Transcriptomic Insight into Terpenoid Biosynthesis and Functional Characterization of Three Diterpene Synthases in Scutellaria barbata
Abstract
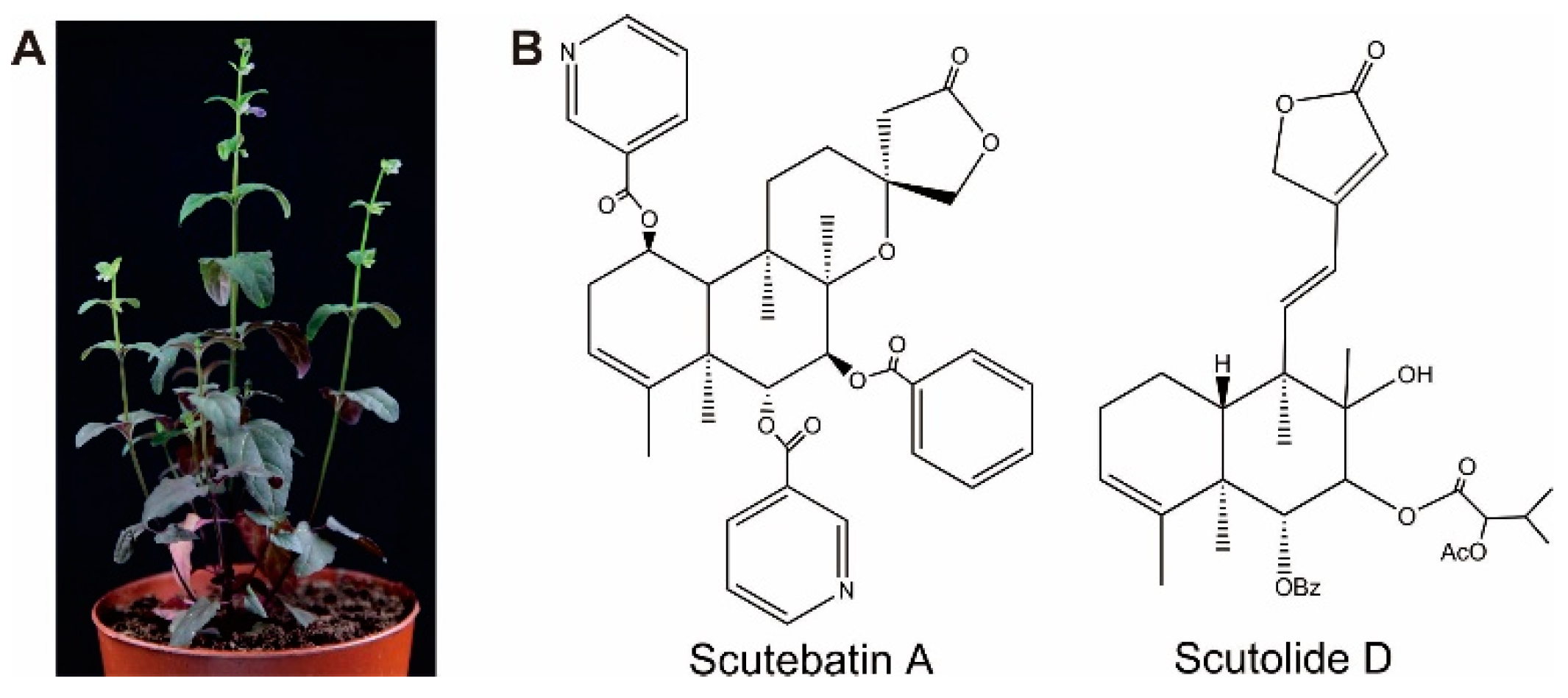
1. Introduction
2. Results
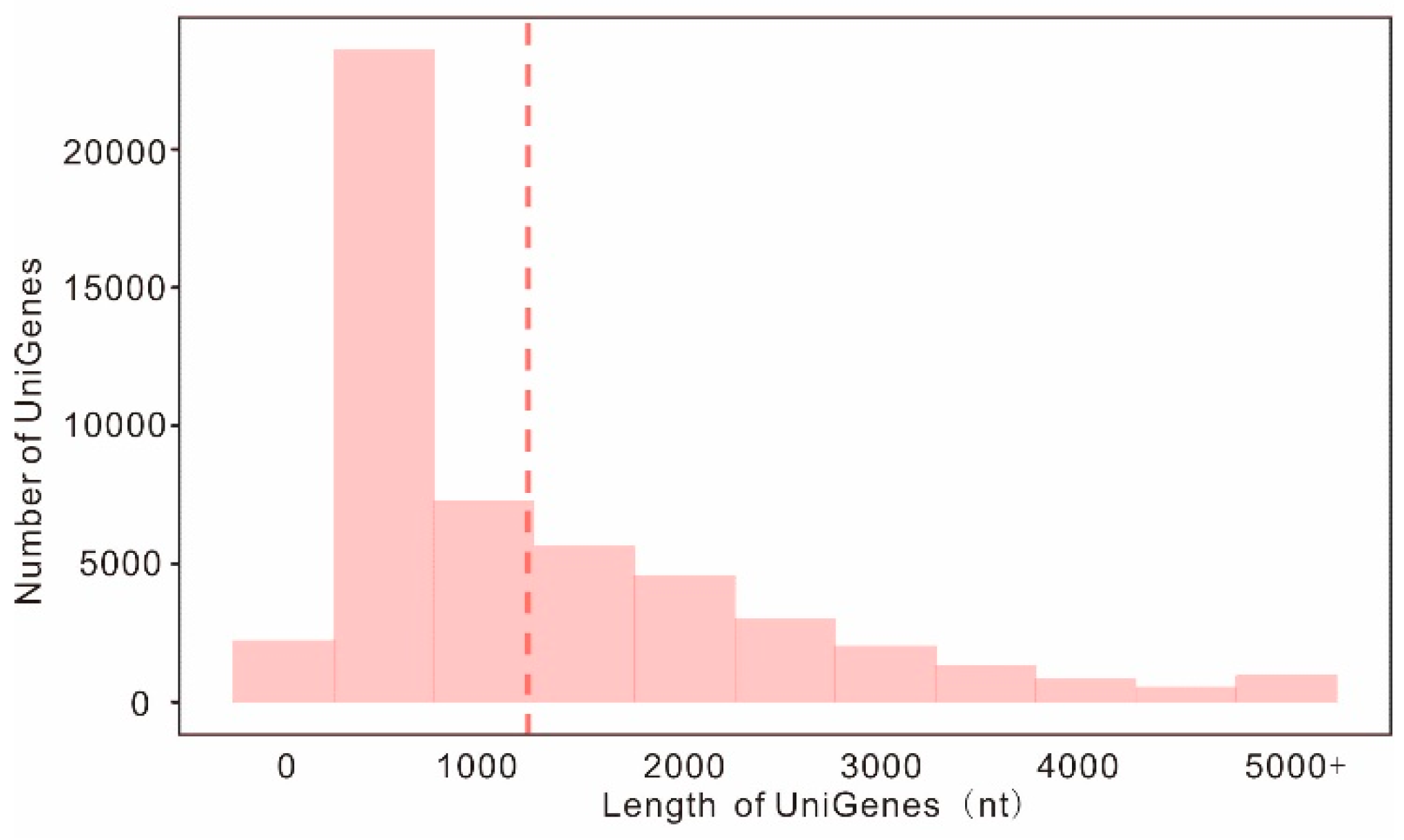
2.1. Transcriptome Sequencing, Assembly, and Annotation
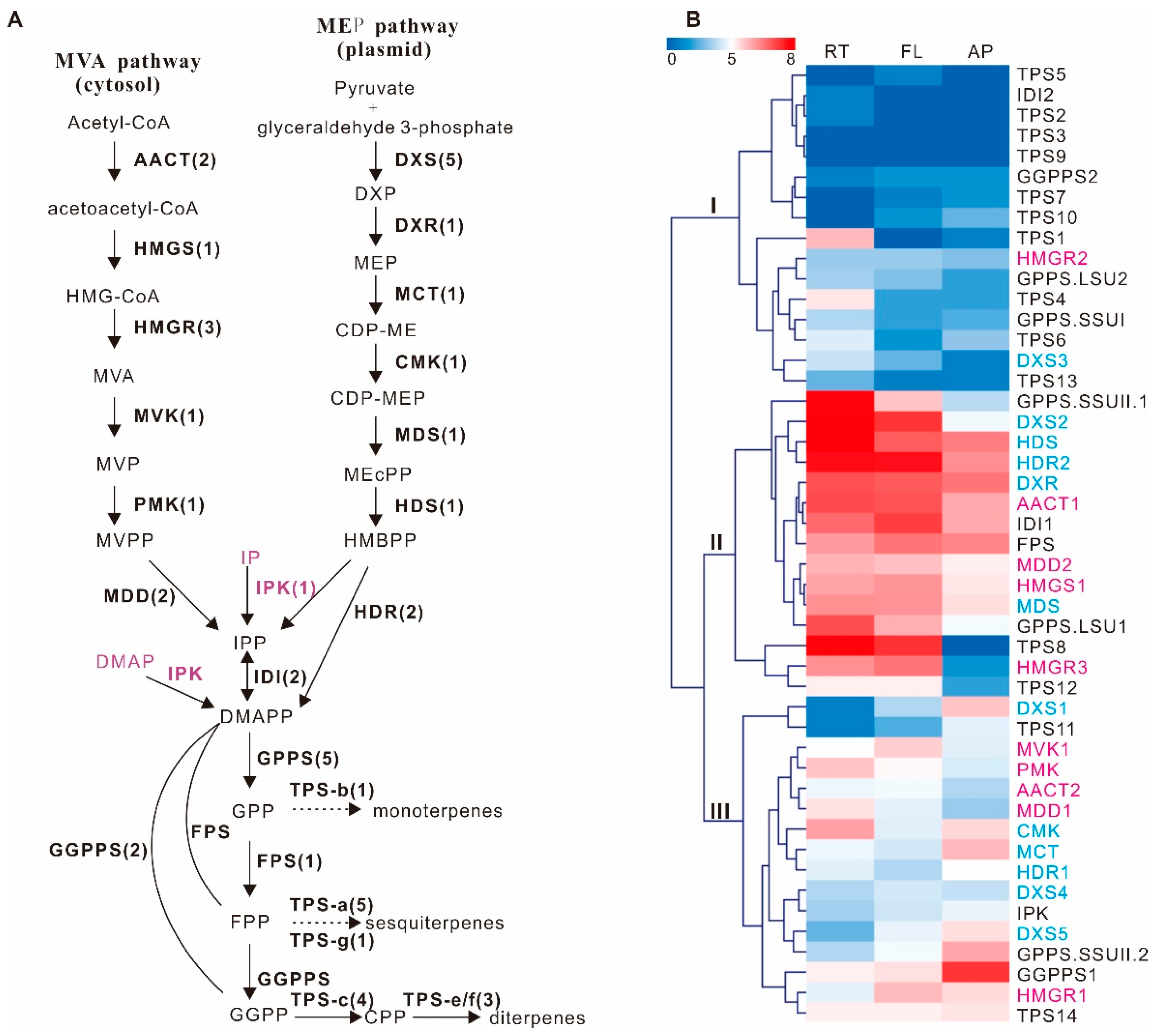
2.2. Identification of 33 Unigenes Involved in Terpenoid Backbone Biosynthesis
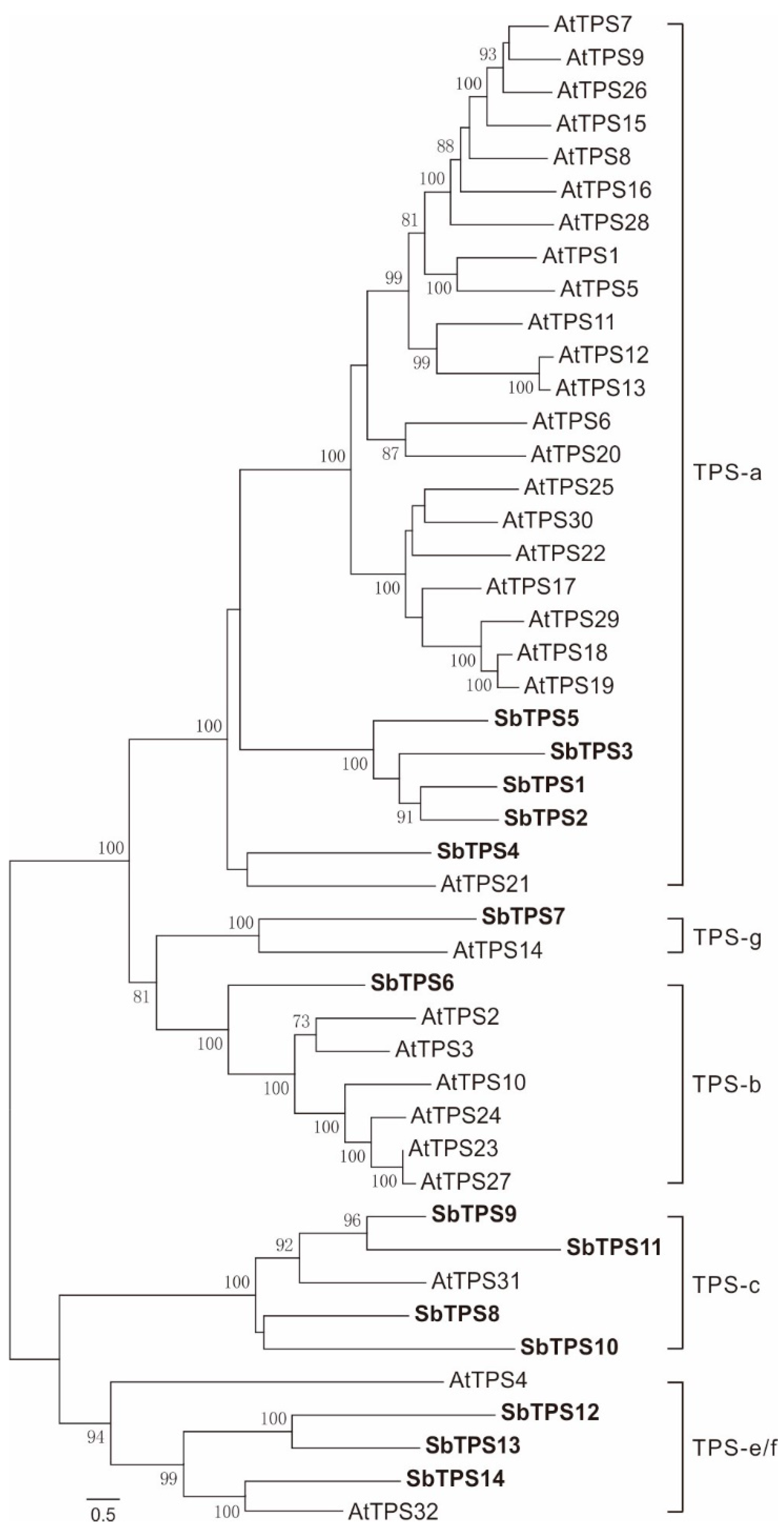
2.3. Identification of 14 Terpene Synthase Genes
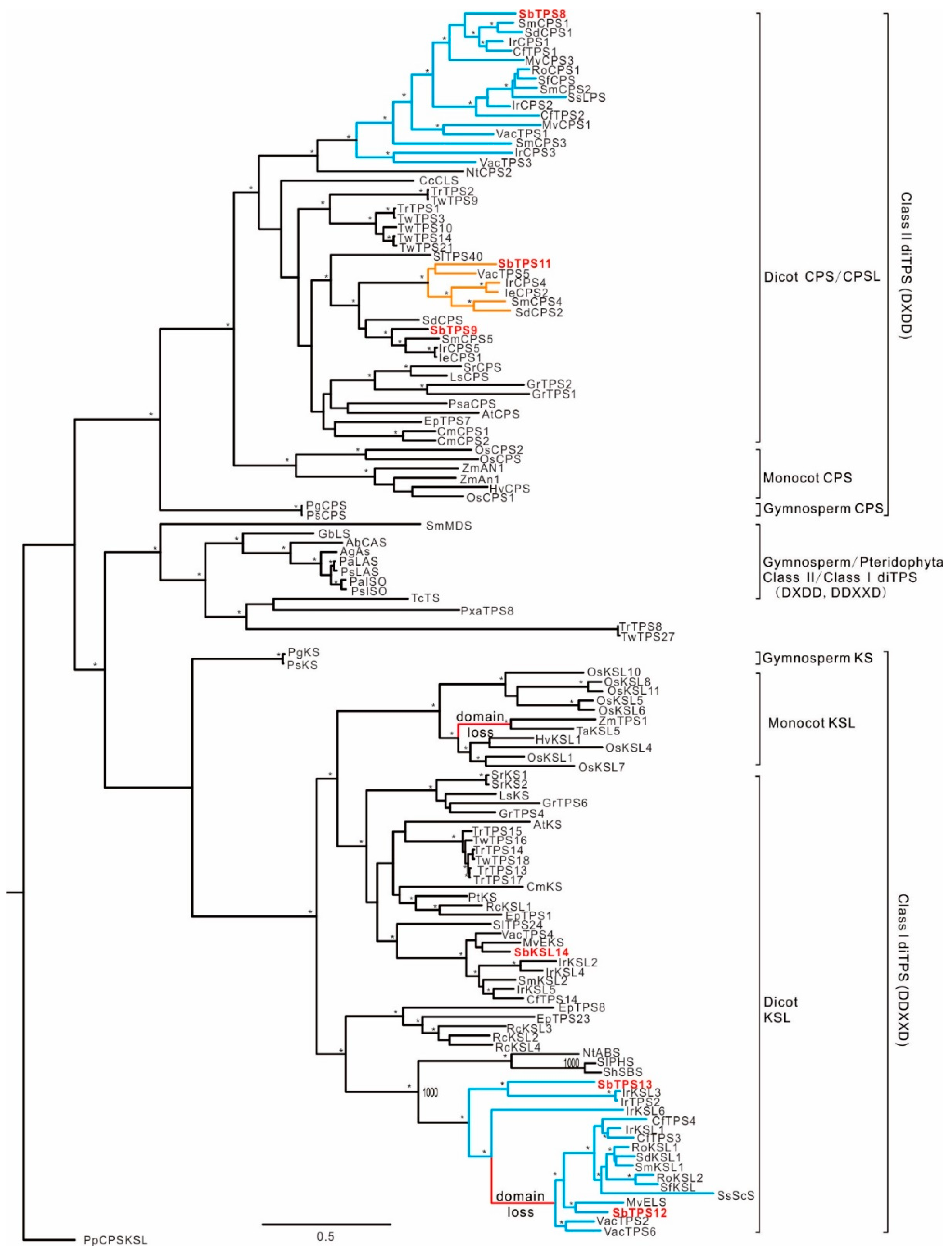
2.4. Phylogenetic Relationship of diTPS within Lamiaceae
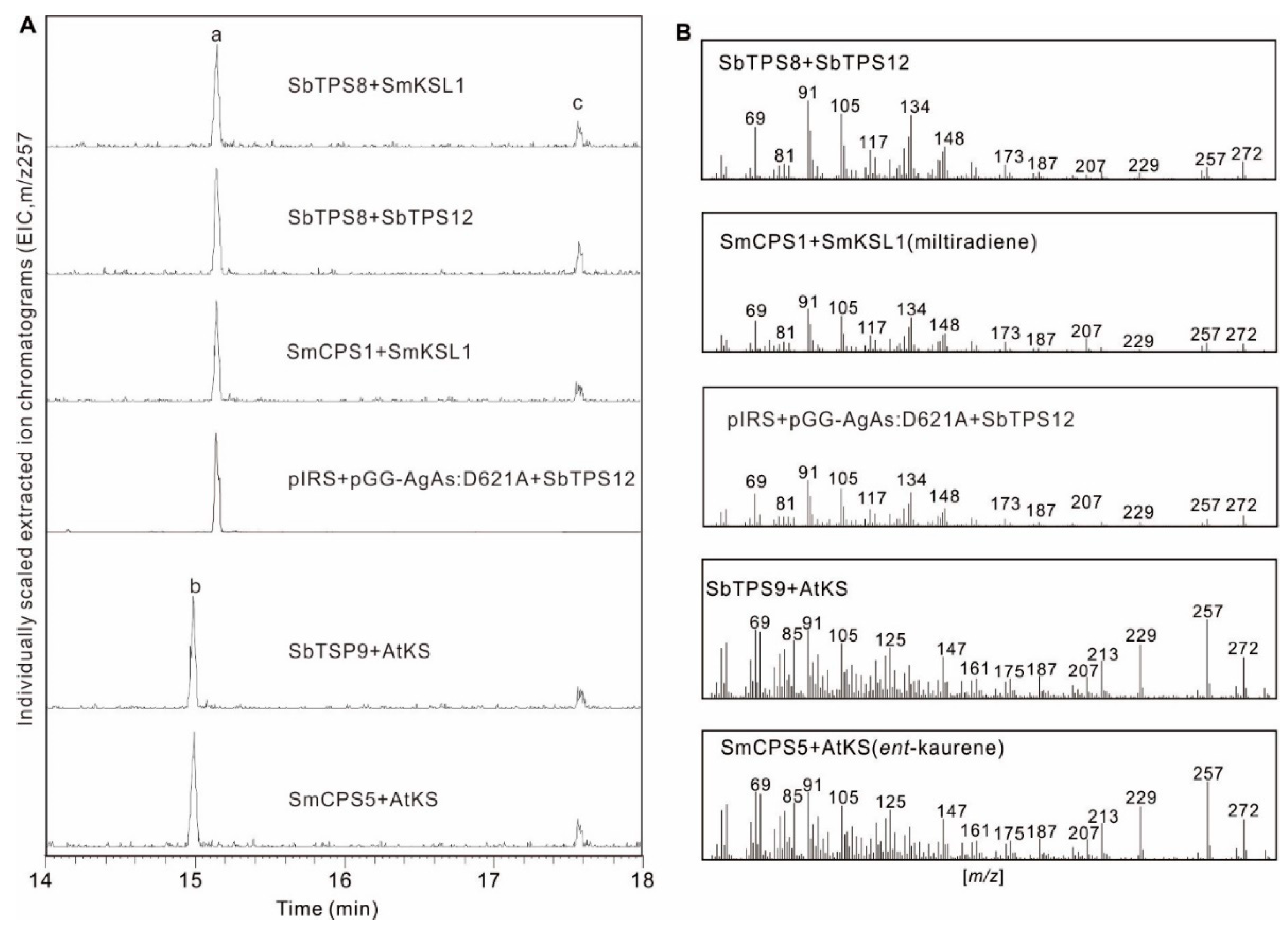
2.5. Functional Characterization of Three Diterpene Synthases
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Isolation and cDNA Library Preparation
4.3. Illumina Sequencing
4.4. RNA-Seq Data Processing
4.5. Transcriptome Annotation
4.6. Phylogenetic Analysis
4.7. Cloning of the Full Length DiTPS Genes
4.8. In Vitro Assays
4.9. Microbial Co-Expression of SbTPS12
4.10. Diterpene Analysis Using GC-MS
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harley, R.M.; Atkins, S.; Budanstev, A.L.; Cantino, P.D.; Conn, B.J.; Grayer, R.; Harley, M.M. Flowering plants, dicotyledons. In The Families and Genera of Vascular Plants; Kubitzki, K., Ed.; Springer: Berlin, Germany, 2004; Volume 6, pp. 167–275. [Google Scholar]
- Alvarenga, S.A.V.; Gastmans, J.P.; Rodrigues, G.D.; Moreno, P.R.H.; Emerenciano, V.D.A. Computer-assisted approach for chemotaxonomic studies—Diterpenes in Lamiaceae. Phytochemistry 2001, 56, 583–595. [Google Scholar] [CrossRef]
- Li, B.; Cantino, P.D.; Olmstead, R.G.; Bramley, G.L.; Xiang, C.L.; Ma, Z.H.; Tan, Y.H.; Zhang, D.X. A large-scale chloroplast phylogeny of the lamiaceae sheds new light on its subfamilial classification. Sci. Rep. 2016, 6, 34343. [Google Scholar] [CrossRef] [PubMed]
- Committee, C.P. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2015; Volume 1, pp. 134–135. [Google Scholar]
