Cold-Responsive Regions of Paradigm Cold-Shock and Non-Cold-Shock mRNAs Responsible for Cold Shock Translational Bias
Abstract
1. Introduction
2. Results
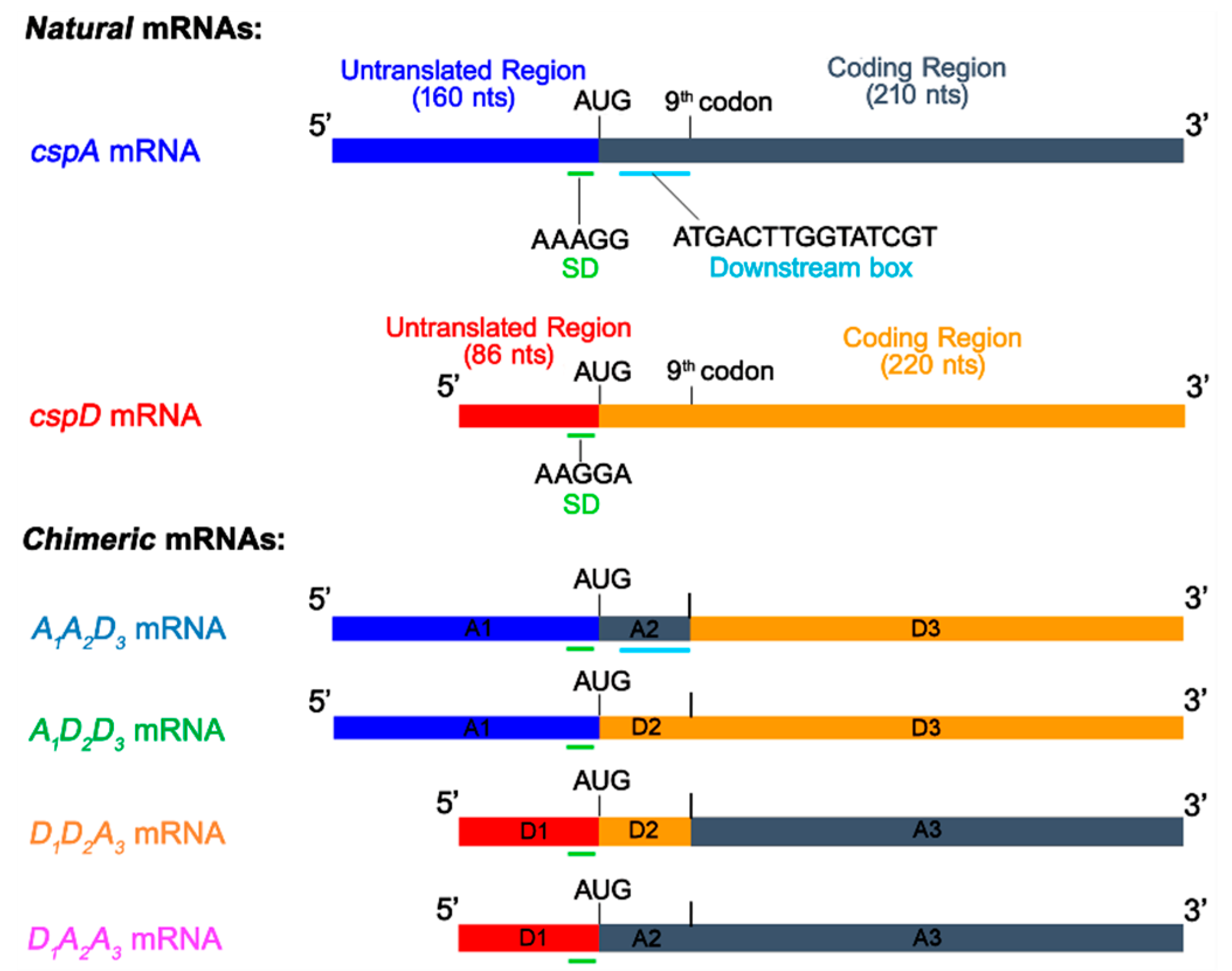
2.1. Construction of Chimeric mRNAs
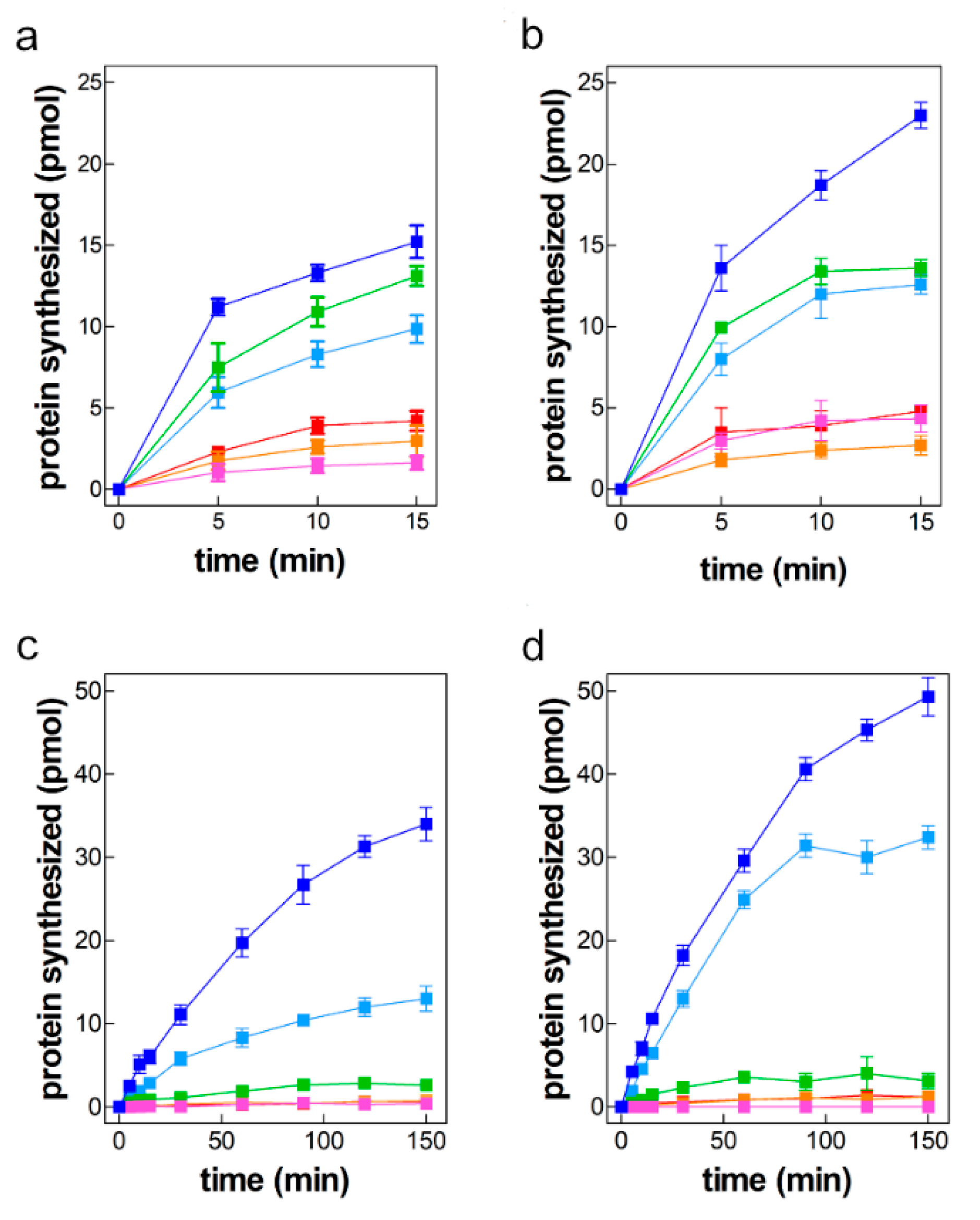
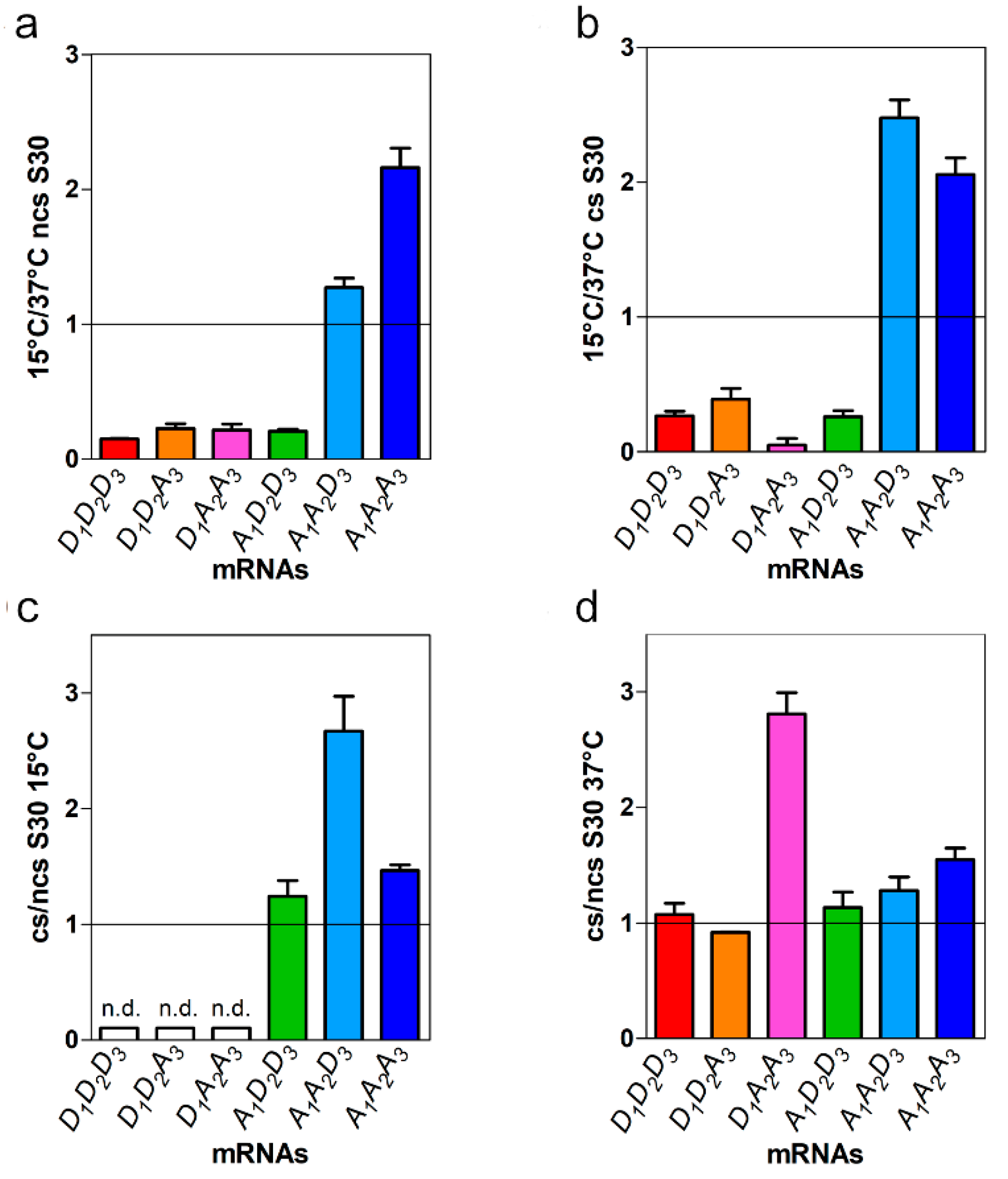
2.2. mRNAs-Dependent Translation in Cell Extracts of Control and Cold-Stressed Cells
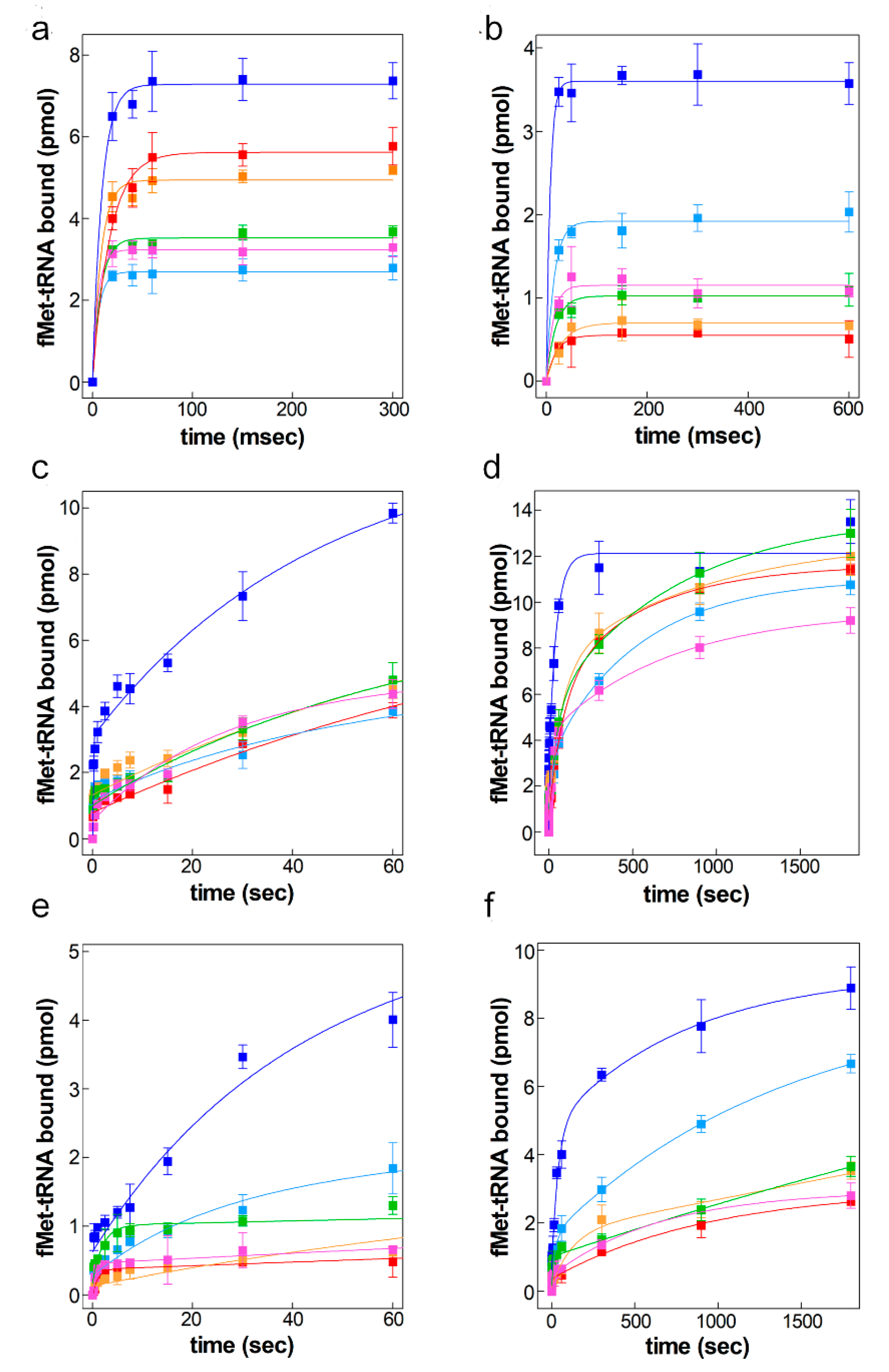
2.3. Kinetic Analyses of 30S Initiation Complex Formation
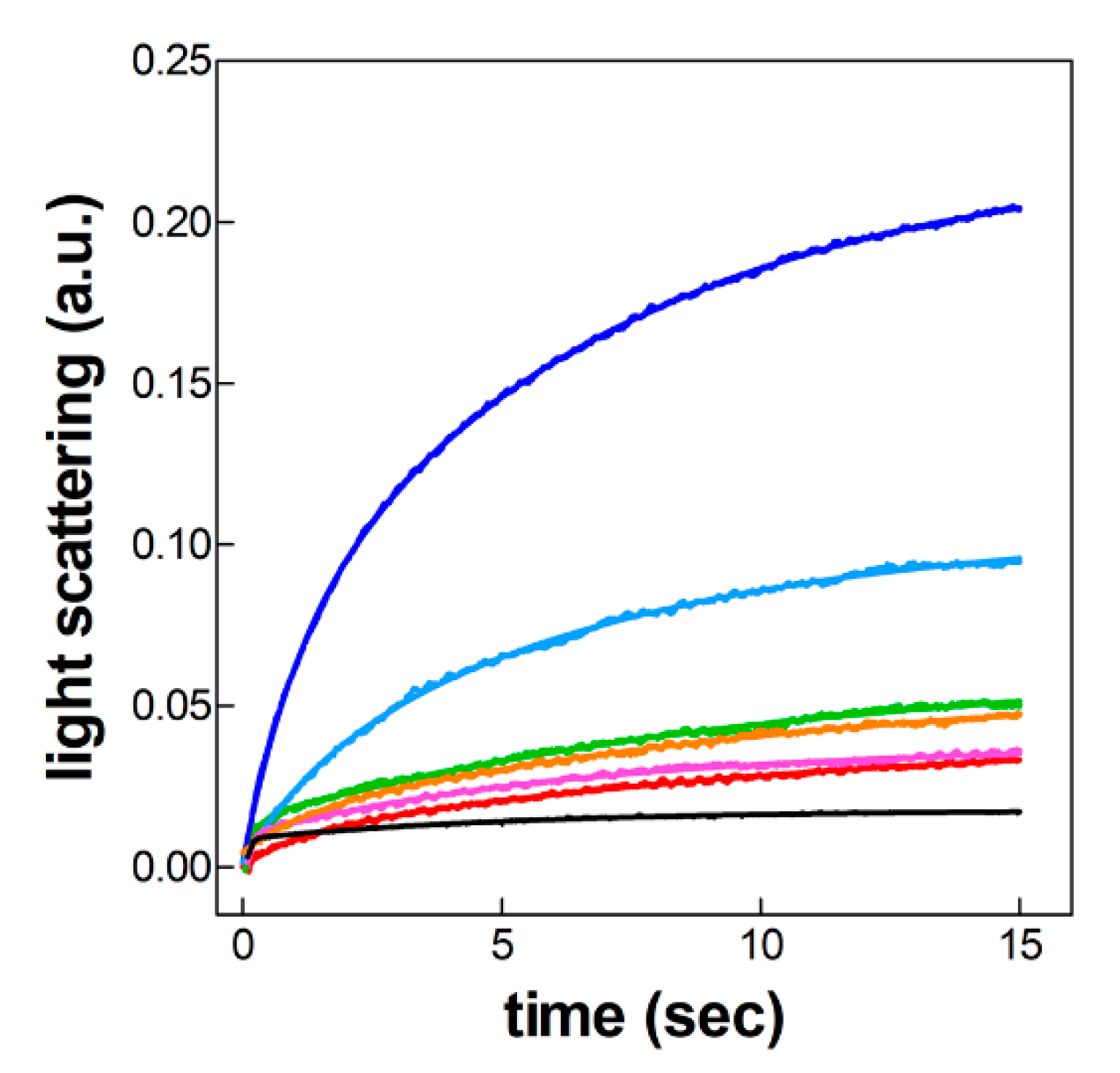
2.4. 50S Docking to the Various 30S Complexes
3. Discussion
4. Materials and Methods
4.1. General Preparations
4.2. Preparation of S30 Fractions
4.3. Construction of pTZ18A1D2D3, pTZ18D1A2A3, pTZ18A1A2D3 and pTZ18D1D2A3
4.4. In Vitro Translation Reactions
4.5. fMet-tRNA Binding Kinetics
4.6. Light Scattering Measurements
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Giuliodori, A.M.; Gualerzi, C.O.; Soto, S.; Vila, J.; Tavío, M.M. Review on bacterial stress topics. Ann. N. Y. Acad. Sci. 2007, 1113, 95–104. [Google Scholar] [CrossRef]
- Storz, G.; Hengge, R. (Eds.) Bacterial Stress Responses, 2nd ed.; ASM Press: Washington, DC, USA, 2011; ISBN 9781555816216. [Google Scholar]
- De Bruijin, F.J. (Ed.) Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; Wiley-Blackwell Publishing: Hoboken, NJ, USA, September 2016; Volume 1–2, ISBN 978-1-119-00488-2. [Google Scholar]
- Holmqvist, E.; Wagner, E.G.H. Impact of bacterial sRNAs in stress responses. Biochem. Soc. Trans. 2017, 45, 1203–1212. [Google Scholar] [CrossRef]
- Falconi, M.; Colonna, B.; Prosseda, G.; Micheli, G.; Gualerzi, C.O. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998, 17, 7033–7043. [Google Scholar] [CrossRef]
