 253–264. [Google Scholar] [CrossRef]
- Aboa, K.; Dogbe, S.Y. Effect of iron toxicity on rice yield in the Amou-Oblo lowland in Togo. In Iron Toxicity in Rice-Based Systems in West Africa; Africa Rice Center (WARDA): Cotonou, Benin, 2006. [Google Scholar]
- Nozoe, T.; Agbisit, R.; Fukuta, Y.; Rodriguez, R.; Yanagihara, S. Characteristics of iron tolerant rice lines developed at IRRI under field conditions. Jpn. Agric. Res. Q. 2008, 42, 187–192. [Google Scholar] [CrossRef]
- Ruengphayak, S.; Ruanjaichon, V.; Saensuk, C.; Phromphan, S.; Tragoonrung, S.; Kongkachuichai, R.; Vanavichit, A. Forward screening for seedling tolerance to Fe toxicity reveals a polymorphic mutation in ferric chelate reductase in rice. Rice 2015, 8, 3. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef] [PubMed]
- Leon, K.; Mery, D.; Pedreschi, F.; Leon, J. Color measurement in L* a* b* units from RGB digital images. Food Res. Int. 2006, 39, 1084–1091. [Google Scholar] [CrossRef]
- Wan, J.-L.; Zhai, H.-Q.; Wan, J.-M.; Yasui, H.; Yoshimura, A. Mapping QTL for traits associated with resistance to ferrous iron toxicity in rice (Oryza sativa L.), using japonica chromosome segment substitution lines. Acta Genet. Sin. 2003, 30, 893–898. [Google Scholar] [PubMed]
- Nugraha, Y.; Utami, D.W.; Rosdianti, I.D.A.; Ardie, S.W.; Ghulammahdi, M.; Suwarno, S.; Aswidinnoor, H. Markers-traits association for iron toxicity tolerance in selected Indonesian rice varieties. Biodivers. J. Biol. Divers. 2016, 17, 753–763. [Google Scholar] [CrossRef]
- Shimizu, A.; Guerta, C.Q.; Gregorio, G.B.; Kawasaki, S.; Ikehashi, H. QTLs for nutritional contents of rice seedlings (Oryza sativa L.) in solution cultures and its implication to tolerance to iron-toxicity. Plant Soil 2005, 275, 57–66. [Google Scholar] [CrossRef]
- Fukuda, A.; Shiratsuchi, H.; Fukushima, A.; Yamaguchi, H.; Mochida, H.; Terao, T.; Ogiwara, H. Detection of chromosomal regions affecting iron concentration in rice shoots subjected to excess ferrous iron using chromosomal segment substitution lines between Japonica and Indica. Plant Prod. Sci. 2012, 15, 183–191. [Google Scholar] [CrossRef]
- Dufey, I.; Draye, X.; Lutts, S.; Lorieux, M.; Martinez, C.; Bertin, P. Novel QTLs in an interspecific backcross Oryza sativa × Oryza glaberrima for resistance to iron toxicity in rice. Euphytica 2015, 204, 609–625. [Google Scholar] [CrossRef]
- Mendoza, R.D.; Moliñawe, J.A.; Gregorio, G.B.; Guerta, C.Q.; Brar, D.S. Genetic variability of tolerance for iron toxicity in different species of Oryza and their derivatives. In Advances in Rice Genetics: (In 2 Parts); World Scientific: Singapore, 2003; pp. 154–157. [Google Scholar]
- Crestani, M.; Da Silva, J.A.G.; de Souza, V.Q.; Hartwig, I.; de Souza Luche, H.; de Sousa, R.O.; De Carvalho, F.I.F.; De Oliveira, A.C.; Luche, H.D.S.; de Sousa, R.O.; et al. Irrigated rice genotype performance under excess iron stress in hydroponic culture. Crop Breed. Appl. Biotechnol. 2009, 9, 85–93. [Google Scholar] [CrossRef]
- Shimizu, A.; Guerta, C.Q.; Gregorio, G.B.; Ikehashi, H. Improved mass screening of tolerance to iron toxicity in rice by lowering temperature of culture solution. J. Plant Nutr. 2005, 28, 1481–1493. [Google Scholar] [CrossRef]
- Ouyang, Y.; Zeng, F.; Zhuang, J.; Yu, S.; Zhu, L.; Jin, Q.; Zhang, G. Genetic analysis of genotype× iron nutrition interaction on coleoptile elongation rate in rice (Oryza sativa L.). Euphytica 2007, 156, 311. [Google Scholar] [CrossRef]
- Matthus, E.; Wu, L.B.; Ueda, Y.; Höller, S.; Becker, M.; Frei, M. Loci, genes, and mechanisms associated with tolerance to ferrous iron toxicity in rice (Oryza sativa L.). Theor. Appl. Genet. 2015, 128, 2085–2098. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, K.; Pang, Y.; Naveed, S.A.; Zhao, X.; Wang, X.; Wang, Y.; Dingkuhn, M.; Pasuquin, J.; Li, Z. QTL mapping and candidate gene analysis of ferrous iron and zinc toxicity tolerance at seedling stage in rice by genome-wide association study. BMC Genom. 2017, 18, 828. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Mangrauthia, S.K.; Sarla, N. Expression profiling of iron deficiency responsive microRNAs and gene targets in rice seedlings of Madhukar × Swarna recombinant inbred lines with contrasting levels of iron in seeds. Plant Soil 2015, 396, 137–150. [Google Scholar] [CrossRef]
- Chrisnawati, L. STS marker associated with iron toxicity tolerance in rice. J. Trop. Life Sci. 2016, 6, 59–64. [Google Scholar]
