Genetic Diversity and Phylogenetic Analysis Among Multidrug-Resistant Pseudomonas spp. Isolated from Solid Waste Dump Sites and Dairy Farms
Abstract
1. Introduction
2. Methods
2.1. Site Selection, Sample Collection, and Processing
2.2. Phenotypic Characterization
Morphology, Gram Staining, and Biochemical Tests
2.3. Bacterial Motility Test
2.4. Antimicrobial Susceptibility Test
2.5. Biofilm Assay
2.6. Genomic DNA Extraction and 16S Ribosomal RNA (16S rRNA)-PCR Amplification
2.7. Amplicon Purification, Sequencing, and Bioinformatics Analysis
2.8. BLAST (Basic Local Alignment Search Tool) Analysis
2.9. Multiple Sequence Alignment (MSA) and Phylogenetic Analysis
2.10. RAPD-PCR Analysis
2.11. Protein Extraction and Estimation
2.12. SDS—PAGE Analysis
3. Results and Discussion
3.1. Morphological and Phenotypic Characterization
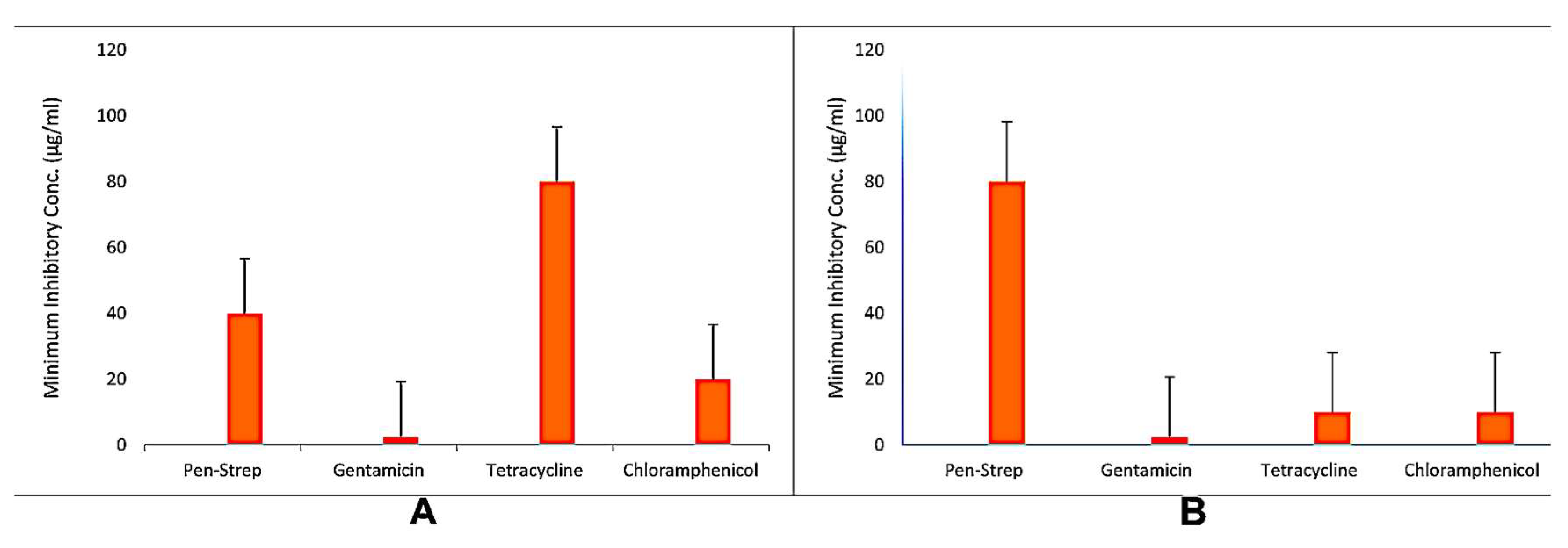
3.2. Antibiotic Resistance Profiling
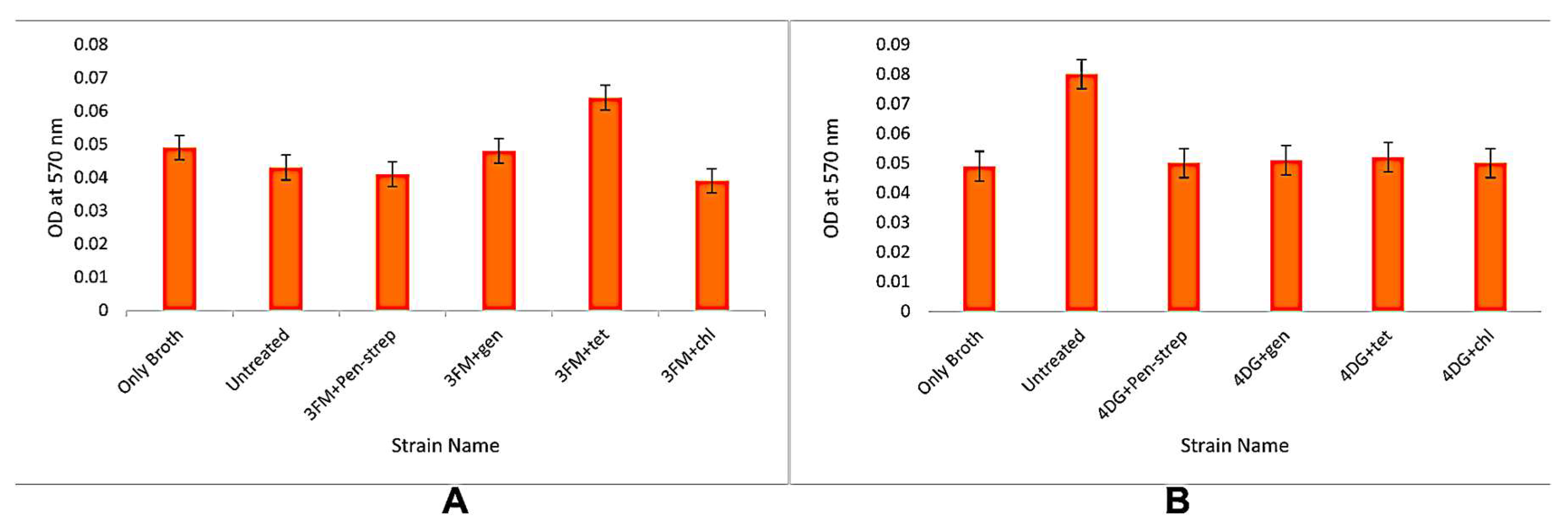
3.3. Biofilm Formation Assays
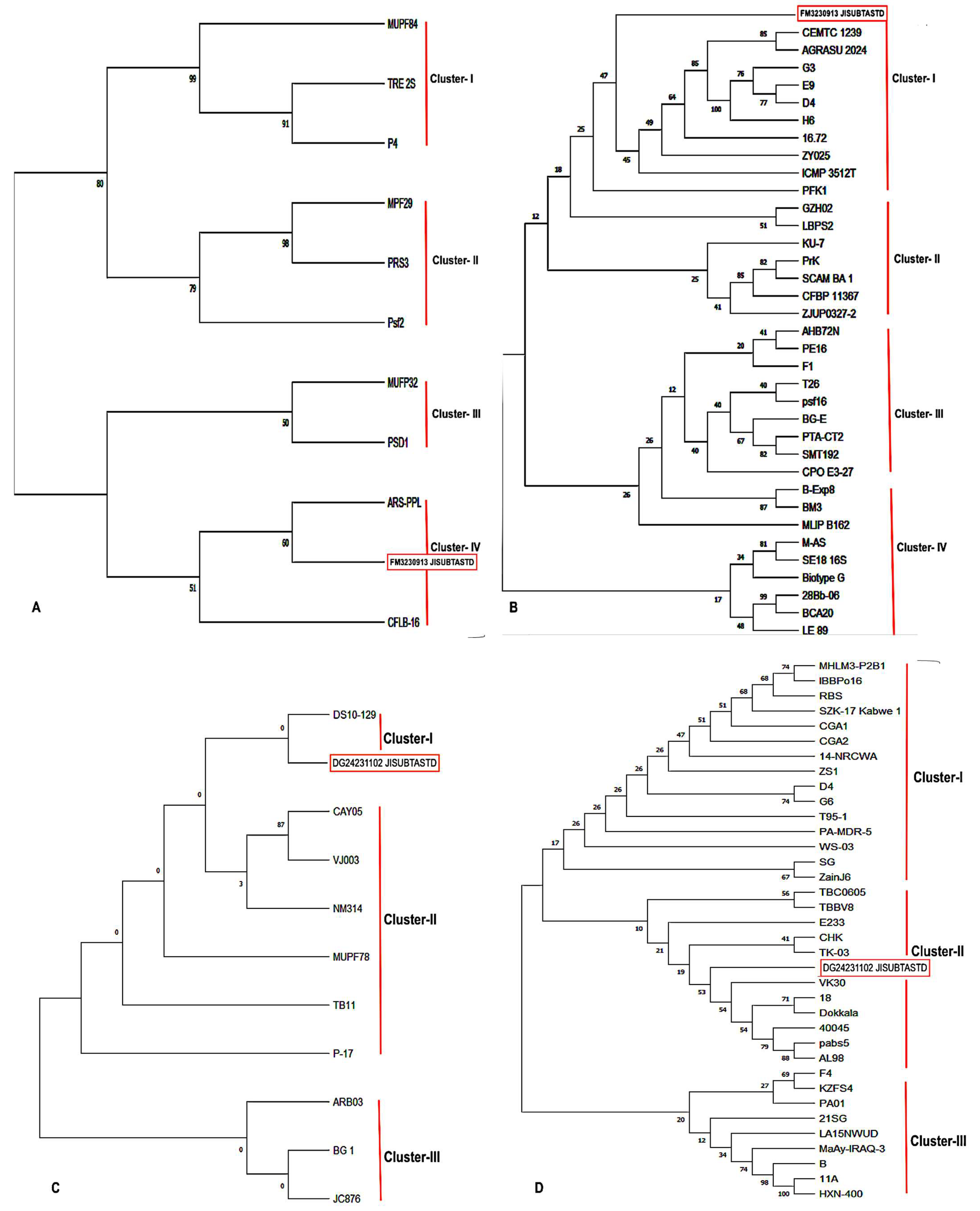
3.4. Molecular Characterization and Phylogenetic Analysis
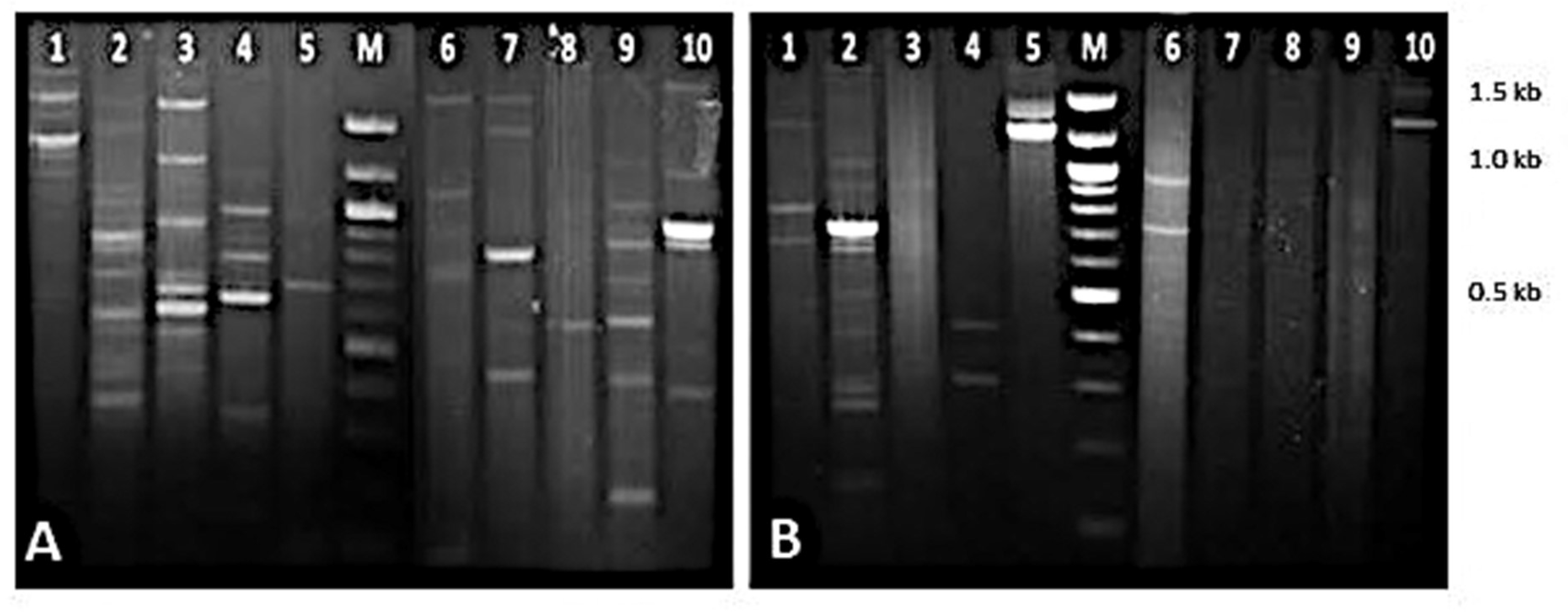
3.5. Genetic Diversity and RAPD-PCR Profiling
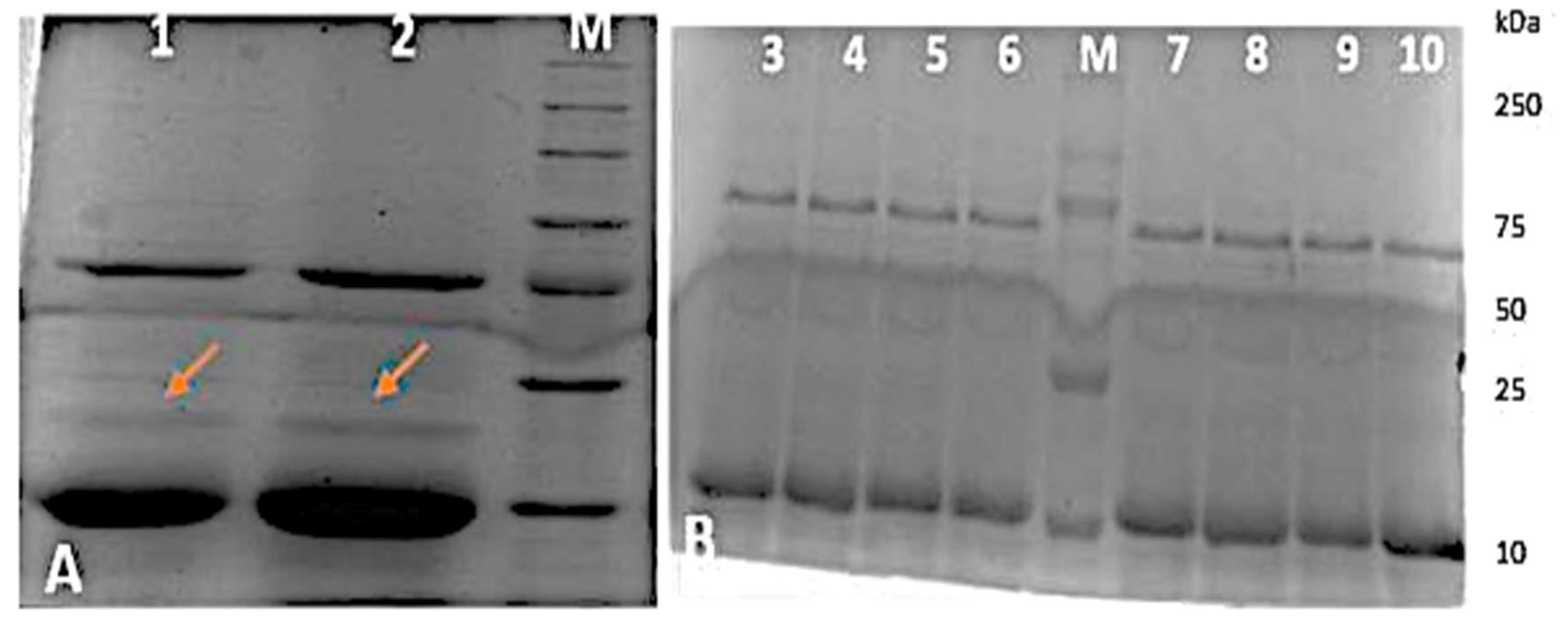
3.6. Antibiotic Resistance Protein Profiling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria-A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Taneja, N.; Sharma, M. Antimicrobial