Responses of HSP70 Gene to Vibrio parahaemolyticus Infection and Thermal Stress and Its Transcriptional Regulation Analysis in Haliotis diversicolor
Abstract
:1. Introduction
2. Results
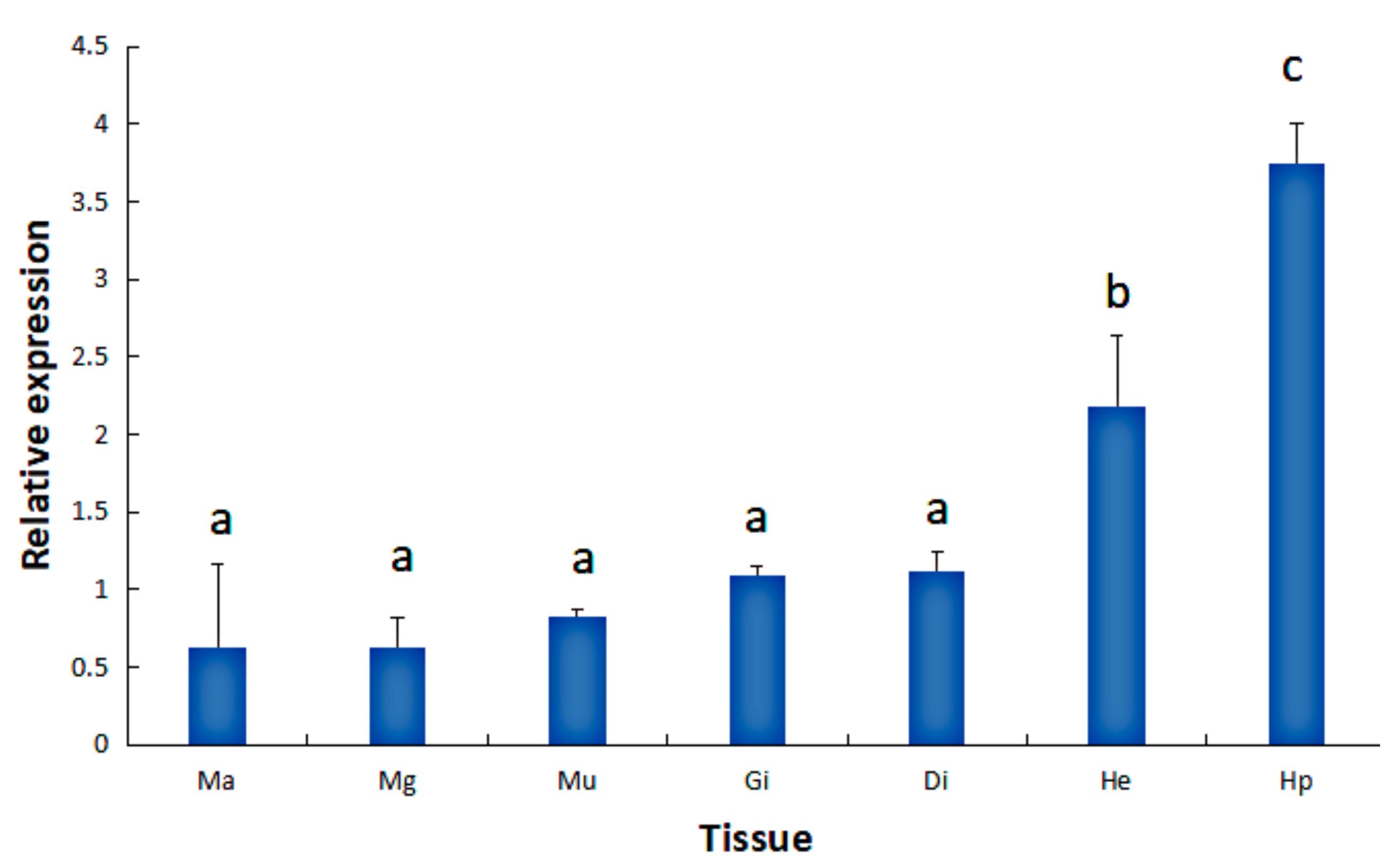
2.1. Tissue Expression of the HdHSP70 Gene
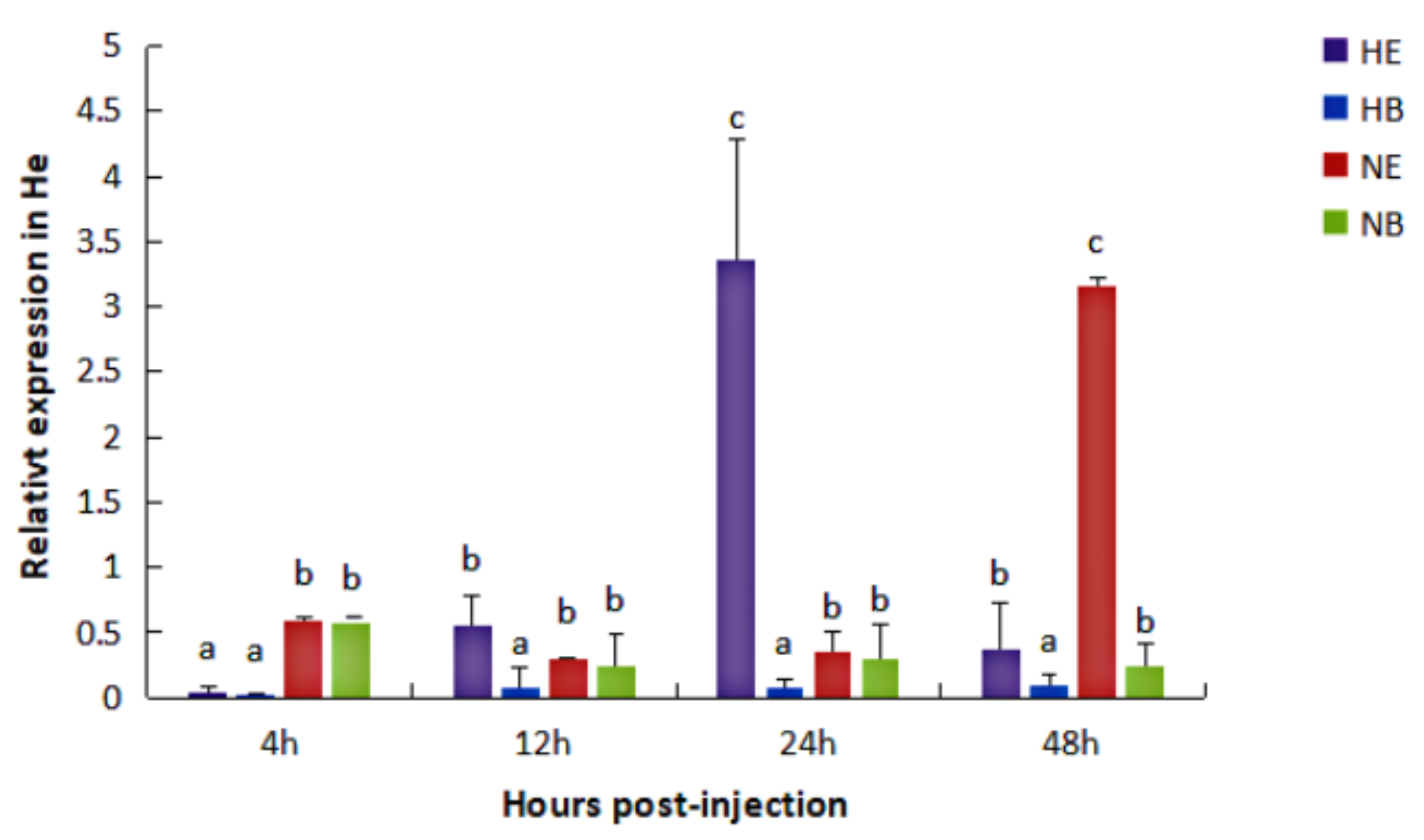
2.2. Expression of the HdHSP70 Gene under Different Stresses
2.3. The Sequence Analysis of the 5′-Upstream Regulatory Region of HdHSP70 Gene
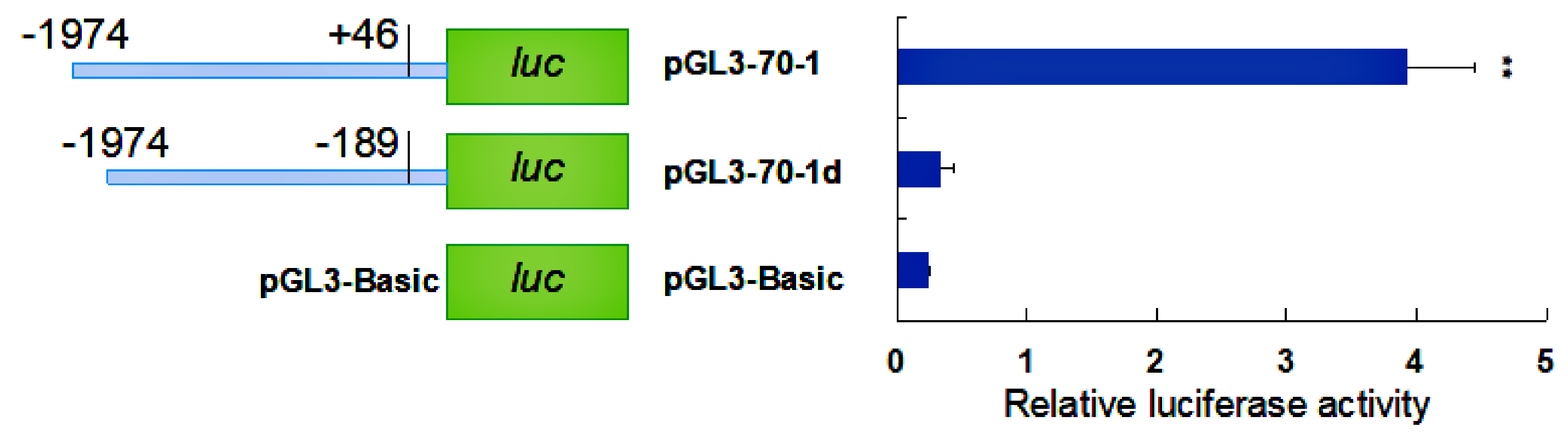
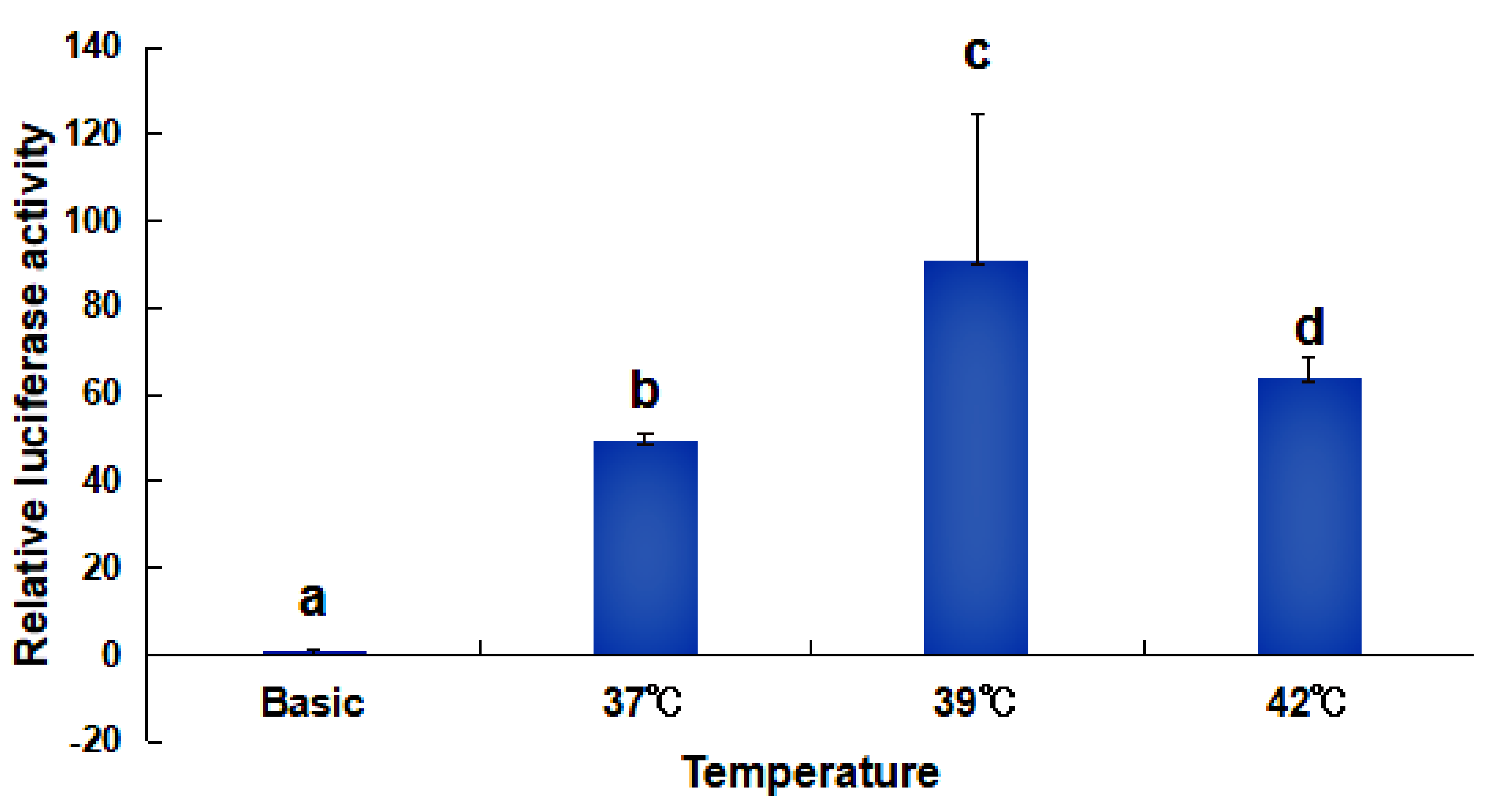
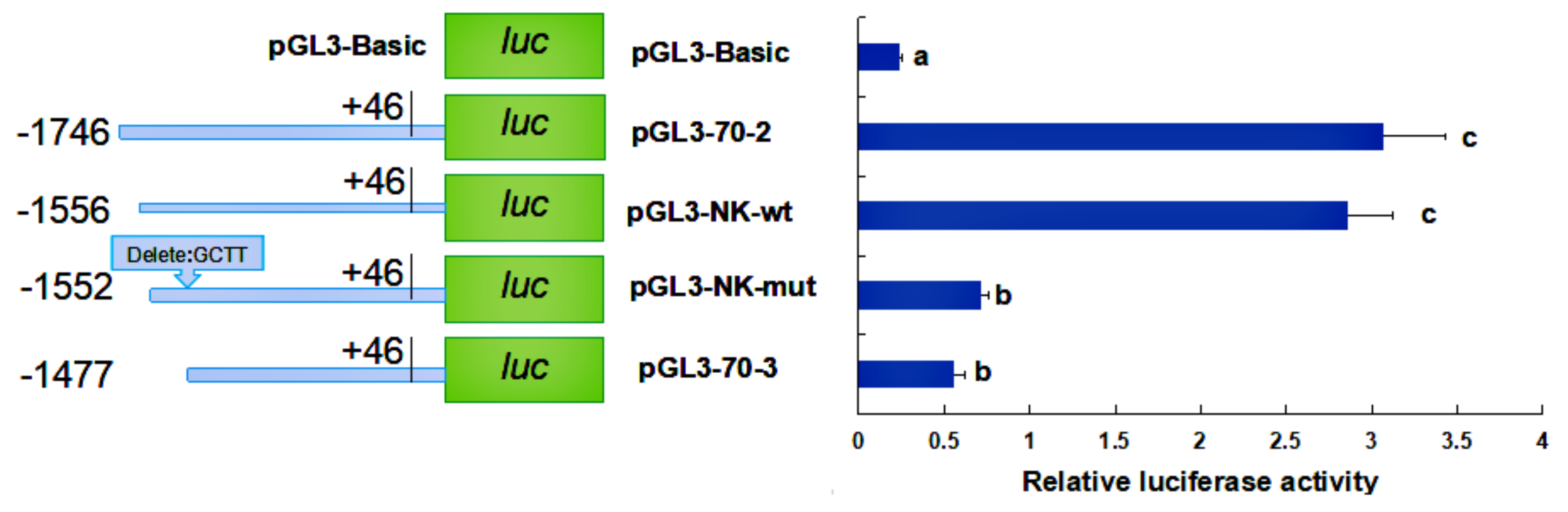
2.4. Activity Analysis of the Promoter Region of HdHSP70 Gene
3. Discussion
3.1. The Expression of HdHSP70 mRNA in Different Tissues and Different Stresses
3.2. Analysis of the 5′-Flanking Sequence of the HSP70 Gene
3.3. Analysis of the Activity of the HdHSP70 Promoter
4. Materials and Methods
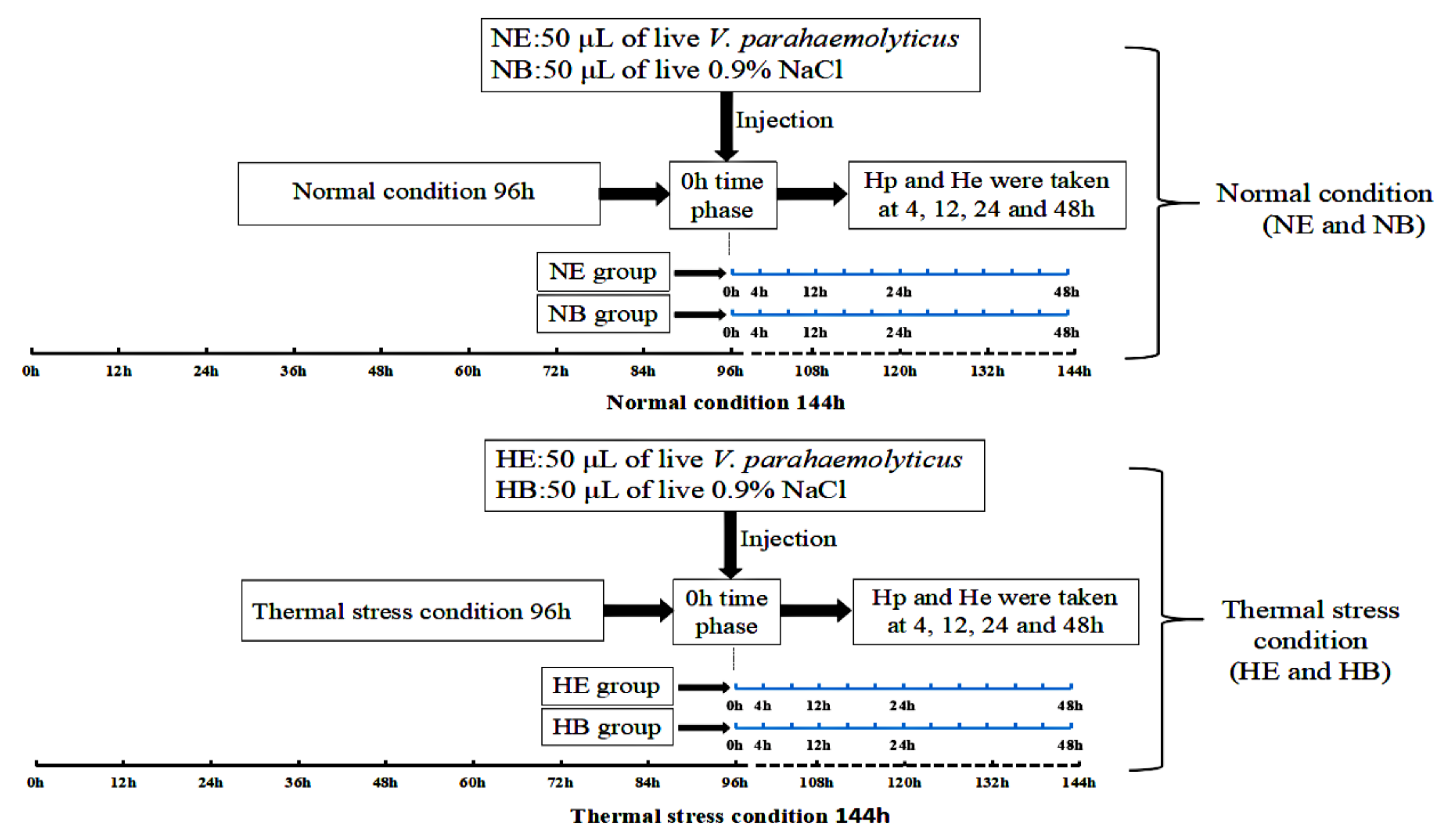
4.1. Animals and Preparation of Samples
4.2. Isolation and Reverse Transcription of Total RNA
4.3. Analysis of HdHSP70 Gene Expression After Stresses by qRT-PCR
4.4. Cloning and Bioinformatic Analyses of the 5′-Flanking Regions of HdHSP70 Gene
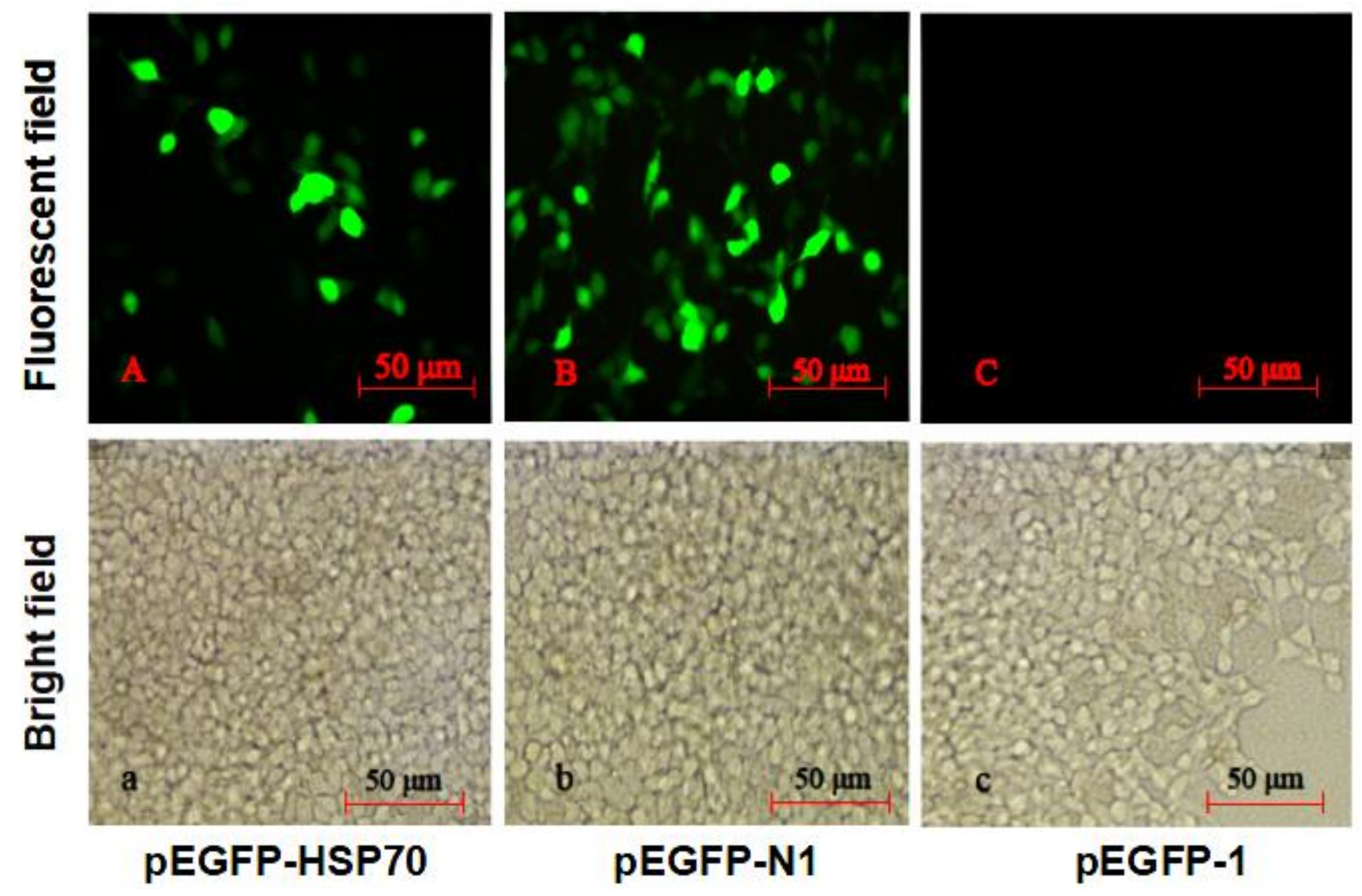
4.5. HdHSP70 Promoter Activity in pEGFP-1 Vector
4.6. Construction and Identification of Missing Fragment Expression Vector
4.7. Mutation of Transcription Factor Binding Sites
4.8. Transient Transfection and Activity Assays of the Luciferase Reporter Plasmids
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ke, C.H.; You, W.W. Advances in genetics and breeding of Haliotis diversicolor. J. Xiamen Univ. 2011, 50, 425–430. (In Chinese) [Google Scholar]
- Wang, L.X.; Wang, Z.Y.; Ke, C.H.; Zhou, S.H. Genetic relationship in Halitis diversicolor. J. Xiamen Univ. 2005, 44, 98–101. (In Chinese) [Google Scholar]
- Travers, M.A.; Basuyaux, O.; Goïc, N.L.; Huchette, S.; Nicolas, J.L.; Koken, M.; Paillard, C. Influence of temperature and spawning effort on Haliotis tuberculata mortalities caused by Vibrio harveyi: An example of emerging vibriosis linked to global warming. Glob. Chang. Biol. 2009, 15, 1365–1376. [Google Scholar] [CrossRef]
- Shiel, B.P.; Hall, N.E.; Cooke, I.R.; Robinson, N.A.; Strugnell, J.M. Epipodial tentacle gene expression and predetermined resilience to summer mortality in the commercially important greenlip abalone, Haliotis laevigata. Mar. Biotechnol. 2017, 19, 191–205. [Google Scholar] [CrossRef]
- Cheng, W.T.; Hsiao, I.S.; Hsu, C.H.; Chen, J.C. Change in water temperature on the immune response of Taiwan abalone Haliotis diversicolor supertexta and its susceptibility to Vibrio parahaemolyticus. Dis. Aquat. Org. 2004, 60, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.C.; Chen, Y.C.; Huang, C.Y.; Lee, K.K. Virulence of Vibrio parahaemolyticus isolated from cultured small abalone, Haliotis diversicolor supertexta, with withering syndrome. Lett. Appl. Microbiol. 2010, 31, 433–437. [Google Scholar] [CrossRef]