- Sadana, U.S.; Nayyar, V.K. Amelioration of iron deficiency in rice and transformations of soil iron in coarse textured soils of Punjab, India. J. Plant Nutr. 2000, 23, 2061–2069. [Google Scholar] [CrossRef]
- Silva, M.M.; Vale, M.G.R.; Damin, I.C.F.; Welz, B.; Mandaji, M.; Fett, J.P. Method development for the determination of iron in milligram amounts of rice plants (Oryza sativa L.) from cultivation experiments using graphite furnace atomic absorption spectrometry. Anal. Bioanal. Chem. 2003, 377, 165–172. [Google Scholar] [CrossRef]
- Singh, N.; Kayal, N.; Gupta, P.K.; Agrawal, A.K. Monitoring the trace metals concentration in rice by flame atomic absorption spectrometer and inductively coupled plasma atomic emission spectrometer. J. Environ. Sci. Eng. 2010, 52, 33–36. [Google Scholar]
- Jahan, G.S.; Hassan, L.; Begum, S.N.; Islam, S.N. Identification of iron rich rice genotypes in Bangladesh using chemical analysis. J. Bangladesh Agric. Univ. 2013, 11, 73–78. [Google Scholar] [CrossRef]
- Lu, L.; Tian, S.; Liao, H.; Zhang, J.; Yang, X.; Labavitch, J.M.; Chen, W. Analysis of metal element distributions in rice (Oryza sativa L.) seeds and relocation during germination based on X-ray fluorescence imaging of Zn, Fe, K, Ca, and Mn. PLoS ONE 2013, 8, e57360. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Nozoye, T.; Kitajima, N.; Fukuda, N.; Hokura, A.; Terada, Y.; Nakai, I.; Ishimaru, Y.; Kobayashi, T.; Nakanishi, H.; et al. In vivo analysis of metal distribution and expression of metal transporters in rice seed during germination process by microarray and X-ray fluorescence imaging of Fe, Zn, Mn, and Cu. Plant Soil 2009, 325, 39–51. [Google Scholar] [CrossRef]
- Kiranmayi, S.L.; Manorama, K.; Tripura Venkata, V.G.N.; Radhika, K.; Cheralu, C.; Roja, V.; Sarla, N. Identification of markers associated with iron and zinc concentration in recombinant inbred lines of brown rice. Indian J. Genet. Plant Breed. 2014, 74, 423–429. [Google Scholar] [CrossRef]
- Gustafsson, J.P.; Pechová, P.; Berggren, D. Modeling metal binding to soils: The role of natural organic matter. Environ. Sci. Technol. 2003, 37, 2767–2774. [Google Scholar] [CrossRef] [PubMed]
- Yokosho, K.; Yamaji, N.; Ueno, D.; Mitani, N.; Ma, J.F. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 2009, 149, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Nerkar, Y.S.; Misal, M.B.; Marekar, R.V. PBN 1, a semidwarf upland rice cultivar tolerant of iron deficiency. Int. Rice Res. Newsl. 1984, 9, 15–16. [Google Scholar]
- Hoan, N.T.; Rao, U.P.; Siddiq, E.A. Genetics of tolerance to iron chlorosis in rice. In Genetic Aspects of Plant Mineral Nutrition; Springer: Dordrecht, Netherland, 1993; pp. 327–333. [Google Scholar]
- Kakei, Y.; Ishimaru, Y.; Kobayashi, T.; Yamakawa, T.; Nakanishi, H.; Nishizawa, N.K. OsYSL16 plays a role in the allocation of iron. Plant Mol. Biol. 2012, 79, 583–594. [Google Scholar] [CrossRef]
- Masuda, H.; Shimochi, E.; Hamada, T.; Senoura, T.; Kobayashi, T.; Aung, M.S.; Ishimaru, Y.; Ogo, Y.; Nakanishi, H.; Nishizawa, N.K. A new transgenic rice line exhibiting enhanced ferric iron reduction and phytosiderophore production confers tolerance to low iron availability in calcareous soil. PLoS ONE 2017, 12, e0173441. [Google Scholar] [CrossRef]
- Lee, S.; An, G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009, 32, 408–416. [Google Scholar] [CrossRef]
- Ogo, Y.; Kobayashi, T.; Itai, R.N.; Nakanishi, H.; Kakei, Y.; Takahashi, M.; Toki, S.; Mori, S.; Nishizawa, N.K. A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J. Biol. Chem. 2008, 283, 13407–13417. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Sarkar, S.; Chakraborty, A.S.; Yelne, R. Phosphate acquisition efficiency and phosphate starvation tolerance locus (PSTOL1) in rice. J. Genet. 2014, 93, 683–688. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Higuchi, K.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3, are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 2003, 36, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ogo, Y.; Itai, R.N.; Nakanishi, H.; Takahashi, M.; Mori, S.; Nishizawa, N.K. The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 19150–19155. [Google Scholar] [CrossRef] [PubMed]
- Ogo, Y.; Kakei, Y.; Nakanishi Itai, R.; Kobayashi, T.; Nakanishi, H.; Nishizawa, N.K. Tissue-specific transcriptional profiling of iron-deficient and cadmium-stressed rice using laser capture microdissection. Plant Signal. Behav. 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Pang, Y.; Wang, C.; Chen, K.; Zhu, Y.; Shen, C.; Ali, J.; Xu, J.; Li, Z. New candidate genes affecting rice grain appearance and milling quality detected by genome-wide and gene-based association analyses. Front. Plant Sci. 2017, 7, 1998. [Google Scholar] [CrossRef]