resistance in the environment: The Indian scenario, Indian. J. Med. Res. 2019, 149, 119. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Regional Office for South—East Asia. Jaipur Declaration on Antimicrobial Resistance. 2011. Available online: www.who.int/iris/handle/10665/205397 (accessed on 15 April 2017).
- Nelson, R.E.; Hatfield, K.M.; Wolford, H.; Samore, M.H.; Scott, R.D.; Reddy, S.C.; Olubajo, B.; Paul, P.; Jernigan, J.A.; Baggs, J. National estimates of healthcare costs associated with multidrug-resistant bacterial infections among hospitalized patients in the united states. Clin. Infect. Dis. 2021, 72, S17–S26. [Google Scholar] [CrossRef]
- In Proceedings of the 61st Conference on Decision and Control (CDC), Cancun, Mexico, 6–9 December 2022; IEEE: Cancun, Mexico. Available online: https://ieeexplore.ieee.org/xpl/conhome/9992315/proceeding (accessed on 20 March 2025).
- Division of Descriptive Research; Indian Council of Medical Research. Annual Report on Antimicrobial Resistance Research and Surveillance Network January 2023 to December 2023. AMR Surveillance Network, Indian Council of Medical Research: New Delhi, India. Available online: https://www.icmr.gov.in/icmrobject/uploads/Documents/1725536060_annual_report_2023.pdf (accessed on 20 March 2025).