- Dai, S.J.; Qu, G.W.; Yu, Q.Y.; Zhang, D.W.; Li, G.S. New neo-clerodane diterpenoids from Scutellaria barbata with cytotoxic activities. Fitoterapia 2010, 81, 7737–7741. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.P.; Qu, G.W.; Yue, X.D.; Li, G.S.; Dai, S.J. Scutelinquanines A–C, three new cytotoxic neo-clerodane diterpenoid from Scutellaria barbata. Phytochem. Lett. 2010, 3, 190–193. [Google Scholar] [CrossRef]
- Yang, G.C.; Hu, J.H.; Li, B.L.; Liu, H.; Wang, J.Y.; Sun, L.X. Six New neo-clerodane diterpenoids from aerial parts of Scutellaria barbata and their cytotoxic activities. Planta Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.Q.; Song, W.B.; Wang, W.Q.; Xuan, L.J. Scubatines A–F, new cytotoxic neo-clerodane diterpenoids from Scutellaria barbata D. Don. Fitoterapia 2017, 119, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Di, Y.T.; Liu, L.L.; Zhang, Q.; Fang, X.; Yang, T.Q.; Hao, X.J.; He, H.P. Cytotoxic neoclerodane diterpenoids from Scutellaria barbata. J. Nat. Prod. 2010, 73, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Yeon, E.T.; Lee, J.W.; Lee, C.; Jin, Q.; Jang, H.; Lee, D.; Ahn, J.S.; Hong, J.T.; Kim, Y.; Lee, M.K.; et al. Neo-clerodane diterpenoids from Scutellaria barbata and their inhibitory effects on lps-induced nitric oxide production. J. Nat. Prod. 2015, 78, 2292–2296. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.Z.; Wang, Q.; Jiang, C.; Morris-Natschke, S.L.; Cui, H.; Wang, Y.; Yan, Y.; Xu, J.; Lee, K.H.; Gu, Q. Neo-clerodane diterpenoids from Scutellaria barbata with activity against Epstein–Barr virus lytic replication. J. Nat. Prod. 2015, 78, 500–509. [Google Scholar] [CrossRef] [PubMed]
- Boachon, B.; Buell, C.R.; Crisovan, E.; Dudareva, N.; Garcia, N.; Godden, G.; Henry, L.; Kamileen, M.O.; Kates, H.R.; Kilgore, M.B.; et al. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol. Plant 2018, 11, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Hemmerlin, A.; Harwood, J.L.; Bach, T.J. A raison d’etre for two distinct pathways in the early steps of plant isoprenoid biosynthesis? Prog. Lipid Res. 2012, 51, 95–148. [Google Scholar] [CrossRef] [PubMed]