- Baruah, K.; Norouzitallab, P.; Linayati, L.; Sorgeloos, P.; Bossier, P. Reactive oxygen species generated by a heat shock protein (Hsp) inducing product contributes to Hsp70 production and Hsp70-mediated protective immunity in Artemia franciscana against pathogenic vibrios. Dev. Comp. Immunol. 2014, 46, 470–479. [Google Scholar] [CrossRef]
- Garrido, C.; Gurbuxani, S.; Ravagnan, L.; Kroemer, G. Heat shock proteins: Endogenous modulators of apoptotic cell death. Biochem. Biophys. Res. Commun. 2001, 286, 433–442. [Google Scholar] [CrossRef]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Ann. Rev Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Chen, T.; Cao, X. Stress for maintaining memory: HSP70 as a mobile messenger for innate and adaptive immunity. Eur. J. Immunol. 2010, 40, 1541–1544. [Google Scholar] [CrossRef] [Green Version]
- Ratcliffe, M.J.H. Developmental and comparative immunology. Vet. Immunol. Immunopathol. 1993, 1, 1–3. [Google Scholar] [CrossRef]
- Gu, J.; Huang, L.X.; Shen, Y.; Huang, L.H.; Feng, Q.L. Hsp70 and small Hsps are the major heat shock protein members involved in midgut metamorphosis in the common cutworm, Spodoptera litura. Insect Mol. Biol. 2012, 21, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.A.; Hightower, L.E. Stress proteins as molecular biomarkers for environmental toxicology. In Stress-Inducible Cellular Responses; Feige, U., Yahara, I., Morimoto, R.I., Polla, B.S., Eds.; Birkhäuser: Basel, Switzerland, 1996; pp. 411–424. [Google Scholar]
- Cheng, W.J.; Li, Q.L.; Sun, Y.M.; Wang, H.M.; Li, J.B.; Zhong, J.F. The research advance of HSP70. J. Anim. Sci. Vet. Med. 2008, 27, 55–57. [Google Scholar]
- Boorstein, W.R.; Ziegelhoffer, T.; Craig, E.A. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 1994, 38, 1–17. [Google Scholar] [CrossRef]
- Karlin, S.; Brocchieri, L. Heat Shock Protein 70 Family: Multiple sequence comparisons, function, and evolution. J. Mol. Evol. 1998, 47, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Chuang, K.H.; Ho, S.H.; Song, Y.L. Cloning and expression analysis of heat shock cognate 70 gene promoter in tiger shrimp (Penaeus monodon). Gene 2007, 405, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.G.; Hepat, R.; Kim, Y. RNA interference of a heat shock protein, Hsp70, loses its protection role in indirect chilling injury to the beet armyworm, Spodoptera exigua. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2014, 168, 90–95. [Google Scholar] [CrossRef]
- Mahroof, R.; Zhu, K.Y.; Subramanyam, B. Changes in expression of heat shock proteins in Tribolium castaneum (Coleoptera: Tenebrionidae) in relation to developmental stage, exposure time, and temperature. Ann. Entomol. Soc. Am. 2005, 98, 100–107. [Google Scholar] [CrossRef]
- Tsan, M.F.; Gao, B. Heat shock proteins and immune system. J. Leukoc. Biol. 2009, 85, 905–910. [Google Scholar] [CrossRef]