- Wang, B.; Li, G.; Zhang, W.-H. Brassinosteroids are involved in Fe homeostasis in rice (Oryza sativa L.). J. Exp. Bot. 2015, 66, 2749–2761. [Google Scholar] [CrossRef]
- Xu, B.; Yu, J.Y.; Xie, T.; Li, Y.L.; Liu, M.J.; Guo, J.X.; Li, H.L.; Yu, Y.; Zheng, C.Y.; Chen, Y.H. Brassinosteroids and iron plaque affect arsenic and cadmium uptake by rice seedlings grown in hydroponic solution. Biol. Plant 2018, 62, 362–368. [Google Scholar] [CrossRef]
- Wan, J.J.; Zhai, H.; Wan, J.J.; Ikehashi, H. Detection and analysis of QTLs for ferrous iron toxicity tolerance in rice, Oryza sativa L. Euphytica 2003, 131, 201–206. [Google Scholar] [CrossRef]
- De Dorlodot, S.; Lutts, S.; Bertin, P. Effects of ferrous iron toxicity on the growth and mineral composition of an interspecific rice. J. Plant Nutr. 2005, 28, 1–20. [Google Scholar] [CrossRef]
- Sarla, N.; Swamy, B.P.M. Oryza glaberrima: A source for the improvement of Oryza sativa. Curr. Sci. 2005, 955–963. [Google Scholar]
- Nyamangyoku, I.O.; Bertin, P. Mechanisms of resistance to ferrous iron toxicity in cutivated rices: Oryza sativa L., Oryza glaberrima Steud. and interspecific hybrids. Int. J. Agron. Plant Prod. 2013, 4, 2570–2591. [Google Scholar]
- Ricachenevsky, F.K.; Sperotto, R.A. Into the wild: Oryza species as sources for enhanced nutrient accumulation and metal tolerance in rice. Front. Plant Sci. 2016, 7, 974. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.-L.; Zhai, H.-Q.; Wan, J.-M. Mapping of QTLs for ferrous iron toxicity tolerance in rice (Oryza sativa L.). Yi Chuan Xue Bao 2005, 32, 1156–1166. [Google Scholar] [PubMed]
- Wu, P.; Luo, A.; Zhu, J.; Yang, J.; Huang, N.; Senadhira, D. Molecular markers linked to genes underlying seedling tolerance for ferrous iron toxicity. Plant Soil 1997, 196, 317–320. [Google Scholar] [CrossRef]
- Shimizu, A. QTL analysis of genetic tolerance to iron toxicity in rice (Oryza sativa L.) by quantification of bronzing score. J. New Seeds 2009, 10, 171–179. [Google Scholar] [CrossRef]
- Dufey, I.; Hiel, M.-P.; Hakizimana, P.; Draye, X.; Lutts, S.; Koné, B.; Dramé, K.N.; Konaté, K.A.; Sie, M.; Bertin, P. Multienvironment quantitative trait loci mapping and consistency across environments of resistance mechanisms to ferrous iron toxicity in rice. Crop Sci. 2012, 52, 539–550. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Wada, Y.; Watanabe, S.; Matsuhashi, S.; Takahashi, M. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef]
- Ogo, Y.; Nakanishi Itai, R.; Nakanishi, H.; Kobayashi, T.; Takahashi, M.; Mori, S.; Nishizawa, N.K. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 2007, 51, 366–377. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Takahashi, R.; Bashir, K.; Shimo, H.; Senoura, T.; Sugimoto, K.; Ono, K.; Yano, M.; Ishikawa, S.; Arao, T. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Sci. Rep. 2012, 2, 286. [Google Scholar] [CrossRef]
- Chandel, G.; Banerjee, S.; Vasconcelos, M.; Grusak, M.A. Characterization of the root transcriptome for iron and zinc homeostasis-related genes in indica rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 2010, 19, 145–152. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Bashir, K.; Fujimoto, M.; An, G.; Itai, R.N.; Tsutsumi, N.; Nakanishi, H.; Nishizawa, N.K. Rice-specific mitochondrial iron-regulated gene (MIR) plays an important role in iron homeostasis. Mol. Plant 2009, 2, 1059–1066. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Nozoye, T.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. Exploiting new tools for iron bio-fortification of rice. Biotechnol. Adv. 2013, 31, 1624–1633. [Google Scholar] [CrossRef]
- Stein, R.J.; Ricachenevsky, F.K.; Fett, J.P. Differential regulation of the two rice ferritin genes (OsFER1 and OsFER2). Plant Sci. 2009, 177, 563–569. [Google Scholar] [CrossRef]
- Briat, J.-F.; Ravet, K.; Arnaud, N.; Duc, C.; Boucherez, J.; Touraine, B.; Cellier, F.; Gaymard, F. New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann. Bot. 2009, 105, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ogo, Y.; Aung, M.S.; Nozoye, T.; Itai, R.N.; Nakanishi, H.; Yamakawa, T.; Nishizawa, N.K. The spatial expression and regulation of transcription factors IDEF1 and IDEF2. Ann. Bot. 2010, 105, 1109–1117. [Google Scholar] [CrossRef]