- Pulido, P.; Perello, C.; Rodriguez-Concepcion, M. New insights into plant isoprenoid metabolism. Mol. Plant 2012, 5, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Vranova, E.; Coman, D.; Gruissem, W. Network analysis of the mva and mep pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef] [PubMed]
- Dellas, N.; Thomas, S.T.; Manning, G.; Noel, J.P. Discovery of a metabolic alternative to the classical mevalonate pathway. eLife 2013, 2, e00672. [Google Scholar] [CrossRef] [PubMed]
- Henry, L.K.; Gutensohn, M.; Thomas, S.T.; Noel, J.P.; Dudareva, N. Orthologs of the archaeal isopentenyl phosphate kinase regulate terpenoid production in plants. Proc. Natl. Acad. Sci. USA 2015, 112, 10050–10055. [Google Scholar] [CrossRef] [PubMed]
- Henry, L.K.; Thomas, S.T.; Widhalm, J.R.; Lynch, J.H.; Davis, T.C.; Kessler, S.A.; Bohlmann, J.; Noel, J.P.; Dudareva, N. Contribution of isopentenyl phosphate to plant terpenoid metabolism. Nat. Plants 2018, 4, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Peters, R.J. Two rings in them all: The labdane-related diterpenoids. Nat. Prod. Rep. 2010, 27, 1521–1530. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Hillwig, M.L.; Huang, L.Q.; Cui, G.H.; Wang, X.Y.; Kong, J.Q.; Yang, B.; Peters, R.J. A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org. Lett. 2009, 11, 5170–5173. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Chen, Q.Q.; Jin, Q.P.; Gao, J.; Zhao, P.J.; Lu, S.; Zeng, Y. IeCPS2 is potentially involved in the biosynthesis of pharmacologically active isodon diterpenoids rather than gibberellin. Phytochemistry 2012, 76, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Caniard, A.; Zerbe, P.; Legrand, S.; Cohade, A.; Valot, N.; Magnard, J.L.; Bohlmann, J.; Legendre, L. Discovery and functional characterization of two diterpene synthases for sclareol biosynthesis in Salvia sclarea (L.) and their relevance for perfume manufacture. BMC Plant Biol. 2012, 12, 119. [Google Scholar] [CrossRef] [PubMed]