- Gornati, R.; Papis, E.; Rimoldi, S.; Terova, G.; Saroglia, M.; Bernardini, G. Rearing density influences the expression of stress-related genes in sea bass (Dicentrarchus labrax, L.). Gene 2004, 341, 111–118. [Google Scholar] [CrossRef]
- Cheng, J.; Xun, X.; Kong, Y.; Wang, S.; Yang, Z.; Li, Y.; Kong, D.; Wang, S.; Zhang, L.; Hu, X. Hsp70 gene expansions in the scallop Patinopecten yessoensis and their expression regulation after exposure to the toxic dinoflagellate Alexandrium catenella. Fish Shellfish Immunol. 2016, 58, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Sun, Y.; Lei, L.M.; Xie, S.T. Molecular identification and expression of heat shock cognate 70 (HSC 70) in the pacific white shrimp Litopenaeus vannamei. Mol. Biol. 2008, 42, 234–242. [Google Scholar] [CrossRef]
- Wang, W.; Deng, D.F.; Riu, N.D.; Moniello, G.; Hung, S.S.O. Heat shock protein 70 (HSP70) responses in tissues of white sturgeon and green sturgeon exposed to different stressors. North Am. J. Aquac. 2013, 75, 164–169. [Google Scholar] [CrossRef]
- Franzellitti, S.; Fabbri, E. Differential HSP70 gene expression in the Mediterranean mussel exposed to various stressors. Biochem. Biophys. Res. Commun. 2005, 336, 1157–1163. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.S.; Geissler, P.; Waller, C.; Fraser, K.P.; Barnes, D.K.; Peck, L.S. Low heat-shock thresholds in wild Antarctic inter-tidal limpets (Nacella concinna). Cell Stress Chaperones 2008, 13, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiu, L.; Zhou, Z.; Song, L. Research progress on the mollusc immunity in China. Dev. Comp. Immunol. 2013, 39, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Faisal, M. Identification of the molecules involved in zebra mussel (Dreissena polymorpha) hemocytes host defense. Comp. Biochem. Physiol Part B Biochem. Mol. Biol. 2009, 154, 143–149. [Google Scholar] [CrossRef]
- Hao, D.Q.; Zhao, L.; Chen, L.M.; Li, W.B. Comparison and application of promoter clone methods. J. Northeast Agric. Univ. 2010, 41, 154–160. (In Chinese) [Google Scholar]
- Hahn, S.; Young, E.T. Transcriptional regulation in Saccharomyces cerevisiae: Transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 2011, 189, 705–736. [Google Scholar] [CrossRef]
- Latchman, D.S. Transcription factors: An overview. J. Biochem. Cell Biol. 1993, 74, 417–422. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 2000, 34, 77–137. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.J.; Tjian, R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 1989, 245, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Oda, S.; Mikami, S.; Urushihara, Y.; Murata, Y.; Kamei, Y.; Deguchi, T.; Kitano, T.; Fujimori, K.E.; Yuba, S.; Todo, T. Identification of a functional medaka heat shock promoter and characterization of its ability to induce exogenous gene expression in medaka in vitro and in vivo. Zool. Sci. 2010, 27, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Stephanou, A.; Isenberg, D.A.; Nakajima, K.; Latchman, D.S. Signal transducer and activator of transcription-1 and heat shock factor-1 interact and activate the transcription of the Hsp-70 and Hsp-90beta gene promoters. J. Biol. Chem. 1999, 274, 1723–1728. [Google Scholar] [CrossRef]