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The Mortality Burden of Multidrug-resistant Pathogens in India: A Retrospective, Observational Study. Clin. Infect. Dis. 2019, 69, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Palleroni, N.J. Pseudomonas. In Bergey’s Manual of Systematics of Archaea and Bacteria, 1st ed.; Whitman, W.B., Ed.; Wiley: Hoboken, NJ, USA, 2015; p. 1. [Google Scholar] [CrossRef]
- Saha, M.; Sarkar, A. Review on multiple facets of drug resistance: A rising challenge in the 21st century. J. Xenobio. 2021, 11, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2012, 10, 1310. [Google Scholar] [CrossRef]
- Samreen Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental antimicrobial resistance and its drivers: A potential threat to public health. J. Global Antimicrobial. Resistance 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Mulet, M.; Lalucat, J.; García-Valdés, E. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 2010, 12, 1513–1530. [Google Scholar] [CrossRef]
- Scales, B.S.; Dickso, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, genomics, and clinical significance of the Pseudomonas fluorescens species complex, an unappreciated colonizer of humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef] [PubMed]
- Raposo, A.; Ferez, E.; Faria, C.T.D.; Ferrus, M.A.; Carrascosa, C. Food spoilage by Pseudomonas spp.: An overview. In Foodborne Pathogens and Antibiotic Resistance, 1st ed.; Singh, O.V., Ed.; Wiley: Hoboken, NJ, USA, 2016; pp. 41–71. [Google Scholar] [CrossRef]
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbe Profiles collection. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Del Olmo, A.; Calzada, J.; Nuñez, M. The blue discoloration of fresh cheeses: A worldwide defect associated with specific contamination by Pseudomonas fluorescens. Food Cont. 2018, 86, 359–366. [Google Scholar] [CrossRef]
- Bédard, E.; Prévost, M.; Déziel, E. Pseudomonas aeruginosa in premise plumbing of large buildings. Microbiol. Open. 2016, 5, 937–956. [Google Scholar] [CrossRef] [PubMed]
- Maurice, N.M.; Bedi, B.; Sadikot, R.T. Pseudomonas aeruginosa biofilms: Host response and clinical implications in lung infections. Am. J. Respp. Cell Mol. Biol. 2018, 58, 428–439. [Google Scholar] [CrossRef]
- Boyd, A.; Chakrabarty, A.M. Pseudomonas aeruginosa biofilms: Role of the alginate exopolysaccharide. J. Ind. Microbiol. 1995, 15, 162–168. [Google Scholar] [CrossRef]
- Elfadadny, A.; Ragab, R.F.; AlHarbi, M.; Badshah, F.; Ibáñez-Arancibia, E.; Farag, A.; Hendawy, A.O.; De Los Ríos-Escalante, P.R.; Aboubakr, M.; Zakai, S.A.; et al. Antimicrobial resistance of Pseudomonas aeruginosa: Navigating clinical impacts, current resistance trends, and innovations in breaking therapies. Front. Microbiol. 2024, 15, 1374466. [Google Scholar] [CrossRef]
- Ghssein, G.; Ezzeddine, Z. A Review of Pseudomonas aeruginosa Metallophores: Pyoverdine, Pyochelin and Pseudopaline. Biology 2022, 11, 1711. [Google Scholar] [CrossRef]
- Ackerley, D.F.; Caradoc-Davies, T.T.; Lamont, I.L. Substrate Specificity of the Nonribosomal Peptide Synthetase PvdD from Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 2848–2855. [Google Scholar] [CrossRef]
- Reimmann, C.; Serino, L.; Beyeler, M.; Haa, D. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 1998, 144, 3135–3148. [Google Scholar] [CrossRef]
- Laffont, C.; Brutesco, C.; Hajjar, C.; Cullia, G.; Fanelli, R.; Ouerdane, L.; Cavelier, F.; Arnoux, P. Simple rules govern the diversity of bacterial nicotianamine-like metallophores. Biochem. J. 2019, 476, 2221–2233. [Google Scholar] [CrossRef] [PubMed]
- Soil Science Division Staff, USDA. Soil Survey Manual. 2017. Available online: www.nrcs.usda.gov/sites/default/files/2022-09/The-Soil-Survey-Manual.pdf (accessed on 20 March 2025).
- Baron, E.J.; Peterson, L.R.; Finegold, S.M. Bailey and Scott’s Diagnostic Microbiology, 9th ed.; Mosby-Year Book Inc.: St. Louis, MI, USA, 1994. [Google Scholar]
- Arai, T.; Otake, M.; Enomoto, S.; Goto, S.; Kuwahara, S. Determination of Pseudomonas aeruginosa by Biochemical Test Methods. II. Acylamidase Test, a Modified Biochemical Test for the Identification of Pseudomonas aeruginosa. Jpn. J. Microbiol. 1970, 14, 279–284. [Google Scholar] [CrossRef]
- Bergey, D.H.; Holt, J.G. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Williams and Wilkins: Baltimore, Maryland, 1994; Available online: https://www.biodiversitylibrary.org/page/11178148 (accessed on 12 February 2025).