- Schalk, M.; Pastore, L.; Mirata, M.A.; Khim, S.; Schouwey, M.; Deguerry, F.; Pineda, V.; Rocci, L.; Daviet, L. Toward a biosynthetic route to sclareol and amber odorants. J. Am. Chem. Soc. 2012, 134, 18900–18903. [Google Scholar] [CrossRef] [PubMed]
- Bruckner, K.; Bozic, D.; Manzano, D.; Papaefthimiou, D.; Pateraki, I.; Scheler, U.; Ferrer, A.; de Vos, R.C.H.; Kanellis, A.K.; Tissier, A. characterization of two genes for the biosynthesis of abietane-type diterpenes in rosemary (Rosmarinus officinalis) glandular trichomes. Phytochemistry 2014, 101, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Pateraki, I.; Andersen-Ranberg, J.; Hamberger, B.; Heskes, A.M.; Martens, H.J.; Zerbe, P.; Bach, S.S.; Moller, B.L.; Bohlmann, J.; Hamberger, B. Manoyl oxide (13r), the biosynthetic precursor of forskolin, is synthesized in specialized root cork cells in Coleus forskohlii. Plant Physiol. 2014, 164, 1222–1236. [Google Scholar] [CrossRef] [PubMed]
- Pelot, K.A.; Mitchell, R.; Kwon, M.; Hagelthorn, D.M.; Wardman, J.F.; Chiang, A.; Bohlmann, J.; Ro, D.; Zerbe, P. Biosynthesis of the psychotropic plant diterpene salvinorin A: Discovery and characterization of the Salvia divinorum clerodienyl diphosphate synthase. Plant J. 2017, 89, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Bozic, D.; Papaefthimiou, D.; Bruckner, K.; de Vos, R.C.H.; Tsoleridis, C.A.; Katsarou, D.; Papanikolaou, A.; Pateraki, I.; Chatzopoulou, F.M.; Dimitriadou, E.; et al. Towards elucidating carnosic acid biosynthesis in lamiaceae: Functional characterization of the three first steps of the pathway in Salvia fruticosa and Rosmarinus officinalis. PLoS ONE 2015, 10, e0124106. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.H.; Duan, L.X.; Jin, B.L.; Qian, J.; Xue, Z.Y.; Shen, G.A.; Snyder, J.H.; Song, J.Y.; Chen, S.L.; Huang, L.Q. Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza. Plant Physiol. 2015, 169, 1607–1618. [Google Scholar] [CrossRef] [PubMed]
