Specific Immobilization of Escherichia coli Expressing Recombinant Glycerol Dehydrogenase on Mannose-Functionalized Magnetic Nanoparticles
Abstract
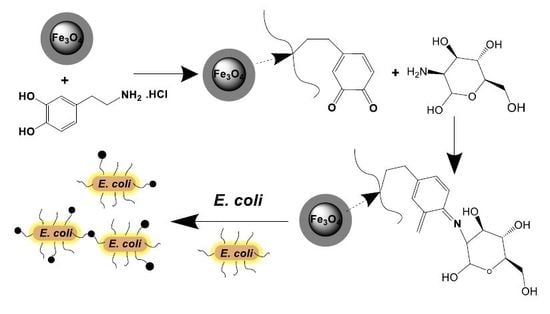
:1. Introduction
2. Results and Discussion
2.1. Characterization of the Nanoparticles
2.2. Optimization of the Immobilization Conditions
2.3. Effect of pH and Temperature on the Free and Immobilized Cells
2.4. Thermal Stability and Reusability of the Free and Immobilized Cells
2.5. Synthesis of 1,3-Dihydroxyacetone (DHA)
3. Materials and Methods
3.1. Materials
3.2. Preparation of Magnetic Nanocarriers Functionalized with Mannose
3.3. Characterization of the Magnetic Nanoparticles
3.4. Immobilization of E. coli Cells onto the Fe3O4@OA@DP–Mannose Nanoparticles
3.5. Desorption of E. coli from the Fe3O4@OA@DP–Mannose Nanoparticles
3.6. Activity Assay
3.7. Effect of Buffer Concentration, pH, and Temperature on the Cells
3.8. Thermal Stability and Reusability of the Immobilized Cells
3.9. Synthesis of 1,3-Dihydroxyacetone (DHA) with the Immobilized Cells
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Matsumoto, T.; Takahashi, S.; Kaieda, M.; Ueda, M.; Tanaka, A.; Fukuda, H.; Kondo, A. Yeast whole-cell biocatalyst constructed by intracellular overproduction of Rhizopus oryzae lipase is applicable to biodiesel fuel production. Appl. Microbiol. Biotechnol. 2001, 57, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.S.; Kumar, V.; Mardina, P.; Li, J.; Lestari, R.; Kalia, V.C.; Lee, J.K. Methanol production from simulated biogas mixtures by co-immobilized Methylomonas methanica and Methylocella tundrae. Bioresour. Technol. 2018, 263, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.Y.; Cong, W.; Xu, M.Q.; Ling, X.M.; Shen, J.J.; Zhang, Y.W. Using ConcanavalinA as a Spacer for Immobilization of E. coli onto Magnetic Nanoparticles. Int. J. Biol. Macromol. 2017, 104, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Krajčovič, T.; Bučko, M.; Vikartovská, A.; Lacík, I.; Uhelská, L.; Chorvát, D.; Neděla, V.; Tihlaříková, E.; Gericke, M.; Heinze, T.; et al. Polyelectrolyte Complex Beads by Novel Two-Step Process for Improved Performance of Viable Whole-Cell Baeyer-Villiger Monoxygenase by Immobilization. Catalysts 2017, 7, 353. [Google Scholar] [CrossRef]
- Jyoti; Bhatia, K.; Chauhan, K.; Attri, C.; Seth, A. Improving stability and reusability of Rhodococcus pyridinivorans NIT-36 nitrilase by whole cell immobilization using chitosan. Int. J. Biol. Macromol. 2017, 103, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.S.; Choi, S.H.; Kang, Y.C.; Lee, J.-K. Eco-Friendly Composite of Fe3O4-Reduced Graphene Oxide Particles for Efficient Enzyme Immobilization. ACS Appl. Mater. Interfaces 2017, 9, 2213–2222. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-S.; Patel, S.K.S.; Selvaraj, C.; Jung, W.-S.; Pan, C.-H.; Kang, Y.C.; Lee, J.-K. A highly efficient sorbitol dehydrogenase from Gluconobacter oxydans G624 and improvement of its stability through immobilization. Sci. Rep. 2016, 6, 33438. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.S.; Singh, R.K.; Kumar, A.; Jeong, J.-H.; Jeong, S.H.; Kalia, V.C.; Kim, I.-W.; Lee, J.-K. Biological methanol production by immobilized Methylocella tundrae using simulated biohythane as a feed. Bioresour. Technol. 2017, 241, 922–927. [Google Scholar] [CrossRef]