- Hucker, G.J.; Conn, H.J. Methods of Gram Staining. J. Bact. 1923, 8, 343–348. [Google Scholar]
- CLSI supplement M100. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; Clinical and Laboratory Standards Institute (CLSI): Malvern, PA, USA, 2023. [Google Scholar]
- IBM SPSS Statistics 25 Step by Step: A Simple Guide and Reference, 15th ed.; George, D., Mallery, P., Eds.; Routledge: New York, NY, USA, 2018; p. 404. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Budvytiene, I.; Sandlund, J.; Färnert, A.; Banaei, N. Simple real-time PCR and amplicon sequencing method for identification of plasmodium species in human whole blood. J. Clin. Microbiol. 2015, 53, 2251–2257. [Google Scholar] [CrossRef] [PubMed]
- Adikesavalu, H.; Patra, A.; Banerjee, S.; Sarkar, A.; Abraham, T.J. Phenotypic and molecular characterization and pathology of Flectobacillus roseus causing flectobacillosis in captive held carp Labeo rohita (Ham.) fingerlings. Aquaculture 2015, 439, 60–65. [Google Scholar] [CrossRef]
- Burks, C. DNA sequence assembly. IEEE Eng. Med. Biol. Mag. 1994, 13, 771–773. [Google Scholar] [CrossRef]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, W20–W25. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2005, 33, D34–D38. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Barido-Sottani, J.; Schwery, O.; Warnock, R.C.M.; Zhang, C.; Wright, A.M. Practical guidelines for Bayesian phylogenetic inference using Markov chain Monte Carlo (Mcmc). Open Res. Europe. 2024, 3, 204. [Google Scholar] [CrossRef]
- Zhang, R.; Drummond, A. Improving the performance of Bayesian phylogenetic inference under relaxed clock models. BMC Evol. Biol. 2020, 20, 54. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analyt. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, A.A.; Kamer, A.M.A.; Al-Monofoy, K.B.; Al-Madboly, L.A. Pseudomonas aeruginosa’s greenish-blue pigment pyocyanin: Its production and biological activities. Microb. Cell Fact. 2023, 22, 110. [Google Scholar] [CrossRef]
- Murray, T.S.; Kazmierczak, B.I. Pseudomonas aeruginosa exhibits sliding motility in the absence of type iv pili and flagella. J. Bacteriol. 2008, 190, 2700–2708. [Google Scholar] [CrossRef]
- Zago, A.; Chugani, S. Pseudomonas. In Encyclopedia of Microbiology, 3rd ed.; Schaechter, M., Ed.; Academic Press: Cambridge, MA, USA, 2009; pp. 245–260. [Google Scholar] [CrossRef]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The minimum inhibitory concentration of antibiotics: Methods, interpretation, clinical relevance. Pathogens 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). The Minimum Inhibitory Concentrations (MICs) of the Antibacterial Agents Were Determined via Broth Dilution. Eucast Discuss. Doc. E. Dis. 5.1. 2023, 9, 1–7. Available online: www.clinicalmicrobiologyandinfection.com/article/S1198-743X(14)64063-5/fulltext (accessed on 12 February 2025).
- Livermore, D.M. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: Our worst nightmare? Clin. Infect. Dis. 2002, 34, 634–640. [Google Scholar] [CrossRef]
- Strateva, T.; Yordanov, D. Pseudomonas aeruginosa- a phenomenon of bacterial resistance. J. Med. Microbiol. 2009, 58, 1133–1148. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside modifying enzymes. Drug Resist. Updates 2010, 13, 151–171. [Google Scholar] [CrossRef]
- Duineveld, B.M.; Kowalchuk, G.A.; Keijzer, A.; van Elsas, J.D.; van Veen, J.A. Analysis of bacterial communities in the rhizosphere of chrysanthemum by denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Applied Environ. Microbiology 2001, 67, 172–178. [Google Scholar] [CrossRef]
- Kiyaga, S.; Kyany’a, C.; Muraya, A.W.; Smith, H.J.; Mills, E.G.; Kibet, C.; Mboowa, G.; Musila, L. Genetic Diversity, Distribution, and Genomic Characterization of Antibiotic Resistance and Virulence of Clinical Pseudomonas aeruginosa Strains in Kenya. Front. Microbiol. 2022, 14, 835403. [Google Scholar] [CrossRef] [PubMed]