- Chen, B.; Jia, T.L.; Ma, R.H.; Zhang, B.; Kang, L. Evolution of Hsp70 gene expression: A role for changes in AT-richness within promoters. PLoS ONE 2011, 6, e20308. [Google Scholar] [CrossRef] [PubMed]
- Sistonen, L.; Sarge, K.D.; Morimoto, R.I. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce hsp70 gene transcription. Mol. Cell. Biol. 1994, 14, 2087–2099. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, Y.T.; Cai, X.H.; Zou, Z.H.; Wang, G.D.; Wang, S.H.; Wang, Y.L.; Zhang, Z.P. Identification and expression analysis of immune-related genes linked to Rel/NF-κB signaling pathway under stresses and bacterial challenge from the small abalone Haliotis diversicolor. Fish Shellfish Immunol. 2014, 41, 200–208. [Google Scholar] [CrossRef]
- Huang, Y.T.; Cai, X.H.; Zou, Z.H.; Wang, S.H.; Wang, G.D.; Wang, Y.L.; Zhang, Z.P. Molecular cloning, characterization and expression analysis of three heat shock responsive genes from Haliotis diversicolor. Fish Shellfish Immunol. 2014, 36, 590–599. [Google Scholar] [CrossRef]
- Sun, Y.L.; Xin, Z.; Wang, G.D.; Shi, L.; Zeng, X.Y.; Wang, Y.L.; Zhang, Z.P. PI3K-AKT signaling pathway is involved in hypoxia/thermal-induced immunosuppression of small abalone Haliotis diversicolor. Fish Shellfish Immunol. 2016, 59, 492–508. [Google Scholar] [CrossRef]
- Norouzitallab, P.; Baruah, K.; Muthappa, D.M.; Bossier, P. Non-lethal heat shock induces HSP70 and HMGB1 protein production sequentially to protect Artemia franciscana against Vibrio campbellii. Fish Shellfish Immunol. 2015, 42, 395–399. [Google Scholar] [CrossRef]
- Sung, Y.Y.; Damme, E.J.M.V.; Sorgeloos, P.; Bossier, P. Non-lethal heat shock protects gnotobiotic Artemia franciscana larvae against virulent Vibrios. Fish Shellfish Immunol. 2007, 22, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Junprung, W.; Supungul, P.; Tassanakajon, A. HSP70 and HSP90 are involved in shrimp Penaeus vannamei tolerance to AHPND-causing strain of Vibrio parahaemolyticus after non-lethal heat shock. Fish Shellfish Immunol. 2016, 60, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Makrinos, D.L.; Bowden, T.J. Natural environmental impacts on teleost immune function. Fish Shellfish Immunol. 2016, 53, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Aleng, N.A.; Sung, Y.Y.; Macrae, T.H.; Wahid, M.E.A. Non-lethal heat shock of the Asian green mussel, Perna viridis, promotes Hsp70 synthesis, induces thermotolerance and protects against Vibrio infection. PLoS ONE 2015, 10, e0135603. [Google Scholar] [CrossRef] [PubMed]