- Connolly, E.L.; Guerinot, M.L. Iron stress in plants. Genome Biol. 2002, 3, reviews1024. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Kobayashi, T.; Takahashi, M.; Nagasaka, S.; Usuda, K.; Kakei, Y.; Ishimaru, Y.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL18 is a rice iron (III)–deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Mol. Biol. 2009, 70, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Ishimaru, Y.; Senoura, T.; Shimo, H.; Ishikawa, S.; Arao, T.; Nakanishi, H.; Nishizawa, N.K. The OsNRAMP1 iron transporter is involved in Cd accumulation in rice. J. Exp. Bot. 2011, 62, 4843–4850. [Google Scholar] [CrossRef]
- Narayanan, N.N.; Vasconcelos, M.W.; Grusak, M.A. Expression profiling of Oryza sativa metal homeostasis genes in different rice cultivars using a cDNA macroarray. Plant Physiol. Biochem. 2007, 45, 277–286. [Google Scholar] [CrossRef]
- Bashir, K.; Takahashi, R.; Nakanishi, H.; Nishizawa, N.K. The road to micronutrient biofortification of rice: Progress and prospects. Front. Plant Sci. 2013, 4, 1–7. [Google Scholar] [CrossRef]
- Bashir, K.; Takahashi, R.; Akhtar, S.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. The knockdown of OsVIT2 and MIT affects iron localization in rice seed. Rice. 2013, 6, 31. [Google Scholar] [CrossRef]
- Shin, S.-Y.; Jeong, J.S.; Lim, J.Y.; Kim, T.; Park, J.H.; Kim, J.-K.; Shin, C. Transcriptomic analyses of rice (Oryza sativa) genes and non-coding RNAs under nitrogen starvation using multiple omics technologies. BMC Genom. 2018, 19, 532. [Google Scholar] [CrossRef]
- Xie, W.; Wang, G.; Yuan, M.; Yao, W.; Lyu, K.; Zhao, H.; Yang, M.; Li, P.; Zhang, X.; Yuan, J. Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc. Natl. Acad. Sci. USA 2015, 112, E5411–E5419. [Google Scholar] [CrossRef]
- Baldoni, E.; Bagnaresi, P.; Locatelli, F.; Mattana, M.; Genga, A. Comparative leaf and root transcriptomic analysis of two rice Japonica cultivars reveals major differences in the root early response to osmotic stress. Rice. 2016, 9, 25. [Google Scholar] [CrossRef]
- Wakasa, Y.; Oono, Y.; Yazawa, T.; Hayashi, S.; Ozawa, K.; Handa, H.; Matsumoto, T.; Takaiwa, F. RNA sequencing-mediated transcriptome analysis of rice plants in endoplasmic reticulum stress conditions. BMC Plant Biol. 2014, 14, 101. [Google Scholar] [CrossRef]
- Ogo, Y.; Itai, R.N.; Nakanishi, H.; Inoue, H.; Kobayashi, T.; Suzuki, M.; Takahashi, M.; Mori, S.; Nishizawa, N.K. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 2006, 57, 2867–2878. [Google Scholar] [CrossRef]
- Ogo, Y.; Kakei, Y.; Itai, R.N.; Kobayashi, T.; Nakanishi, H.; Takahashi, H.; Nakazono, M.; Nishizawa, N.K. Spatial transcriptomes of iron-deficient and cadmium-stressed rice. New Phytol. 2014, 201, 781–794. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Bashir, K.; Nishizawa, N.K. Zn uptake and translocation in rice plants. Rice. 2011, 4, 21–27. [Google Scholar] [CrossRef]
- Wu, L. Genetic and Physiological Analyses of the Tolerance Mechanisms to Ferrous Iron Toxicity in Rice (Oryza sativa L.). Ph.D. Thesis, University of Bonn, Bonn, Germany, 2016. [Google Scholar]
- Mendoza-Soto, A.B.; Sánchez, F.; Hernández, G. MicroRNAs as regulators in plant metal toxicity response. Front. Plant Sci. 2012, 3, 105. [Google Scholar] [CrossRef]
- Li, Y.; Lin, L.; Li, Z.; Ye, X.; Xiong, K.; Aryal, B.; Xu, Z.; Paroo, Z.; Liu, Q.; He, C. Iron homeostasis regulates the activity of the microRNA pathway through poly (C)-binding protein 2. Cell Metab. 2012, 15, 895–904. [Google Scholar] [CrossRef]
- Paul, S.; Gayen, D.; Datta, S.K.; Datta, K. Analysis of high iron rice lines reveals new miRNAs that target iron transporters in roots. J. Exp. Bot. 2016, 67, 5811–5824. [Google Scholar] [CrossRef]
- Waters, B.M.; McInturf, S.A.; Stein, R.J. Rosette iron deficiency transcript and microRNA profiling reveals links between copper and iron homeostasis in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 5903–5918. [Google Scholar] [CrossRef]
- Ivanov, R.; Brumbarova, T.; Bauer, P. Fitting into the harsh reality: Regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 2012, 5, 27–42. [Google Scholar] [CrossRef]
- Junior, A.T.D.; Farias, D.D.; dos Santos, R.S.; do Amaral, M.N.; Arge, L.W.P.; Oliveira, D.D.C.; de Oliveira, A.C. The quest for more tolerant rice: How high concentrations of iron affect alternative splicing. Transcriptomics 2015, 3, 2. [Google Scholar]
- Chen, L.; Ding, C.; Zhao, X.; Xu, J.; Mohammad, A.A.; Wang, S.; Ding, Y. Differential regulation of proteins in rice (Oryza sativa L.) under iron deficiency. Plant Cell Rep. 2015, 34, 83–96. [Google Scholar] [CrossRef]