- Prosseda, G.; Falconi, M.; Giangrossi, M.; Gualerzi, C.O.; Micheli, G.; Colonna, B. The virF promoter in Shigella: More than just a curved DNA stretch. Mol. Microbiol. 2004, 51, 523–537. [Google Scholar] [CrossRef]
- Jones, P.G.; VanBogelen, R.A.; Neidhardt, F.C. Induction of proteins in response to low temperature in Escherichia coli. J. Bacteriol. 1987, 169, 2092–2095. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.G.; Inouye, M. The cold-shock response: A hot topic. Mol. Microbiol. 1994, 11, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Gualerzi, C.O.; Giuliodori, A.M.; Pon, C.L. Transcriptional and post-transcriptional control of cold shock genes. J. Mol. Biol. 2003, 331, 527–539. [Google Scholar] [CrossRef]
- Weber, M.H.; Marahiel, M.A. Bacterial cold shock responses. Sci. Prog. 2003, 86, 9–75. [Google Scholar] [CrossRef] [PubMed]
- Giuliodori, A.M. Cold-shock response in Escherichia coli: A model system to study post-transcriptional regulation. In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; de Bruijn, F.J., Ed.; Wiley Blackwell: Hoboken, NJ, USA, 2016; pp. 859–872. ISBN 9781119004813. [Google Scholar]
- Gualerzi, C.O.; Giuliodori, A.M.; Brandi, A.; Di Pietro, F.; Piersimoni, L.; Fabbretti, A.; Pon, C.L. Translation initiation at the root of the cold-shock translational bias. In Ribosomes Structure Function and Dynamics; Rodnina, M.V., Wintermeyer, W., Green, R., Eds.; Springer: Wien, Austria, 2011; pp. 143–154. ISBN 9783709102152. [Google Scholar]