- Trikka, F.A.; Nikolaidis, A.; Ignea, C.; Tsaballa, A.; Tziveleka, L.A.; Ioannou, E.; Roussis, V.; Stea, E.A.; Bozic, D.; Argiriou, A.; Kanellis, A.K.; Kampranis, S.C.; Makris, A.M. Combined metabolome and transcriptome profiling provides new insights into diterpene biosynthesis in S. pomifera glandular trichomes. BMC Genom. 2015, 16, 935. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Tong, Y.R.; Cheng, Q.Q.; Hu, Y.T.; Zhang, M.; Yang, J.; Teng, Z.Q.; Gao, W.; Huang, L.Q. Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway. Sci. Rep. 2016, 6, 23057. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Berim, A.; Dayan, F.E.; Gang, D.R. A (–)-kolavenyl diphosphate synthase catalyzes the first step of salvinorin a biosynthesis in Salvia divinorum. J. Exp. Bot. 2017, 68, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.L.; Cui, G.H.; Guo, J.; Tang, J.F.; Duan, L.X.; Lin, H.X.; Shen, Y.; Chen, T.; Zhang, H.B.; Huang, L.Q. Functional diversification of kaurene synthase-like genes in Isodon rubescens. Plant Physiol. 2017, 174, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Zerbe, P.; Chiang, A.; Dullat, H.; O’Neil-Johnson, M.; Starks, C.; Hamberger, B.; Bohlmann, J. Diterpene synthases of the biosynthetic system of medicinally active diterpenoids in Marrubium vulgare. Plant J. 2014, 79, 914–927. [Google Scholar] [CrossRef] [PubMed]
- Heskes, A.M.; Sundram, T.C.M.; Boughton, B.A.; Jensen, N.B.; Hansen, N.L.; Crocoll, C.; Cozzi, F.; Rasmussen, S.; Hamberger, B.; Hamberger, B.; et al. Biosynthesis of bioactive diterpenoids in the medicinal plant Vitex agnus-castus. Plant J. 2018, 93, 943–958. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.B.; Song, J.Y.; Luo, H.M.; Zhang, Y.J.; Li, Q.S.; Zhu, Y.J.; Xu, J.; Li, Y.; Song, C.; Wang, B.; et al. Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant 2016, 9, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.C.; Liang, C.; Li, S.G.; Liu, M.; Jia, M.J.; Xu, X.N.; Wang, X.B.; Hua, H.M.; Sun, L.X. Neoclerodane diterpenoids from aerial parts of Scutellaria barbata. Phytochem. Lett. 2017, 19, 1–6. [Google Scholar] [CrossRef]