- Xu, W.F.; Shi, W.M. Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: Analysis by real-time RT–PCR. Ann. Bot. 2006, 98, 965–974. [Google Scholar] [CrossRef]
- Chen, X.; Zhou, D.-X. Rice epigenomics and epigenetics: Challenges and opportunities. Curr. Opin. Plant Biol. 2013, 16, 164–169. [Google Scholar] [CrossRef]
- Galindo-González, L.; Sarmiento, F.; Quimbaya, M. Shaping plant adaptability, genome structure and gene expression through transposable element epigenetic control: Focus on methylation. Agronomy 2018, 8, 180. [Google Scholar] [CrossRef]
- Shi, J.; Dong, A.; Shen, W.-H. Epigenetic regulation of rice flowering and reproduction. Front. Plant Sci. 2015, 5, 803. [Google Scholar] [CrossRef]
- Tikhodeyev, O.N. The mechanisms of epigenetic inheritance: How diverse are they? Biol. Rev. 2018, 93, 1987–2005. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; He, K.; Ma, Y.; Su, N.; He, H.; Stolc, V.; Tongprasit, W.; Jin, W.; Jiang, J. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell 2008, 20, 259–276. [Google Scholar] [CrossRef]
- Bocchini, M.; Bartucca, M.L.; Ciancaleoni, S.; Mimmo, T.; Cesco, S.; Pii, Y.; Albertini, E.; Del Buono, D. Iron deficiency in barley plants: Phytosiderophore release, iron translocation, and DNA methylation. Front. Plant Sci. 2015, 6, 514. [Google Scholar] [CrossRef]
- Choi, C.-S.; Sano, H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genom. 2007, 277, 589–600. [Google Scholar] [CrossRef]
- Cao, D.H.; Gao, X.; Liu, J.; Kimatu, J.N.; Geng, S.J.; Wang, X.P.; Zhao, J.; Shi, D.C. Methylation sensitive amplified polymorphism (MSAP) reveals that alkali stress triggers more DNA hypomethylation levels in cotton (Gossypium hirsutum L.) roots than salt stress. Afr. J. Biotechnol. 2011, 10, 18971–18980. [Google Scholar]
- Wang, W.; Zhao, X.; Pan, Y.; Zhu, L.; Fu, B.; Li, Z. DNA methylation changes detected by methylation-sensitive amplified polymorphism in two contrasting rice genotypes under salt stress. J. Genet. Genom. 2011, 38, 419–424. [Google Scholar] [CrossRef]
- El-Jendoubi, H.; Vázquez, S.; Calatayud, Á.; Vavpetič, P.; Vogel-Mikuš, K.; Pelicon, P.; Abadía, J.; Abadía, A.; Morales, F. The effects of foliar fertilization with iron sulfate in chlorotic leaves are limited to the treated area. A study with peach trees (Prunus persica L. Batsch) grown in the field and sugar beet (Beta vulgaris L.) grown in hydroponics. Front. Plant Sci. 2014, 5, 2. [Google Scholar] [CrossRef]
- Mortvedt, J.J. Correcting iron deficiencies in annual and perennial plants: Present technologies and future prospects. Plant Soil 1991, 130, 273–279. [Google Scholar] [CrossRef]
- Prado, R.M.; Alcantara-Vara, E. Tolerance to iron chlorosis in non-grafted quince seedlings and in pear grafted onto quince plants. J. Soil Sci. Plant Nutr. 2011, 11, 119–128. [Google Scholar] [CrossRef]
- Sarkar, D.; Mandal, B.; Kundu, M.C. Increasing use efficiency of boron fertilisers by rescheduling the time and methods of application for crops in India. Plant Soil 2007, 301, 77–85. [Google Scholar] [CrossRef]
- Fageria, N.K.; Filho, M.P.B.; Moreira, A.; Guimarães, C.M. Foliar fertilization of crop plants. J. Plant Nutr. 2009, 32, 1044–1064. [Google Scholar] [CrossRef]
- Shao, G.; Chen, M.; Wang, D.; Xu, C.; Mou, R.; Cao, Z.; Zhang, X. Using iron fertilizer to control Cd accumulation in rice plants: A new promising technology. Sci. China Ser. C Life Sci. 2008, 51, 245–253. [Google Scholar] [CrossRef]
- Vert, G.A.; Briat, J.-F.; Curie, C. Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol. 2003, 132, 796–804. [Google Scholar] [CrossRef]
- Fernández, V.; Ebert, G. Foliar iron fertilization: A critical review. J. Plant Nutr. 2005, 28, 2113–2124. [Google Scholar] [CrossRef]
- Mandal, L.N.; Haldar, M. Influence of phosphorus and zinc application on the availability of zinc, copper, iron, manganese, and phosphorus in waterlogged rice soils. Soil Sci. 1980, 130, 251–257. [Google Scholar] [CrossRef]
- Alam, S.M. Influence of phosphorus on the growth and nutrient contents of the rice plant. J. Plant Nutr. 1983, 6, 291–299. [Google Scholar] [CrossRef]
- Ramírez, L.M.; Claassen, N.; Ubiera, A.A.; Werner, H.; Moawad, A.M. Effect of phosphorus, potassium and zinc fertilizers on iron toxicity in wetland rice (Oryza sativa L.). Plant Soil 2002, 239, 197–206. [Google Scholar] [CrossRef]
- Ismunadji, M.; Ardjasa, W.S. Potash fertilization for lowland rice can prevent iron toxicity losses. Better Crop. Int. 1989, 3, 12–14. [Google Scholar]