- Brandi, A.; Pietroni, P.; Gualerzi, C.O.; Pon, C.L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol. Microbiol. 1996, 19, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, D.; Azar, I.; Oppenheim, A.B.; Brandi, A.; Pon, C.L.; Gualerzi, C.O. Role of Escherichia coli cspA promoter sequences and adaptation of translational apparatus in the cold-shock response. Mol. Gen. Genet. 1997, 256, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Giuliodori, A.M.; Brandi, A.; Gualerzi, C.O.; Pon, C.L. Preferential translation of cold-shock mRNAs during cold adaptation. RNA 2004, 10, 265–276. [Google Scholar] [CrossRef]
- Giuliodori, A.M.; Brandi, A.; Giangrossi, M.; Gualerzi, C.O.; Pon, C.L. Cold-stress induced de novo expression of infC and role of IF3 in cold-shock translational bias. RNA 2007, 13, 1355–1365. [Google Scholar] [CrossRef]
- Giangrossi, M.; Brandi, A.; Giuliodori, A.M.; Gualerzi, C.O.; Pon, C.L. Cold-shock induced de novo transcription and translation of infA and role of IF1 during cold adaptation. Mol. Microbiol. 2007, 64, 807–821. [Google Scholar] [CrossRef] [PubMed]
- Piersimoni, L.; Giangrossi, M.; Marchi, P.; Brandi, A.; Gualerzi, C.O.; Pon, C.L. De novo synthesis and assembly of rRNA into ribosomal subunits during cold acclimation in Escherichia coli. J. Mol. Biol. 2016, 428, 1558–1573. [Google Scholar] [CrossRef] [PubMed]
- Giuliodori, A.M.; Di Pietro, F.; Marzi, S.; Masquida, B.; Wagner, R.; Romby, P.; Gualerzi, C.O.; Pon, C.L. The cspA mRNA is a thermosensor that modulates translation of the cold-shock protein CspA. Mol. Cell 2010, 37, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Brandi, A.; Spurio, R.; Gualerzi, C.O.; Pon, C.L. Massive presence of the Escherichia coli ‘major cold-shock protein’ CspA under non-stress conditions. EMBO J. 1999, 18, 1653–1659. [Google Scholar] [CrossRef] [PubMed]
- Brandi, A.; Pon, C.L. Expression of Escherichia coli cspA during early exponential growth at 37 °C. Gene 2012, 492, 382–388. [Google Scholar] [CrossRef]