- Zininga, T.; Ramatsui, L.; Shonhai, A. Heat Shock Proteins as Immunomodulants. Molecules 2018, 23, 2846. [Google Scholar] [CrossRef] [PubMed]
- Song, X.Y.; Zhang, H.; Wang, L.L.; Zhao, J.M.; Mu, C.K.; Song, L.S.; Qiu, L.M.; Liu, X.L. A galectin with quadruple-domain from bay scallop Argopecten irradians is involved in innate immune response. Dev. Comp. Immunol. 2011, 35, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Gueguen, Y.; Cadoret, J.P.; Flament, D.; Barreauroumiguière, C.; Girardot, A.L.; Garnier, J.; Hoareau, A.; Bachère, E.; Escoubas, J.M. Immune gene discovery by expressed sequence tags generated from hemocytes of the bacteria-challenged oyster, Crassostrea gigas. Gene 2003, 303, 139–145. [Google Scholar] [CrossRef]
- Li, Y.T.; Zhang, T.; Zhang, X.; Wang, G.D.; Wang, Y.L.; Zhang, Z.P. Heat shock cognate 70 gene in Haliotis diversicolor: Responses to pathogen infection and environmental stresses and its transcriptional regulation analysis. Cell Stress Chaperones 2017, 90, 1–12. [Google Scholar] [CrossRef]
- Currie, S.; Moyes, C.D.; Tufts, B.L. The effects of heat shock and acclimation temperature on hsp70 and hsp30 mRNA expression in rainbow trout: In vivo and in vitro comparisons. J. Fish Biol. 2000, 56, 398–408. [Google Scholar] [CrossRef]
- Hofmann, G.; Somero, G. Evidence for protein damage at environmental temperatures: Seasonal changes in levels of ubiquitin conjugates and hsp70 in the intertidal mussel Mytilus trossulus. J. Exp. Biol. 1995, 198, 1509–1518. [Google Scholar]
- Chapple, J.P.; Smerdon, G.R.; Berry, R.J.; Hawkins, A.J.S. Seasonal changes in stress-70 protein levels reflect thermal tolerance in the marine bivalve Mytilus edulis L. J. Exp. Mar. Biol. Ecol. 1998, 229, 53–68. [Google Scholar] [CrossRef]
- Tirard, C.T.; Grossfeld, R.M.; Levine, J.F.; Kennedy-Stoskopf, S. Effect of hyperthermia in vitro on stress protein synthesisand accumulation in oyster haemocytes. Fish Shellfish Immunol. 1995, 5, 9–25. [Google Scholar] [CrossRef]
- Roberts, D.A.; Hofmann, G.E.; Somero, G.N. Heat-shock protein expression in Mytilus californianus: Acclimatization (seasonal and tidal-height comparisons) and acclimation effects. Biol Bull. 1997, 192, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Encomio, V.G.; Chu, F.E. Seasonal variation of heat shock protein 70 in eastern oysters (Crassostera virginica) infected with Perkinsus marinus (Dermo). J. Shellfish Res. 2007, 24, 167–175. [Google Scholar]