- Patel, S.K.S.; Kondaveeti, S.; Otari, S.V.; Pagolu, R.T.; Jeong, S.H.; Kim, S.C.; Cho, B.-K.; Kang, Y.C.; Lee, J.-K. Repeated batch methanol production from a simulated biogas mixture using immobilized Methylocystis bryophila. Energy 2018, 145, 477–485. [Google Scholar] [CrossRef]
- Patel, S.K.S.; Kumar, P.; Singh, M.; Lee, J.-K.; Kalia, V.C. Integrative approach to produce hydrogen and polyhydroxybutyrate from biowaste using defined bacterial cultures. Bioresour. Technol. 2015, 176, 136–141. [Google Scholar] [CrossRef]
- Yang, L.; Xin, J.; Zhang, Z.; Yan, H.; Wang, J.; Sun, E.; Hou, J.; Jia, X.; Lv, H. TPGS-modified liposomes for the delivery of ginsenoside compound K against non-small cell lung cancer: Formulation design and its evaluation in vitro and in vivo. J. Pharm. Pharmacol. 2016, 68, 1109–1118. [Google Scholar] [CrossRef]
- Wu, Y.-S.; Ngai, S.-C.; Goh, B.-H.; Chan, K.-G.; Lee, L.-H.; Chuah, L.-H. Anticancer Activities of Surfactin and Potential Application of Nanotechnology Assisted Surfactin Delivery. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rui, M.; Xin, Y.; Li, R.; Ge, Y.; Feng, C.; Xu, X. Targeted Biomimetic Nanoparticles for Synergistic Combination Chemotherapy of Paclitaxel and Doxorubicin. Mol. Pharm. 2017, 14, 107–123. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Morsy, R.; El-Zawawy, N.; Fareed, M.; Bedaiwy, M. Synthesized zinc peroxide nanoparticles (ZnO2-NPs): A novel antimicrobial, anti-elastase, anti-keratinase, and anti-inflammatory approach toward polymicrobial burn wounds. Int. J. Nanomed. 2017, 12, 6059–6073. [Google Scholar] [CrossRef]
- Wang, G.; Wang, J.J.; Chen, X.L.; Du, L.; Li, F. Quercetin-loaded freeze-dried nanomicelles: Improving absorption and anti-glioma efficiency in vitro and in vivo. J. Controll. Release 2016, 235, 276–290. [Google Scholar] [CrossRef]
- Shi, F.; Zhao, Y.; Firempong, C.K.; Xu, X. Preparation, characterization and pharmacokinetic studies of linalool-loaded nanostructured lipid carriers. Pharm. Biol. 2016, 54, 2320–2328. [Google Scholar] [CrossRef]
- Peng, W.; Jiang, X.; Zhu, Y.; Omari-Siaw, E.; Deng, W.; Yu, J.; Xu, X.; Zhang, W. Oral delivery of capsaicin using MPEG-PCL nanoparticles. Acta Pharmacol. Sin. 2015, 36, 139–148. [Google Scholar] [CrossRef]
- Zhang, H.; Firempong, C.K.; Wang, Y.; Xu, W.; Wang, M.; Cao, X.; Zhu, Y.; Tong, S.; Yu, J.; Xu, X. Ergosterol-loaded poly(lactide-co-glycolide) nanoparticles with enhanced in vitro antitumor activity and oral bioavailability. Acta Pharmacol. Sin. 2017, 37, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Grate, J.W.; Wang, P. Nanostructures for enzyme stabilization. Chem. Eng. Sci. 2006, 61, 1017–1026. [Google Scholar] [CrossRef]
- Xiong, F.; Hu, K.; Yu, H.; Zhou, L.; Song, L.; Zhang, Y.; Shan, X.; Liu, J.; Gu, N. A Functional Iron Oxide Nanoparticles Modified with PLA-PEG-DG as Tumor-Targeted MRI Contrast Agent. Pharm. Res. 2017, 34, 1683–1692. [Google Scholar] [CrossRef]