- Goldstein, J.; Pollitt, N.S.; Inouye, M. Major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 1990, 87, 283–287. [Google Scholar] [CrossRef]
- La Teana, A.; Brandi, A.; Falconi, M.; Spurio, R.; Pon, C.L.; Gualerzi, C.O. Identification of a cold shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc. Natl. Acad. Sci. USA 1991, 88, 10907–10911. [Google Scholar] [CrossRef]
- Yamanaka, K.; Fang, L.; Inouye, M. The CspA family in Escherichia coli: Multiple gene duplication for the stress adaptation. Mol. Microbiol. 1998, 27, 247–255. [Google Scholar] [CrossRef]
- Yamanaka, K.; Inouye, M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J. Bacteriol. 1997, 179, 5126–5130. [Google Scholar] [CrossRef]
- Kim, Y.; Wang, X.; Zhang, X.S.; Grigoriu, S.; Page, R.; Peti, W.; Wood, T.K. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 2010, 12, 1105–1121. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Wood, T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010, 391, 209–213. [Google Scholar] [CrossRef]
- Uppal, S.; Shetty, D.M.; Jawali, N. Cyclic AMP receptor protein regulates cspD, a bacterial toxin gene, in Escherichia coli. J. Bacteriol. 2014, 196, 1569–1577. [Google Scholar] [CrossRef]
- Giuliodori, A.M.; Spedalieri, G.; Fabbretti, A.; Gualerzi, C.O. Cold shock translational bias. Discrimination of non-cold-shock mRNA by IF3. 2019; in preparation. [Google Scholar]
- Sprengart, M.L.; Fuchs, E.; Porter, A.G. The downstream box: An efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996, 15, 665–674. [Google Scholar] [CrossRef]
- Sprengart, M.L.; Porter, A.G. Functional importance of RNA interactions in selection of translation initiation codons. Mol. Microbiol. 1997, 24, 19–28. [Google Scholar] [CrossRef]