- Naidu, B.S.; Mahadevappa, M.; Inamdar, S.S.; Maharudrappa, K. Performance of IR36 in Karnataka, India. Int. Rice Res. Newsl. 1981, 6, 4. [Google Scholar]
- Sikirou, M. Agro-Morphological Characterization of Lowland Rice Collection for Tolerance to Iron Toxicity; Faculty of Agricultural Sciences (FSA): Abomey-Calavi, Benin, 2009; p. 68. [Google Scholar]
- Yamauchi, M.; Peng, X.X. Iron toxicity and stress-induced ethylene production in rice leaves. Plant Soil 1995, 173, 21–28. [Google Scholar] [CrossRef]
- Muhrizal, S.; Shamshuddin, J.; Fauziah, I.; Husni, M.A.H. Changes in iron-poor acid sulfate soil upon submergence. Geoderma 2006, 131, 110–122. [Google Scholar] [CrossRef]
- Shamshuddin, J.; Elisa, A.A.; Shazana, M.A.R.S.; Fauziah, I.C. Rice defense mechanisms against the presence of excess amount of Al3+ and Fe2+ in the water. Aust. J. Crop Sci. 2013, 7, 314. [Google Scholar]
- Sahrawat, K.L.; Sika, M. Comparative tolerance of Oryza sativa and O. glaberrima rice cultivars for iron toxicity in West Africa. Int. Rice Res. Notes 2002, 27, 30–31. [Google Scholar]
- Linares, O.F. African rice (Oryza glaberrima): History and future potential. Proc. Natl. Acad. Sci. USA 2002, 99, 16360–16365. [Google Scholar] [CrossRef]
- Engel, K.; Asch, F.; Becker, M. Classification of rice genotypes based on their mechanisms of adaptation to iron toxicity. J. Plant Nutr. Soil Sci. 2012, 175, 871–881. [Google Scholar] [CrossRef]
- Gregorio, G.B.; Senadhira, D.; Mendoza, R.D.; Manigbas, N.L.; Roxas, J.P.; Guerta, C.Q. Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 2002, 76, 91–101. [Google Scholar] [CrossRef]
- Singh, B.N. Breeding Improved Varieties for the Irrigated and Rainfed Lowlands; Res. Summ. WARDA Annu. Rep.: Bouaké, Côte d’Ivoire, 1991; pp. 66–70. [Google Scholar]
- Nugraha, Y.; Rumanti, I.A. Breeding for rice variety tolerant to iron toxicity. Iptek Tanam. Pangan 2017, 12, 9–24. [Google Scholar]
- Suhartini, T.; Syafaruddin, S. Genetic estimation of Fe toxicity tolerance in lowland rice through diallel analysis. Zuriat 1996, 7, 33–40. [Google Scholar]
- Suhartini, T.; Makarim, A.K. Teknik seleksi genotipe padi toleran keracunan besi. J. Penelit. Pertan. Tanam. Pangan 2009, 28, 125–130. [Google Scholar]
- Virmani, S.S. Varietal tolerance of rice to iron toxicity in Liberia. Int. Rice Res. Newsl. Int. Rice Res. Inst. 1977, 2, 4–5. [Google Scholar]
| Method | Treatment | Concentration | Fe Compound | Genetic Resources | Adjusted pH | Days After Stress Imposed | References | |
|---|---|---|---|---|---|---|---|---|
| Genotypes | Populations | |||||||
| HNS | FT | 250 mg L−1 | FeSO4 | 135 | DHs | 4.5 | 4 weeks | [117] |
| HNS | FT | 250 mg L−1 | FeSO4 | 96 | BC1F9 | 4.5 | 1 week | [151] |
| HNS | FT | 400 mg L−1 | FeSO4 | 18 | Inbred | 5.5 | 2 weeks | [152] |
| HNS | FT | 0.09 mM | Fe-EDTA | 10 | Inbred | 5.0 | 1 week | [153] |
| HNS | FT | 100 μM | FeCl3 | 8 | Diverse accessions | 5.6 | 2 weeks | [98] |
| HNS | FD | 1 μM | FeCl3 | 8 | Diverse accessions | 4.6 | 2 weeks | [98] |
| HNS | FD | Without Fe | Fe-NaEDTA | 4 | Inbred | 6.5 | 10 days | [7] |
| HNS | FD | 0.1 mmol L−1 | Fe-EDTA | 2 | Inbred | 6.9 | 4 weeks | [128] |
| HNS | FT | 250 mg L−1 | FeSO4 | 164, | RILs | 4.5 | 2 weeks | [31] |
| HNS | FT | 5 mM | FeSO4 | 39 | CSSLs | 4.5 | 2 weeks | [154] |
| HNS | FT | 400 mg L−1 | FeSO4 | 2 | Inbred | 5.5 | 3 days | [85] |
| HNS | FT | 250 mg L−1 | FeSO4 | 220 | DHs | 4.5 | 3 weeks | [155] |
| HNS | FD | Without Fe | - | 2 | Inbred | 5.1 | 3 days | [59] |
| HNS | FT | 500 mg L−1 | FeSO4 | 2 | Inbred | 5.1 | 5 days | [59] |
| HNS | FD | 0.01 μM | Fe-EDTA | 2 | Inbred | 5.9 | 2 weeks | [82] |
| HNS | FT | 0 to 200 mM | FeSO4 | 51 | Inbred | 6.8 | 3 weeks | [26] |
| HNS | FT | 400 ppm | FeSO4 | 161 | Inbred | 5.5 | 4 days | [156] |
| HNS | FT | 0 to 640 mg L−1 | FeSO4 | 6 | Inbred | 5.1 | 3 days | [157] |
| HNS | FT | 7 μM | FeSO4 | 2 | Inbred | 4.0 | 1 week | [17] |
| HNS | FT | 1000 ppm | FeSO4 | 2 | Inbred | 5.5 | 10 days | [67] |
| HNS | FT | 0 to 3000 mg L−1 | FeSO4 | 14 | Inbred | 5.0 | 4 weeks | [42] |
| HNS | FT | 600 mg L−1 & 5 mg L−1 | FeSO4 and Fe-EDTA | 97 | F8 | 5.0 | 2 weeks | [158] |
| HNS | FT | 300 mg L−1 | FeSO4 | 20 | Diverse accessions | 3.0–4.5 | 3 days | [145] |
| HNS | FT | 1.79, 7.16, & 14.32 mM | FeSO4 | 244 | RILs | 5.0 | 12 hours | [159] |
| HNS | FT | 1000 ppm | FeSO4 | 329 | Diverse accessions | 5.5 | 2 weeks | [160] |
| HNS | FT | 300 mg L−1 | FeSO4 | 211 | Inbred | 5.0 | 5 days | [161] |