- Shen, S.; Wu, L.; Liu, J.; Xie, M.; Shen, H.; Qi, X.; Yan, Y.; Ge, Y.; Jin, Y. Core–shell structured Fe3O4@TiO2-doxorubicin nanoparticles for targeted chemo-sonodynamic therapy of cancer. Int. J. Pharm. 2015, 486, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Kilonzo, P.; Margaritis, A.; Bergougnou, M. Effects of surface treatment and process parameters on immobilization of recombinant yeast cells by adsorption to fibrous matrices. Bioresour. Technol. 2011, 102, 3662–3672. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.W.; Prabhu, P.; Lee, J.K. Alginate immobilization of recombinant Escherichia coli whole cells harboring L-arabinose isomerase for L-ribulose production. Bioprocess Biosyst. Eng. 2010, 33, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Ayer, M.; Klok, H.A. Cell-mediated delivery of synthetic nano- and microparticles. J. Controll. Release 2017. [Google Scholar] [CrossRef] [PubMed]
- Ni, K.; Lu, H.; Wang, C.; Black, K.C.; Wei, D.; Ren, Y.; Messersmith, P.B. A novel technique for in situ aggregation of Gluconobacter oxydans using bio-adhesive magnetic nanoparticles. Biotechnol. Bioeng. 2012, 109, 2970–2977. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, J.; Jiang, S.; Liu, Z.; Gu, W.; Yu, H.; Li, Y. Porous starch based self-assembled nano-delivery system improves the oral absorption of lipophilic drug. Int. J. Pharm. 2013, 444, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Akashi, M.; Maruyama, I.; Fukudome, N.; Yashima, E. Immobilization of human thrombomodulin on glass beads and its anticoagulant activity. Bioconj. Chem. 1992, 3, 363. [Google Scholar] [CrossRef]
- Guedri, H.; Durrieu, C. A self-assembled monolayers based conductometric algal whole cell biosensor for water monitoring. Microchim. Acta 2008, 163, 179–184. [Google Scholar] [CrossRef]
- Chouteau, C.; Dzyadevych, S.; Durrieu, C.; Chovelon, J.M. A bi-enzymatic whole cell conductometric biosensor for heavy metal ions and pesticides detection in water samples. Biosens. Bioelectron. 2005, 21, 273–281. [Google Scholar] [CrossRef]
- Lagarde, F.; Jaffrezic-Renault, N. Cell-based electrochemical biosensors for water quality assessment. Anal. Bioanal. Chem. 2011, 400, 947. [Google Scholar] [CrossRef]
- Miller, E.; Garcia, T.; Hultgren, S.; Oberhauser, A.F. The mechanical properties of E. coli type 1 pili measured by atomic force microscopy techniques. Biophys. J. 2006, 91, 3848–3856. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Dong, W.; Zhou, J.; Xu, X.; Li, F. Triggering effect of N-acetylglucosamine on retarded drug release from a lectin-anchored chitosan nanoparticles-in-microparticles system. Int. J. Pharm. 2013, 449, 37–43. [Google Scholar] [CrossRef]
- Yazgan, I.; Noah, N.M.; Toure, O.; Zhang, S.; Sadik, O.A. Biosensor for selective detection of E. coli in spinach using the strong affinity of derivatized mannose with fimbrial lectin. Biosens. Bioelectron. 2014, 61, 266–273. [Google Scholar] [CrossRef]
- Yang, W.; Pan, C.Y.; Luo, M.D.; Zhang, H.B. Fluorescent mannose-functionalized hyperbranched poly(amido amine)s: Synthesis and interaction with E. coli. Biomacromolecules 2010, 11, 1840–1846. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.Y.; Zhou, Q.L.; Wang, X.Y.; Zhang, J.X.; Xue, L.; Wang, R.; Zhang, J.X.; Zhang, Y.W. Immobilization of Lipase Onto Dopamine Functionalized Magnetic Nanoparticles. Nanosci. Nanotechnol. Lett. 2016, 8, 251–254. [Google Scholar] [CrossRef]