- Mitta, M.; Fang, L.; Inouye, M. Deletion analysis of cspA of Escherichia coli: Requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol. Microbiol. 1997, 26, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Etchegaray, J.P.; Inouye, M. DB or not DB in translation? Mol. Microbiol. 1999, 33, 438–439. [Google Scholar] [CrossRef]
- Etchegaray, J.P.; Inouye, M. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J. Biol. Chem. 1999, 274, 10079–10085. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, K.; Anastasov, N.; Kaberdin, V.; Francis, K.P.; Scherer, S. The AGUAAA motif in cspA1/A2 mRNA is important for adaptation of Yersinia enterocolitica to grow at low temperature. Mol. Microbiol. 2003, 50, 1629–1645. [Google Scholar] [CrossRef]
- Gualerzi, C.; Risuleo, G.; Pon, C.L. Initial rate kinetic analysis of the mechanism of initiation complex formation and the role of initiation factor IF-3. Biochemistry 1977, 16, 1684–1689. [Google Scholar] [CrossRef] [PubMed]
- Milon, P.; Carotti, M.; Konevega, A.L.; Wintermeyer, W.; Rodnina, M.V.; Gualerzi, C.O. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 2010, 11, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Fabbretti, A.; Schedlbauer, A.; Brandi, L.; Kaminichi, T.; Giuliodori, A.M.; Galofaro, R.; Ochoa-Lizarralde, B.; Takemoto, C.; Yokoyama, S.; Connell, S.R.; et al. Inhibition of translation initiation complex formation by GE81112 unravels a 16S rRNA structural switch involved in P-site decoding. Proc. Natl. Acad. Sci. USA 2016, 113, E2286–E2295. [Google Scholar] [CrossRef] [PubMed]