- Tran, T.K.A.; Macfarlane, G.R.; Kong, R.Y.C.; O’Connor, W.A.; Yu, R.M.K. Mechanistic insights into induction of vitellogenin gene expression by estrogens in Sydney rock oysters, Saccostrea glomerata. Aquat. Toxicol. 2016, 174, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Su, J.; Wan, Q.; Su, J. CpA/CpG methylation of CiMDA5 possesses tight association with the resistance against GCRV and negatively regulates mRNA expression in grass carp, Ctenopharyngodon idella. Dev. Comp. Immunol. 2015, 48, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Sarda, S.; Das, A.; Vinson, C.; Hannenhalli, S. Distal CpG islands can serve as alternative promoters to transcribe genes with silenced proximal-promoters. Genome Res. 2017, 27, 553–566. [Google Scholar] [CrossRef]
- O’Connell, M.; Wright, J.M. Microsatellite DNA in fishes. Rev. Fish Biol. Fish. 1997, 7, 331–363. [Google Scholar] [CrossRef]
- Kube, D.; Laser, H.; Von, K.A.; Tesch, H. The AT-rich region between-54 to-66 is important for the promoter activity of interleukin-10 in Epstein-Barr virus positive Burkitt’s lymphoma cells. J. Genes Immun. 1999, 1, 105–114. [Google Scholar] [CrossRef]
- Delaney, S.K.; Orford, S.J.; Martinharris, M.; Timmis, J.N. The fiber specificity of the cotton FSltp4 gene promoter is regulated by an AT-rich promoter region and the AT-hook transcription factor GhAT1. Plant Cell Physiol. 2007, 48, 1426–1437. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Seidman, M.; Howard, B.H.; Gorman, C.M. Enhanced gene expression by the poly (dT-dG). poly (dC-dA) sequence. Mol. Cell. Biol. 1984, 4, 2622–2630. [Google Scholar] [CrossRef] [PubMed]
- Tae, H.J.; Luo, X.; Kim, K.H. Roles of CCAAT/enhancer-binding protein and its binding site on repression and derepression of acetyl-CoA carboxylase gene. J. Biological Chem. 1994, 269, 10475–10484. [Google Scholar]
- Shimajiri, S.; Arima, N.; Tanimoto, A.; Murata, Y.; Hamada, T.; Wang, K.Y.; Sasaguri, Y. Shortened microsatellite d (CA) 21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 1999, 455, 70–74. [Google Scholar] [CrossRef]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, L.F.; Wei, H.; Lin, J.R.; Zhong, B.X. Identification of Bombyx mori hsp70 promoter and its function. Chin. J. Cell Biol. 2011, 33, 503–509. [Google Scholar]
- Kust, N.; Rybalkina, E.; Mertsalov, I.; Savchenko, E.; Revishchin, A.; Pavlova, G. Functional analysis of Drosophila HSP70 promoter with different HSE numbers in human cells. PLoS ONE 2014, 9, e101994. [Google Scholar] [CrossRef]
- Bienz, M. Transient and developmental activation of heat-shock genes. Trends Biochem. Sci. 1985, 10, 157–161. [Google Scholar] [CrossRef]
- Tsutsumiishii, Y.; Tadokoro, K.; Hanaoka, F.; Tsuchida, N. Response of heat shock element within the human HSP70 promoter to mutated p53 genes. Cell Growth Differ. 1995, 6, 1–8. [Google Scholar]
- Bienz, M. Xenopus hsp70 genes are constitutively expressed in injected oocytes. Embo J. 1984, 3, 2477–2483. [Google Scholar] [CrossRef]
- Pei, H.L.; Hu, H.G.; Zhang, X.G.; Su, C.G.; Song, X.X. Cloning and functional analysis of the heat-inducible promoter AtHSP70b. Chin. Agric. Sci. Bull. 2007, 23, 82–86. (In Chinese) [Google Scholar]