| HNS | FD | Without Fe | - | 2 | Inbred | 6.0 | 1 week | [162] |
| HNS | FT | 25, 50, & 75 mg L−1 | FeSO4 | 2 | Inbred | 5.5 | 3 weeks | [28] |
| HNS | FT | 300 ppm | Fe-EDTA | 4500 | Mutants | 3.0 | 10 days | [148] |
| HNS | FT | 200 mg L−1 | Fe-EDTA | 4 | Inbred | 5.6 | 3 weeks | [76] |
| HNS | FT | 125 mg L−1 | FeSO4 | 1 | Inbred | 5.0 | 2 weeks | [46] |
| HNS | FT | 400 mg L−1 | FeSO4 | 23 | Inbred | 5.6 | 2 weeks | [39] |
| Field experiment | FT | HTS | - | 2 | Inbred | 6.7 | 3 weeks | [28] |
| Field experiment | FT | 2030 mg kg−1 | - | 2 | Inbred | 3.9 | 3 weeks | [85] |
| Field experiment | FT | 40 to 140 mg L−1 | FeSO4 | 2 | NILs | 5.0 | 3 weeks | [147] |
| Field experiment | FT | 750 ppm | FeSO4 | 5 | DHs | 5.0 | 3 weeks | [163] |
| Field experiment | FD | 7.2 2 mg kg−1 | FeSO4 | 1 | Inbred | 8.2 | 6 weeks | [164] |
| Pot experiment | FD | 14.0 mg kg−1 | FeSO4 | 1 | Inbred | 8.2 | 5 weeks | [164] |
| Pot experiment | FT | HTS | FeSO4 | 5 | Inbred | 4.5 | 3 weeks | [84] |
| Pot experiment | FT | HTS | - | 172 | Inbred | 5.0 | 3 weeks | [88] |
| Pot experiment | FT | HTS | - | 40 | Inbred | 5.1 | 2 weeks | [88] |
| S. No. | Genes | Location | Function | References |
|---|---|---|---|---|
| 1 | OsNAS3 | Shoot ▲ | Involved in mugineic acid pathways (MAs) to transport from soil to root | [180] |
| 2 | OsTOM1 | Root ▲ | Mediates the efflux of DMA into rhizosphere and followed by formation of Fe3+-MA complexes | [96] |
| OSIRT1 and 2 | Root ▲ | Integral membrane Fe2+ transporter | [45,195,196] | |
| 3 | OsIRO2 | Root and shoot | DMA biosynthesis | [196] |
| 4 | OsNAS1-2, Os NAAT1, and OsDMAS1 | Root and shoot ▲ | Fe acquisition and translocation | [45,180] |
| 5 | OsNRAMP1 | Root and shoot ▲ | Transporation of Fe from roots to aerial parts, including rice grains | [46,197] |
| 6 | OsYSL2, 15, 16 | Root ▲ | Phenolics efflux mechanisms, which can bind with Fe3+ for uptake and long-distance Fe transport | [45,46] |
| 7 | OsZIP4 | Root ▲ | Zinc-transporting protein involves Fe transport and homeostasis | [46] |
| 8 | OsYSL2 | Root ▲ | Fe-NA transporter and Fe accumulation in seeds and translocation of Fe into the grain | [108] |
| 9 | OsFRO2 | Root ▲ | Absorption and uptake of Fe from soil to roots | [24,198] |
| 10 | OsFRDL1 | Shoot and flower ▲ | Long-distance Fe transport and homeostasis | [108] |
| 11 | OsPIC1 | Chloroplast ▲ | Fe transport from root to chloroplast | [107] |
| 12 | OsMIR | Shoot ▲ | Fe homeostasis | [199] |
| OsNRAMP5 | Shoot ▲ | Fe translocation | [197] | |
| 13 | OsDMAS1 | Root and shoot ◈ | Vacuolar Fe transporter from roots to shoots | [20,107,200] |
| 14 | OsVIT1,2 | Leaves and seeds ◈ | Vacuolar Fe transporter | [45,106,107] |
| 15 | OsFER1 | Aleurone layer ◈ | Vacuolar Fe transporter and homeostasis | [201] |
| 16 | OsFER1 and OsFER2 | Leaves ◈ | Increased in leaves under excess ferrous iron conditions | [19,59,201,202] |
| 17 | OsIRT1, OsNAS1-3, OsNAAT1, OsFRO2, and OsDMAS1 | Root and shoot  | Biosynthesis of chelates and PS, expressed in phloem companion cells to contribute to iron translocation and transport of Fe from roots to shoots | [24,45,114,203] |
| 18 | OsFRDL1 | Root and shoot  | Fe3+-citrate complex localized on pericycle cells of roots to contribute to iron translocation to shoots | [172] |
| 19 | OsFRO2 | Root  | Changing Fe oxidation state | [201,204] |
| 20 | OsWRKY55, OsWRKY46, OsWRKY64, and OsWRKY113 | Root and shoot  | Repress Fe translocation to shoots by regulating root elongation | [120] |
| 21 | OsIDEF1, OsIDEF2 | Leaves and roots  | Metal transporters and through the binding process of TF IDE-binding factor 1 and 2 genes | [74,104] |
| 22 | OsYSL2, OsYSL15, and OsYSL18 | Root  | Fe-chelate transporter, expressed in phloem companion cells, to contribute to iron translocation through the phloem | [45,203] |
| 23 | OsNAS1, OsNAS2, OsNAAT1, OsDMAS1, and OsYSL15 | Root  | Regulate PS-mediated Fe uptake and homeostasis | [181,196] |
| 24 | OsYSL16 | Epidermis and cortex  | Acquisition and inter-organ transport of Fe xylem-to-phloem | [94,205] |
| 25 | OsYSL18 | Lamina joints, flower  | Fe3+-DMA transporter involved in DMA-mediated Fe dispersal in reproductive organs | [205] |
| 26 | OsYSL5-7, -14, and -17 | Epidermis, cortex, and stele  | Fe transport from root to shoot and grain | [45] |
| 27 | OsYSL12 | Cortex and stele  | Fe transport from root to shoot and grain | [45] |
| 28 | OsNRAMP1 | Shoot  | Fe transport from root to shoot and highly expressed in Fe deficiency | [206] |
 Fe uptake.