- Hillwig, M.L.; Xu, M.M.; Toyomasu, T.; Tiernan, M.S.; Wei, G.; Cui, G.H.; Huang, L.Q.; Peters, R.J. Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J. 2011, 68, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Andersen-Ranberg, J.; Kongstad, K.T.; Nielsen, M.T.; Jensen, N.B.; Pateraki, I.; Bach, S.S.; Hamberger, B.; Zerbe, P.; Staerk, D.; Bohlmann, J.; et al. Expanding the landscape of diterpene structural diversity through stereochemically controlled combinatorial biosynthesis. Angew. Chem. Int. Ed. 2016, 55, 2142–2146. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.R.; Potter, K.C.; Peters, R.J. Extreme promiscuity of a bacterial and a plant diterpene synthase enables combinatorial biosynthesis. Metab. Eng. 2016, 37, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Cyr, A.; Wilderman, P.R.; Determan, M.; Peters, R.J. A modular approach for facile biosynthesis of labdane-related diterpenes. J. Am. Chem. Soc. 2007, 129, 6684–6685. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, W.; Wei, H.L.; He, L.Q.; Chen, J.H.; Zhang, B.H.; Zhu, S.J. Species-specific expansion and molecular evolution of the 3-hydroxy-3-methylglutaryl coenzyme a reductase (hmgr) gene family in plants. PLoS ONE 2014, 9, e94172. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D.; Lee, S. Terpene specialized metabolism in arabidopsis thaliana. In Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2011; Volume 9, p. e0143. [Google Scholar]