- Wintermeyer, W.; Gualerzi, C. Effect of E. coli initiation factors on the kinetics of NAcPhe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry 1983, 22, 690–694. [Google Scholar] [CrossRef] [PubMed]
- Gualerzi, C.O.; Pon, C.L. Initiation of mRNA translation in bacteria: Structural and dynamic aspects. Cell. Mol. Life Sci. 2015, 72, 4341–4367. [Google Scholar] [CrossRef] [PubMed]
- Milon, P.; Konevega, A.L.; Gualerzi, C.O.; Rodnina, M.V. Kinetic checkpoint at a late step in translation initiation. Mol. Cell 2008, 30, 712–720. [Google Scholar] [CrossRef]
- Yamanaka, K.; Mitta, M.; Inouye, M. Mutation analysis of the 5′-untranslated region of the cold shock cspA mRNA of Escherichia coli. J. Bacteriol. 1999, 181, 6284–6291. [Google Scholar] [PubMed]
- O’Connor, M.; Asai, T.; Squires, C.L.; Dahlberg, A.E. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc. Natl. Acad. Sci. USA 1999, 96, 8973–8978. [Google Scholar] [CrossRef]
- La Teana, A.; Brandi, A.; O’Connor, M.; Freddi, S.; Pon, C.L. Translation during cold adaptation does not involve mRNA-rRNA base pairing through the downstream box. RNA 2000, 6, 1393–1402. [Google Scholar] [CrossRef]
- Giuliodori, A.M.; Duval, M.; Belardinelli, R.; Garofalo, R.; Schenckbecher, E.; Ennifar, E.; Marzi, S. The RNA- chaperone protein CspA promotes the progression of the ribosomes to stimulate translation during cold acclimation. 2019; in preparation. [Google Scholar]
- Kim, S.H.; Heo, M.A.; Kim, Y.J.; Kim, S.Y.; Neelamegam, R.; Lee, S.G. The effect of the cspA 5′-untranslated region on recombinant protein production at low temperature. Biotechnol. Bioprocess Eng. 2008, 13, 366–371. [Google Scholar] [CrossRef]