- Bukholm, G.; Tannaes, T.; Kjelsberg, A.B.B.; Smith-Erichsen, N. An outbreak of multidrug-resistant Pseudomonas aeruginosa associated with increased risk of patient death in an intensive care unit. Infect. Cont. Hospp. Epidemiol. 2002, 23, 441–446. [Google Scholar] [CrossRef]
- Luo, J.; Dong, B.; Wang, K.; Cai, S.; Liu, T.; Cheng, X.; Lei, D.; Chen, Y.; Li, Y.; Cong, J.; et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. Plos One 2017, 12, e0176883. [Google Scholar] [CrossRef]
- Topa, S.H.; Subramoni, S.; Palombo, E.A.; Kingshott, P.; Rice, E.A.; Blackall, L.L. Cinnamaldehyde disrupts biofilm formation and swarming motility of Pseudomonas aeruginosa. Microbiology 2018, 164, 1087–1097. [Google Scholar] [CrossRef] [PubMed]
- Cendra, M.D.M.; Torrents, E. Pseudomonas aeruginosa biofilms and their partners in crime. Biotechnol. Adv. 2021, 49, 107734. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Černáková, L.; Kadam, S.S.; Kaushik, K.; Salehi, B.; Bevilacqua, A.; Corbo, M.R.; Antolak, H.; Dybka-Stępień, K.; Leszczewicz, M.; et al. Advances in Chemical and Biological Methods to Identify Microorganisms-From Past to Present. Microorganisms 2019, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef]
- Sharma, S.; Mohler, J.; Mahajan, S.D.; Schwartz, S.A.; Bruggemann, L.; Aalinkeel, R. Microbial Biofilm: A Review on Formation, Infection, Antibiotic Resistance, Control Measures, and Innovative Treatment. Microorganisms 2023, 11, 1614. [Google Scholar] [CrossRef]
- Grooters, K.E.; Ku, J.C.; Richter, D.M.; Krinock, M.J.; Minor, A.; Li, P.; Kim, A.; Sawyer, R.; Li, Y. Strategies for combating antibiotic resistance in bacterial biofilms. Front. Cell. Infect. Microbiol. 2024, 14, 1352273. [Google Scholar] [CrossRef]
- Kamel, N.A.; Tohamy, S.T.; Yahia, I.S.; Aboshanab, K.M. Insights on the performance of phenotypic tests versus genotypic tests for the detection of carbapenemase-producing gram-negative bacilli in resource-limited settings. BMC Microbiol. 2022, 22, 248. [Google Scholar] [CrossRef]
- Clarridge, J.E. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef]
- Woese, C.R. Bacterial evolution. Microbiol. Rev. 1987, 51, 221–271. [Google Scholar] [CrossRef]
- Quiroz-Morales, S.E.; García-Reyes, S.; Ponce-Soto, G.Y.; Servín-González, L.; Soberón-Chávez, G. Tracking the Origins of Pseudomonas aeruginosa Phylogroups by Diversity and Evolutionary Analysis of Important Pathogenic Marker Genes. Diversity 2022, 14, 345. [Google Scholar] [CrossRef]
- Falagas, M.E.; Rafailidis, P.I.; Kofteridis, D.; Virtzili, S.; Chelvatzoglou, F.C.; Papaioannou, V.; Maraki, S.; Samonis, G.; Michalopoulos, A. Risk factors for carbapenem-resistant Klebsiella pneumoniae infections: A matched case control study. J. Antimicrob. Chemother. 2007, 60, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Didelot, X.; Bowden, R.; Wilson, D.J.; Peto, T.E.A.; Crook, D.W. Transforming clinical microbiology with bacterial genome sequencing. Nat. Rev. Genet. 2012, 13, 601–612. [Google Scholar] [CrossRef]
- Singh, A.; Goering, R.V.; Simjee, S.; Foley, S.L.; Zervos, M.J. Application of molecular techniques to the study of hospital infection. Clin. Microbiol. Rev. 2006, 19, 512–530. [Google Scholar] [CrossRef] [PubMed]
- Nanvazadeh, F.; Khosravi, A.D.; Zolfaghari, M.R. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns 2013, 39, 1409–1413. [Google Scholar] [CrossRef]
- Katsanis, S.H.; Katsanis, N. Molecular genetic testing and the future of clinical genomics. Nat. Rev. Genet. 2013, 14, 415–426. [Google Scholar] [CrossRef]
- Chang, Y.; Li, J.; Zhang, L. Genetic diversity and molecular diagnosis of Giardia. Infect. Genet. Evol. 2023, 113, 105482. [Google Scholar] [CrossRef]
- Moore, E.R.B.; Tindall, B.J.; Martins Dos Santos, V.A.P.; Pieper, D.H.; Ramos, J.L.; Palleroni, N.J. Nonmedical: Pseudomonas. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006. [Google Scholar] [CrossRef]