- Li, Y.Q.; Li, J.; Li, H.X.; Wang, X.F.; Zhu, Y.; Ye, F.; Zhang, Z.Z.; Ren, X.L. Construction of an HSP 70B’ promoter-driven heat-inducible vectors pHSP- shTERT and its anti-proliferative effect in breast cancer MCF-7 cells. Chin. J. Cancer Biother. 2014, 21, 130–135. (In Chinese) [Google Scholar]
- Wang, H.D.; Zheng, J.H.; Deng, C.L.; Song, N.; Wu, J.J.; Zhang, S.X.; Ding, Z.; Mao, G.Y.; Lan, G.E.; Zhao, X.B. Study of NF-1 expression in keloid. Chin. J. Aesthet. Med. 2009, 18, 497–499. [Google Scholar]
- Anania, F.A.; Potter, J.J.; Rennietankersley, L.; Mezey, E. Effects of acetaldehyde on nuclear protein binding to the nuclear factor I consensus sequence in the alpha 2(I) collagen promoter. Hepatology 1995, 21, 1640–1648. [Google Scholar] [PubMed]
- Chen, S.J.; Artlett, C.M.; Jimenez, S.A.; Varga, J. Modulation of human α 1(I) procollagen gene activity by interaction with Sp1 and Sp3 transcription factors in vitro. Gene 1998, 215, 101–110. [Google Scholar] [CrossRef]
- Artlett, C.M.; Chen, S.J.; Varga, J.; Jimenez, S.A. Modulation of basal expression of the human α1(I) procollagen gene (COL1A1) by tandem NF-1/Sp1 promoter elements in normal human dermal fibroblasts. Matrix Biol. 1998, 17, 425–434. [Google Scholar] [CrossRef]
- Ouellet, S.; Vigneault, F.; Lessard, M.; Leclerc, S.; Drouin, R.; Guérin, S.L. Transcriptional regulation of the cyclin-dependent kinase inhibitor 1A (p21) gene by NFI in proliferating human cells. Nucleic Acids Res. 2006, 34, 6472–6487. [Google Scholar] [CrossRef]
- Yao-Borengassar, A.; Rogers, L.; Raj, V.; Kadlubar, S. Abstract B23: Sulfotransferase isoform 1A1 (SULT1A1) gene expression is regulated by transcription factor NF1 in human breast cancer cell lines. Cancer Prev. Res. 2014, 5, B23. [Google Scholar] [CrossRef]
- Pahl, H.L. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 2003, 18, 6853–6866. [Google Scholar] [CrossRef]
- Vallabhapurapu, S.; Karin, M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-κB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1, a000034. [Google Scholar] [CrossRef] [PubMed]
- Tanji, T.; Yun, E.Y.; Ip, Y.T. Heterodimers of NF-kappaB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 14715–14720. [Google Scholar] [CrossRef] [PubMed]
- Greten, F.R.; Karin, M. The IKK/NF-kappaB activation pathway-a target for prevention and treatment of cancer. Cancer Lett. 2004, 206, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Ammirante, M.; Rosati, A.; Gentilella, A.; Festa, M.; Petrella, A.; Marzullo, L.; Pascale, M.; Belisario, M.A.; Leone, A.; Turco, M.C. The activity of hsp90α promoter is regulated by NF-κB transcription factors. Oncogene 2008, 27, 1175–1178. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, X.J.; Li, F.H.; Huan, P.; Xiang, J.H. Functional analysis of the promoter of the heat shock cognate 70 gene of the Pacific white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol. 2013, 34, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Wilhide, M.E.; Tranter, M.; Ren, X.; Chen, J.; Sartor, M.A.; Medvedovic, M.; Jones, W.K. Identification of a NF-κB cardioprotective gene program: NF-κB regulation of Hsp70.1 contributes to cardioprotection after permanent coronary occlusion. J. Mol. Cell. Cardiol. 