- Henriquez, M.A.; Soliman, A.; Li, G.Y.; Hannoufa, A.; Ayele, B.T.; Daayf, F. Molecular cloning, functional characterization and expression of potato (Solanum tuberosum) 1-deoxy-d-xylulose 5-phosphate synthase 1 (stdxs1) in response to phytophthora infestans. Plant Sci. 2016, 243, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Simpson, K.; Quiroz, L.F.; Rodriguez-Concepcion, M.; Stenge, C.R. Differential contribution of the first two enzymes of the mep pathway to the supply of metabolic precursors for carotenoid and chlorophyll biosynthesis in carrot (Daucus carota). Front. Plant Sci. 2016, 7, e6373. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.P.; Rohwer, J.M.; Ghirardo, A.; Hammerbacher, A.; Ortiz-Alcaide, M.; Raguschke, B.; Schnitzler, J.P.; Gershenzon, J.; Phillips, M.A. Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in Arabidopsis. Plant Physiol. 2014, 165, 1488–1504. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Huang, F.F.; Li, S.; Wang, Y.; Zhou, C.C.; Shi, M.; Wang, J.; Chen, Y.J.; Wang, Y.; Wang, H.Z.; et al. Molecular cloning and characterization of two 1-deoxy-d-xylulose-5-phosphate synthase genes involved in tanshinone biosynthesis in Salvia miltiorrhiza. Mol. Breed. 2016, 36, 124. [Google Scholar] [CrossRef]
- Shi, M.; Luo, X.; Ju, G.; Li, L.; Huang, S.; Zhang, T.; Wang, H.; Kai, G. Enhanced diterpene tanshinone accumulation and bioactivity of transgenic Salvia miltiorrhiza hairy roots by pathway engineering. J. Agric. Food Chem. 2016, 64, 2523–2530. [Google Scholar] [CrossRef] [PubMed]
- Kai, G.; Xu, H.; Zhou, C.; Liao, P.; Xiao, J.B.; Luo, X.Q.; You, L.J.; Zhang, L. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 2011, 13, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Cui, G.H.; Zhou, S.F.; Zhang, X.N.; Huang, L.Q. Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation. J. Plant Physiol. 2011, 168, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.S.; Chen, M.M.; Liu, Y.; Guo, J.; Zhang, B.; Feng, J.T.; Zhang, X. Effect of over-expressing TwHMGR and TwDXR on the biosynthesis of terpenoids in Tripterygium wilfordii. J. Agric. Biotechnol. 2018, 26, 940–948. [Google Scholar]