- Klemm, P.; Jørgensen, B.J.; Die, I.V.; Han, D.R.; Bergmans, H. The fim genes responsible for synthesis of type 1 fimbriae in Escherichia coli, cloning and genetic organization. Mol. Gen. Genet. MGG 1985, 199, 410. [Google Scholar] [CrossRef]
- Klemm, P.; Christiansen, G. Three FIM genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol. Gen. Genet. MGG 1987, 208, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Krogfelt, K.A.; Bergmans, H.; Klemm, P. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect. Immun. 1990, 58, 1995–1998. [Google Scholar] [CrossRef]
- Aprikian, P.; Tchesnokova, V.; Kidd, B.; Yakovenko, O.; Yarov-Yarovoy, V.; Trinchina, E.; Vogel, V.; Thomas, W.; Sokurenko, E. Interdomain Interaction in the FimH Adhesin of Escherichia coli Regulates the Affinity to Mannose. J. Biol. Chem. 2007, 282, 23437. [Google Scholar] [CrossRef] [PubMed]
- Möckl, L.; Fessele, C.; Despras, G.; Bräuchle, C.; Lindhorst, T.K. En route from artificial to natural: Evaluation of inhibitors of mannose-specific adhesion of E. coli under flow. Biochim. Biophys. Acta 2016, 1860, 2031–2036. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhou, F.; Zhang, X.-Y.; Li, Y.; Wang, X.-Y.; Xu, X.-M.; Zhang, Y.-W. Preparation of Magnetic Fe3O4@SiO2 Nanoparticles for Immobilization of Lipase. J. Nanosci. Nanotechnol. 2014, 14, 3068–3072. [Google Scholar] [CrossRef]
- Liu, C.H.; Li, X.Q.; Jiang, X.P.; Zhuang, M.Y.; Zhang, J.X.; Bao, C.H.; Zhang, Y.W. Preparation of Functionalized Graphene Oxide Nanocomposites for Covalent Immobilization of NADH Oxidase. Nanosci. Nanotechnol. Lett. 2016, 8, 164–167. [Google Scholar] [CrossRef]
- Tao, Q.-L.; Li, Y.; Shi, Y.; Liu, R.-J.; Zhang, Y.-W.; Guo, J. Application of Molecular Imprinted Magnetic Fe3O4@SiO2 Nanoparticles for Selective Immobilization of Cellulase. J. Nanosci. Nanotechnol. 2016, 16, 6055–6060. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Liu, W.; Tao, Q.-L.; Jiang, X.P.; Liu, C.-H.; Zeng, S.; Zhang, Y.-W. Immobilization of Lipase by Adsorption onto Magnetic Nanoparticles in Organic Solvents. J. Nanosci. Nanotechnol. 2016, 16, 601–607. [Google Scholar] [CrossRef] [PubMed]
- Rambaud, C.; Oppenländer, A.; Trommsdorff, H.P.; Vial, J.C. Tunneling dynamics of delocalized protons in hydrogen bonds at low temperatures. J. Lumin. 1990, 45, 310–312. [Google Scholar] [CrossRef]
- Liu, F.; Kozlovskaya, V.; Zavgorodnya, O.; Martinezlopez, C.; Catledge, S.; Kharlampieva, E. Encapsulation of anticancer drug by hydrogen-bonded multilayers of tannic acid. Soft Matter 2014, 10, 9237. [Google Scholar] [CrossRef]
- Sharon, N. Carbohydrates as future anti-adhesion drugs for infectious diseases. Biochim. Biophys. Acta BBA Gen. Subj. 2006, 1760, 527–537. [Google Scholar] [CrossRef]
- Hung, C.-S.; Bouckaert, J.; Hung, D.; Pinkner, J.; Widberg, C.; DeFusco, A.; Auguste, C.G.; Strouse, R.; Langermann, S.; Waksman, G.; et al. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection: FimH mannose-binding pocket. Mol. Microbiol. 2002, 44, 903–915. [Google Scholar] [CrossRef]
- Yang, H.; Shen, Y.; Xu, Y.; Maqueda, A.S.; Zheng, J.; Wu, Q.; Tam, J. A novel strategy for the discrimination of gelatinous Chinese medicines based on enzymatic digestion followed by nano-flow liquid chromatography in tandem with orbitrap mass spectrum detection. Int. J. Nanomed. 2015, 4947–4955. [Google Scholar] [CrossRef]