- Milon, P.; Konevega, A.L.; Peske, F.; Fabbretti, A.; Gualerzi, C.O.; Rodnina, M.V. Transient Kinetics, Fluorescence, and FRET in Studies of Initiation of Translation in Bacteria. Methods Enzymol. 2007, 430, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Ohsawa, H.; Gualerzi, C.O. Chemical modification in situ of Escherichia coli 30S ribosomal proteins by the site-specific reagent pyridoxal phosphate. Inactivation of the aminoacyl-tRNA and mRNA binding sites. J. Biol. Chem. 1983, 258, 150–156. [Google Scholar] [PubMed]
- Rodnina, M.V.; Semenkov, Y.P.; Wintermeyer, W. Purification of fMet-tRNA (fMet) by fast protein liquid chromatography. Anal. Biochem. 1994, 219, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Brandi, L.; Fabbretti, A.; Milon, P.; Carotti, M.; Pon, C.L.; Gualerzi, C.O. Methods for identifying compounds that specifically target translation. Methods Enzymol. 2007, 431, 229–267. [Google Scholar] [CrossRef]

| mRNA | 37 °C | 15 °C |
|---|---|---|
| Kapp (sec−1) | Kapp (sec−1) | |
| cspA | 106 ± 14 | 130 ± 29 |
| A1A2D3 | 160 ± 37 | 66 ± 10 |
| A1D2D3 | 120 ± 27 | 73 ± 24 |
| cspD | 57 ± 6 | 51 ± 9 |
| D1D2A3 | 116 ± 28 | 34 ± 7 |
| D1A2A3 | 175 ± 24 | 53 ± 11 |
| 37 °C | ||||||
| mRNA | K1app (sec−1) | Ymax1 | K2app (sec−1) | Ymax2 | K3app (sec−1) | Ymax3 |
| cspA | 9.7 ± 3.2 | 3.0 ± 0.2 | 0.026 ± 0.003 | 8.4 ± 3.1 | - | - |
| A1A2D3 | 9.2 ± 2.9 | 1.5 ± 0.1 | 0.017 ± 0.007 | 2.2 ± 0.8 | 0.0017 ± 0.0004 | 7.5 ± 0.7 |
| A1D2D3 | 11.9 ± 5.7 | 1.2 ± 0.1 | 0.016 ± 0.003 | 4.5 ± 0.8 | 0.0012 ± 0.0003 | 8.1 ± 0.5 |
| cspD | 12.0 ± 5.7 | 0.96 ± 0.08 | 0.009 ± 0.002 | 5.8 ± 1.3 | 0.00170 ± 0.00065 | 4.96 ± 1.11 |
| D1D2A3 | 11.75 ± 3.89 | 1.64 ± 0.09 | 0.009 ± 0.002 | 6.2 ± 1.0 | 0.0008 ± 0.0005 | 5 ± 1 |
| D1A2A3 | 2.50 ± 1.15 | 0.94 ± 0.01 | 0.027 ± 0.005 | 3.8 ± 0.5 | 0.0012 ± 0.0004 | 5.2 ± 0.5 |
| 15 °C | ||||||