- Wang, Y.; Song, F.; Zhu, J.; Zhang, S.; Yang, Y.; Chen, T.; Tang, B.; Dong, L.; Ding, N.; Zhang, Q.; et al. GSA: Genome sequence archive. Genom. Proteom. Bioinform. 2017, 15, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Bo, L. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar] [PubMed]
- Sonnhammer, E.L.L.; Eddy, S.R.; Durbin, R. Pfam: A comprehensive database of protein families based on seed alignments. Proteins 1997, 28, 405–420. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Morrone, D.; Lowry, L.; Determan, M.K.; Hershey, D.M.; Xu, M.; Peters, R.J. Increasing diterpene yield with a modular metabolic engineering system in E. coli: Comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol. 2010, 85, 1893–1906. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds miltiradiene and ent-kaurene are available from the authors. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Jin, B.; Bu, J.; Guo, J.; Chen, T.; Ma, Y.; Tang, J.; Cui, G.; Huang, L. Transcriptomic Insight into Terpenoid Biosynthesis and Functional Characterization of Three Diterpene Synthases in Scutellaria barbata. Molecules 2018, 23, 2952. https://doi.org/10.3390/molecules23112952
Zhang H, Jin B, Bu J, Guo J, Chen T, Ma Y, Tang J, Cui G, Huang L. Transcriptomic Insight into Terpenoid Biosynthesis and Functional Characterization of Three Diterpene Synthases in Scutellaria barbata. Molecules. 2018; 23(11):2952. https://doi.org/10.3390/molecules23112952
Chicago/Turabian StyleZhang, Huabei, Baolong Jin, Junling Bu, Juan Guo, Tong Chen, Ying Ma, Jinfu Tang, Guanghong Cui, and Luqi Huang. 2018. "Transcriptomic Insight into Terpenoid Biosynthesis and Functional Characterization of Three Diterpene Synthases in Scutellaria barbata" Molecules 23, no. 11: 2952. https://doi.org/10.3390/molecules23112952
APA StyleZhang, H., Jin, B., Bu, J., Guo, J., Chen, T., Ma, Y., Tang, J., Cui, G., & Huang, L. (2018). Transcriptomic Insight into Terpenoid Biosynthesis and Functional Characterization of Three Diterpene Synthases in Scutellaria barbata. Molecules, 23(11), 2952. https://doi.org/10.3390/molecules23112952