Fe uptake.| S. No. | Microarray | Shoot | Root | Reference | ||
|---|---|---|---|---|---|---|
| Down-Regulation | Up-Regulation | Down-Regulation | Up-Regulation | |||
| 1 | 44K | - | - | 325 a | 1068 a | [109] |
| 2 | 110K | 318 a & 1655 b | 258 a & 2480 b | 2655 a & 116 b | 1509 a & 36 b | [15] |
| 3 | 44K | - | 1346 a | - | 80 b | [215] |
| 4 | 44K | 280 b | 519 b | - | - | [46] |
| 5 | 60K | 195 b | 645 b | 2304 b | 1656 b | [118] |
| S. No. | Cultivar/Variety | Tolerance Level | References |
|---|---|---|---|
| 1 | Cauvery, ARC 10372 | T a | [174] |
| 8 | IPB Kapuas 7R, IPB Batola 6R, IPB1 R Dadahup, IPB Batola 5R, Indragiri, Margasari, and A. Tenggulang | T b | [257] |
| 10 | Kapuas | T b | [258] |
| 11 | Tuljapur | T a | [174] |
| 13 | Inpara 2, B13144-1, Cilamaya Muncul, and Margasari | T b | [152] |
| 14 | B13144-1-MR-2 | T b | [259] |
| 15 | Cilamaya, Siam Saba, Mahsuri, Pokkali, and Awan Kuning | T b | [39] |
| 16 | TOX 85C-C1-15-WAS 1, TOX 85C-C1-16-WAS 1, WITA 3, TOX 3069-66-2-1-6, FKR 19, WITA 4, CK 4, CK 73, BW 348-1, TOX 4216-25-2-3-1-3, WAT 1059-B-51-2, WAT 1282-B-3-3, WAT 1131-B-26-2-1-2, Nerica-L19, ARICA 6, ARICA 7, and ARICA 8 | T b | [28,40,88,146,260] |
| 17 | IR61246-3B-15-2-2-3, IR61612-3B-16-2-2-1, IR61640-3B-14-3-3-2, WITA 7, Suakoko 8 (ROK 24), TCA 4, and Azucena | T b | [145] |
| 20 | CK4 and Tox4004-8-1-2-3 | T b | [42] |
| 21 | OG 7206, TOG 6218-B, and TOG 7250-A | T b | [84] |
| 22 | Ghanteswari, Mahanadi, Surendra, Bhanza, Lalat, Daya, Keshan, Rajeswari, Tejaswini, Sankar, Bhuban, Uphar, Khandagiri, Udayagiri, Manika, and Rudra | T b | [26] |
| 23 | Suakoko 8 | T b | [186] |
| 24 | BR IRGA 414, IRGA 419, and BRS AGRISUL | T b | [157] |
| 25 | PBN1 (Prabhavati) | T a | [173] |
| 26 | ISA-40 and PSQ-4 | T b | [245] |
| 27 | Pusa-33 | T a | [98] |
| 28 | EPAGRI 108 | T b | [59] |
| 29 | IR36 | T a | [247] |
| 30 | Dom Sofid | T b | [67] |
| S. No. | Cultivar/Variety | Tolerance Level | References |
|---|---|---|---|
| 1 | Mahsurian | MT b | [259] |
| 2 | WITA 1 and Matkandu | MT b | [78] |
| 3 | Prasanna | MT a | [174] |
| 4 | Akashi | MT a | |
| 5 | WBPH 25 | MT a | |
| 6 | Inpara 3 | MT b | [152] |
| 7 | IET7613 | MT a | [145] |
| 8 | Phalguna | MT b | [145] |
| 9 | PSBRc 18 | MT b | |
| 10 | WITA 1 and WITA 2 | MT b | |
| 11 | Mahsuri | MT b | |
| 12 | CG14 | MT b | [38] |
| 13 | I Kong Pao and Sahel 108 | MT b | |
| 14 | ITA 306 and ITA 320 | MT b | |
| 15 | IR74 and Mahsuri | MT b | [186] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahender, A.; Swamy, B.P.M.; Anandan, A.; Ali, J. Tolerance of Iron-Deficient and -Toxic Soil Conditions in Rice. Plants 2019, 8, 31. https://doi.org/10.3390/plants8020031
Mahender A, Swamy BPM, Anandan A, Ali J. Tolerance of Iron-Deficient and -Toxic Soil Conditions in Rice. Plants. 2019; 8(2):31. https://doi.org/10.3390/plants8020031
Chicago/Turabian StyleMahender, Anumalla, B. P. Mallikarjuna Swamy, Annamalai Anandan, and Jauhar Ali. 2019. "Tolerance of Iron-Deficient and -Toxic Soil Conditions in Rice" Plants 8, no. 2: 31. https://doi.org/10.3390/plants8020031
APA StyleMahender, A., Swamy, B. P. M., Anandan, A., & Ali, J. (2019). Tolerance of Iron-Deficient and -Toxic Soil Conditions in Rice. Plants, 8(2), 31. https://doi.org/10.3390/plants8020031