2011, 51, 82–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Huang, D.P.; Jia, X.W.; Zou, Z.H.; Wang, Y.L.; Zhang, Z.P. Functional analysis of the promoter of the molt-inhibiting hormone (MIH) gene in mud crab Scylla paramamosain. Gener. Comp. Endocrinol. 2017, 259, 131–140. [Google Scholar] [CrossRef]
Sample Availability: Samples of HdHSP70 cDNA and seven target plasmids are available from the authors. |




| Primer Name | Start | End | Length (bp) | Primer Sequence (5′→3′) | Tm (°C) | Used for |
|---|---|---|---|---|---|---|
| HSP70-1 | CTGAGATTTTGAAGAGAATGGC | 56.2 | Genome walking and Tail-PCR | |||
| HSP70-2 | AGAGAGGGAAAGCACACGAT | 56.0 | ||||
| HSP70-3 | TATGGTGAAAGGGAGGTAATCC | 57.7 | ||||
| HSP70-4 | ATGGGGAGTGAGTCTGGTGG | 59.1 | ||||
| HSP70-5 | CAACTCCTGGCAGACTGGGGA | 64.5 | ||||
| pGL3-70-r | CCGCTCGAGTGTGTCTCCGTAGTATACAGTCGT | 56.0 | ||||
| pGL3-70-1 | −1974 | 46 | 2020 | CGGGGTACCGAACAGCCACACTTCACTAACC | 56.5 | HSP70 promoter activity |
| pGL3-70-2 | −1746 | 46 | 1792 | CGGGGTACCGTCTCAAACTCCTACTTTCCTCG | 57.1 | |
| pGL3-70-3 | −1477 | 46 | 1523 | CGGGGTACCTGGGGAGTGAGTCTGGTGGT | 59.5 | |
| pGL3-70-4 | −1163 | 46 | 1209 | CGGGGTACCTCAGGGCAGACACCCAACAG | 61.3 | |
| pGL3-70-5 | −625 | 46 | 671 | CGGGGTACCAGTTAGGAGGATACACAAAGGACA | 57.1 | |
| pGL3-70-6 | −479 | 46 | 525 | CGGGGTACCAAGTAAGGGCAGGGAGCAGG | 60.9 | |
| pGL3-70-7 | −209 | 46 | 255 | CGGGGTACCACTGGGCGTCTGTCTACCTT | 56.2 | |
| pGL3-70-1dr | CCGCTCGAGAAGGTAGACAGACGCCCAGT | 56.1 | ||||
| P-rt-f | CATAGACGAGGGCTCCATGT | 57.3 | qRT-PCR | |||
| P-rt-r | TCATGGCTCGTGTGTTGTTG | 58.1 | ||||
| β-actin-f | CCGTGACCTTACAGACTACCT | 53.6 | ||||
| β-actin-r | TACCAGCGGATTCCATAC | 54.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Z.; Sun, Y.; Zhang, X.; Wang, G.; Li, Y.; Wang, Y.; Zhang, Z. Responses of HSP70 Gene to Vibrio parahaemolyticus Infection and Thermal Stress and Its Transcriptional Regulation Analysis in Haliotis diversicolor. Molecules 2019, 24, 162. https://doi.org/10.3390/molecules24010162
Fang Z, Sun Y, Zhang X, Wang G, Li Y, Wang Y, Zhang Z. Responses of HSP70 Gene to Vibrio parahaemolyticus Infection and Thermal Stress and Its Transcriptional Regulation Analysis in Haliotis diversicolor. Molecules. 2019; 24(1):162. https://doi.org/10.3390/molecules24010162
Chicago/Turabian StyleFang, Zhiqiang, Yulong Sun, Xin Zhang, Guodong Wang, Yuting Li, Yilei Wang, and Ziping Zhang. 2019. "Responses of HSP70 Gene to Vibrio parahaemolyticus Infection and Thermal Stress and Its Transcriptional Regulation Analysis in Haliotis diversicolor" Molecules 24, no. 1: 162. https://doi.org/10.3390/molecules24010162