- Man, R.C.; Ismail, A.F.; Fuzi, S.F.Z.M.; Ghazali, N.F.; Illias, R.M. Effects of Culture Conditions of Immobilized Recombinant Escherichia coli on Cyclodextrin Glucanotransferase (CGTase) Excretion and Cell Stability. Process Biochem. 2016, 51, 474–483. [Google Scholar] [CrossRef]
- Zhang, Y.-W.; Jeya, M.; Lee, J.-K. L-Ribulose production by an Escherichia coli harboring l-arabinose isomerase from Bacillus licheniformis. Appl. Microbiol. Biotechnol. 2010, 87, 1993–1999. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, F.; Zhang, S.P.; Su, Z.G.; Ma, G.H.; Wang, P. Simultaneous production of 1,3-dihydroxyacetone and xylitol from glycerol and xylose using a nanoparticle-supported multi-enzyme system with in situ cofactor regeneration. Bioresour. Technol. 2011, 102, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Zhang, Y.W.; Nguyen, N.P.T.; Jeya, M.; Lee, J.K. Covalent immobilization of β-1,4-glucosidase from Agaricus arvensis onto functionalized silicon oxide nanoparticles. Appl. Microbiol. Biotechnol. 2011, 89, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.P.; Lu, T.T.; Liu, C.H.; Ling, X.M.; Zhuang, M.Y.; Zhang, J.X.; Zhang, Y.W. Immobilization of dehydrogenase onto epoxy-functionalized nanoparticles for synthesis of (R)-mandelic acid. Int. J. Biol. Macromol. 2016, 88, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, M.Y.; Jiang, X.P.; Ling, X.-M.; Xu, M.-Q.; Zhu, Y.-H.; Zhang, Y.W. Immobilization of glycerol dehydrogenase and NADH oxidase for enzymatic synthesis of 1,3-dihydroxyacetone with in situ cofactor regeneration. J. Chem. Technol. Biotechnol. 2018, 93, 2351–2358. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Yang, W.; Wang, L.; Zhu, Z.; Zhang, S.; Zhao, Z.K. Engineering NAD+ availability for Escherichia coli whole-cell biocatalysis: A case study for dihydroxyacetone production. Microb. Cell Fact. 2013, 12, 103. [Google Scholar] [CrossRef]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.-L.; Zhuang, M.-Y.; Shen, J.-J.; Fan, X.-M.; Choi, H.; Lee, J.-K.; Zhang, Y.-W. Specific Immobilization of Escherichia coli Expressing Recombinant Glycerol Dehydrogenase on Mannose-Functionalized Magnetic Nanoparticles. Catalysts 2019, 9, 7. https://doi.org/10.3390/catal9010007
Li F-L, Zhuang M-Y, Shen J-J, Fan X-M, Choi H, Lee J-K, Zhang Y-W. Specific Immobilization of Escherichia coli Expressing Recombinant Glycerol Dehydrogenase on Mannose-Functionalized Magnetic Nanoparticles. Catalysts. 2019; 9(1):7. https://doi.org/10.3390/catal9010007
Chicago/Turabian StyleLi, Fei-Long, Meng-Yao Zhuang, Jia-Jia Shen, Xiao-Man Fan, Hyunsoo Choi, Jung-Kul Lee, and Ye-Wang Zhang. 2019. "Specific Immobilization of Escherichia coli Expressing Recombinant Glycerol Dehydrogenase on Mannose-Functionalized Magnetic Nanoparticles" Catalysts 9, no. 1: 7. https://doi.org/10.3390/catal9010007
APA StyleLi, F.-L., Zhuang, M.-Y., Shen, J.-J., Fan, X.-M., Choi, H., Lee, J.-K., & Zhang, Y.-W. (2019). Specific Immobilization of Escherichia coli Expressing Recombinant Glycerol Dehydrogenase on Mannose-Functionalized Magnetic Nanoparticles. Catalysts, 9(1), 7. https://doi.org/10.3390/catal9010007