| mRNA | K1app (sec−1) | Ymax1 | K2app (sec−1) | Ymax2 | K3app (sec−1) | Ymax3 |
| cspA | - | - | 0.07 ± 0.03 | 3.4 ± 0.8 | 0.002 ± 0.001 | 5.3 ± 0.8 |
| A1A2D3 | 10.0 ± 5.7 | 0.36 ± 0.05 | 0.031 ± 0.007 | 1.35 ± 0.15 | 0.0006 ± 0.0001 | 7.2 ± 0.6 |
| A1D2D3 | 1.1 ± 0.2 | 0.91 ± 0.05 | - | - | 0.0003 ± 0.0002 | 8 ± 5 |
| cspD | 1.5 ± 0.9 | 0.24 ± 0.09 | 0.043 ± 0.003 | 0.5 ± 0.1 | 0.0004 ± 0.0003 | 3.35 ± 1.35 |
| D1D2A3 | 2.34 ± 1.45 | 0.16 ± 0.03 | 0.018 ± 0.004 | 1.1 ± 0.2 | 0.0021 ± 0.0005 | 1.8 ± 0.2 |
| D1A2A3 | 1.00 ± 0.55 | 0.54 ± 0.08 | - | - | 0.0015 ± 0.0005 | 2.4 ± 0.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giuliodori, A.M.; Fabbretti, A.; Gualerzi, C. Cold-Responsive Regions of Paradigm Cold-Shock and Non-Cold-Shock mRNAs Responsible for Cold Shock Translational Bias. Int. J. Mol. Sci. 2019, 20, 457. https://doi.org/10.3390/ijms20030457
Giuliodori AM, Fabbretti A, Gualerzi C. Cold-Responsive Regions of Paradigm Cold-Shock and Non-Cold-Shock mRNAs Responsible for Cold Shock Translational Bias. International Journal of Molecular Sciences. 2019; 20(3):457. https://doi.org/10.3390/ijms20030457
Chicago/Turabian StyleGiuliodori, Anna Maria, Attilio Fabbretti, and Claudio Gualerzi. 2019. "Cold-Responsive Regions of Paradigm Cold-Shock and Non-Cold-Shock mRNAs Responsible for Cold Shock Translational Bias" International Journal of Molecular Sciences 20, no. 3: 457. https://doi.org/10.3390/ijms20030457
APA StyleGiuliodori, A. M., Fabbretti, A., & Gualerzi, C. (2019). Cold-Responsive Regions of Paradigm Cold-Shock and Non-Cold-Shock mRNAs Responsible for Cold Shock Translational Bias. International Journal of Molecular Sciences, 20(3), 457. https://doi.org/10.3390/ijms20030457