- Al-Wrafy, F.; Brzozowska, E.; Górska, S.; Drab, M.; Strus, M.; Gamian, A. Identification and characterization of phage protein and its activity against two strains of multidrug-resistant Pseudomonas aeruginosa. Sci. Rep. 2019, 9, 13487. [Google Scholar] [CrossRef]
- Ezzeddine, Z.; Ghssein, G. Towards new antibiotics classes targeting bacterial metallophores. Microb. Pathog. 2023, 182, 106221. [Google Scholar] [CrossRef] [PubMed]
- Górska, A.; Sloderbach, A.; Marszałł, M.P. Siderophore-drug complexes: Potential medicinal applications of the ‘Trojan horse’ strategy. Trends Pharmacol. Sci. 2014, 35, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Ghosh, M.; Niu, C.; Malouin, F.; Moellmann, U.; Miller, M.J. Iron transport-mediated drug delivery using mixed-ligand siderophore-beta-lactam conjugates. Chem. Biol. 1996, 3, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Seyedsayamdost, M.R.; Traxler, M.F.; Zheng, S.L.; Kolter, R.; Clardy, J. Structure and biosynthesis of amychelin, an unusual mixed-ligand siderophore from Amycolatopsis spp. AA4. J. Am Chem. Soc. 2011, 133, 11434–11437. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, L.; Molin, S. Synergistic activities of an efflux pump inhibitor and iron chelators against Pseudomonas aeruginosa growth and biofilm formation. Antimicrob. Agents Chemother. 2010, 54, 3960–3973. [Google Scholar] [CrossRef]
- Llanes, C.; Hocquet, D.; Vogne, C.; Benali-Baitich, D.; Neuwirth, C.; Plésiat, P. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 2004, 48, 1797–1802. [Google Scholar] [CrossRef]

| Phenotypes | Nutrient Agar | Cetrimide Agar | Pseudomonas Agar | |||
|---|---|---|---|---|---|---|
| P. fluorescens FM3 | P. aeruginosa DG2 | P. fluorescens FM3 | P. aeruginosa DG2 | P. fluorescens FM3 | P. aeruginosa DG2 | |
| Growth | + | + | – | + | + | + |
| Shape | Round | Round | Round | Round | Round | Round |
| Size (mm) | 0.3–0.5 | 0.3–0.5 | – | 1.0–1.2 | 0.5–0.6 | 0.8–1.0 |
| Elevation | Convex | Convex | – | Convex | Convex | Convex |
| Surface | Smooth, shiny | Smooth, shiny | – | Smooth, shiny | Smooth, shiny | Smooth, shiny |
| Margin | Regular | Regular | – | Regular | Regular | Regular |
| Color | Pinkish Red | Gray | – | Green | Pinkish red | Pinkish red |
| Sl. No. | Biochemical Tests | P. fluorescens FM3 | P. aeruginosa DG2 |
|---|---|---|---|
| 1. | Gram staining | – | – |
| 2. | Pigmentation | + | + |
| 3. | Indole | – | – |
| 4. | Simmons citrate | + | + |
| 5. | Methyl red (MR) | – | – |
| 6. | Voges–Proskauer (VP) | – | – |
| 7. | Catalase | + | + |
| 8. | Starch hydrolysis | + | + |
| 9. | Lipid hydrolysis | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the Hellenic Society for Microbiology. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, T.; Das, A.; Das, N.; Mukherjee, R.; Saha, M.; Das, D.; Sarkar, A. Genetic Diversity and Phylogenetic Analysis Among Multidrug-Resistant Pseudomonas spp. Isolated from Solid Waste Dump Sites and Dairy Farms. Acta Microbiol. Hell. 2025, 70, 30. https://doi.org/10.3390/amh70030030
Das T, Das A, Das N, Mukherjee R, Saha M, Das D, Sarkar A. Genetic Diversity and Phylogenetic Analysis Among Multidrug-Resistant Pseudomonas spp. Isolated from Solid Waste Dump Sites and Dairy Farms. Acta Microbiologica Hellenica. 2025; 70(3):30. https://doi.org/10.3390/amh70030030
Chicago/Turabian StyleDas, Tuhina, Arkaprava Das, Neha Das, Rittika Mukherjee, Mousumi Saha, Dipanwita Das, and Agniswar Sarkar. 2025. "Genetic Diversity and Phylogenetic Analysis Among Multidrug-Resistant Pseudomonas spp. Isolated from Solid Waste Dump Sites and Dairy Farms" Acta Microbiologica Hellenica 70, no. 3: 30. https://doi.org/10.3390/amh70030030
APA StyleDas, T., Das, A., Das, N., Mukherjee, R., Saha, M., Das, D., & Sarkar, A. (2025). Genetic Diversity and Phylogenetic Analysis Among Multidrug-Resistant Pseudomonas spp. Isolated from Solid Waste Dump Sites and Dairy Farms. Acta Microbiologica Hellenica, 70(3), 30. https://doi.org/10.3390/amh70030030






