Comparison of Two Different Integration Methods for Quantifying Monoclonal Proteins on Agarose Gel and Capillary Zone Electrophoresis Instruments
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
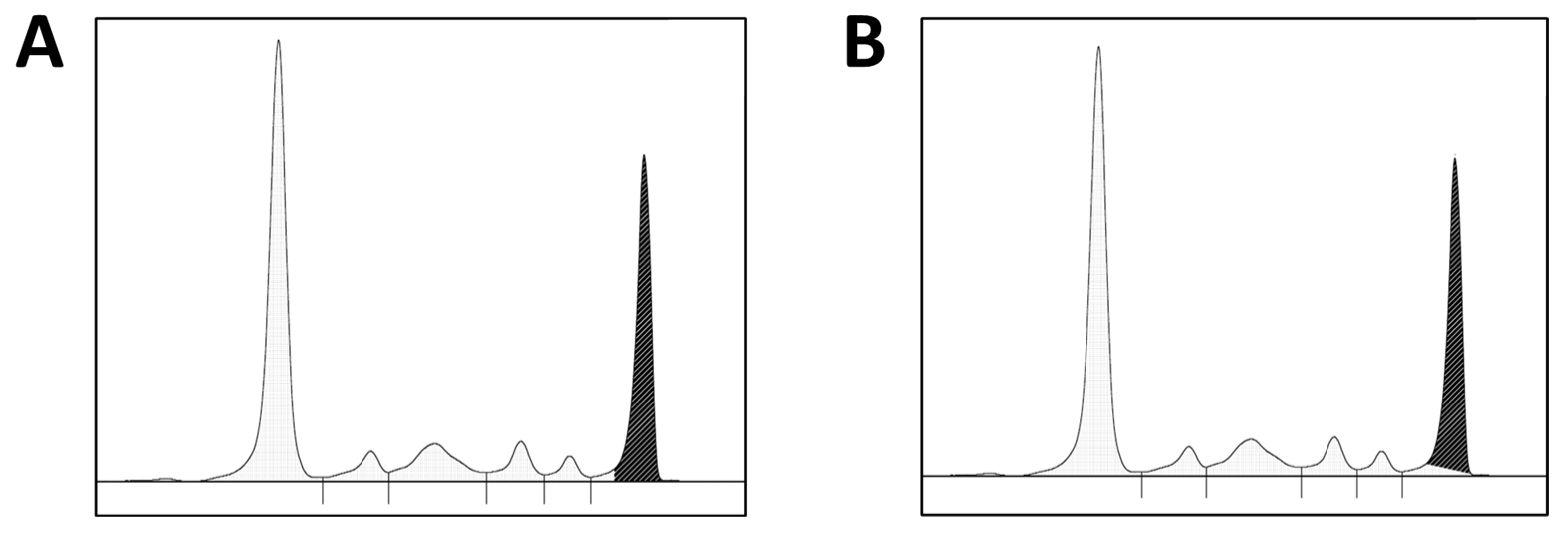
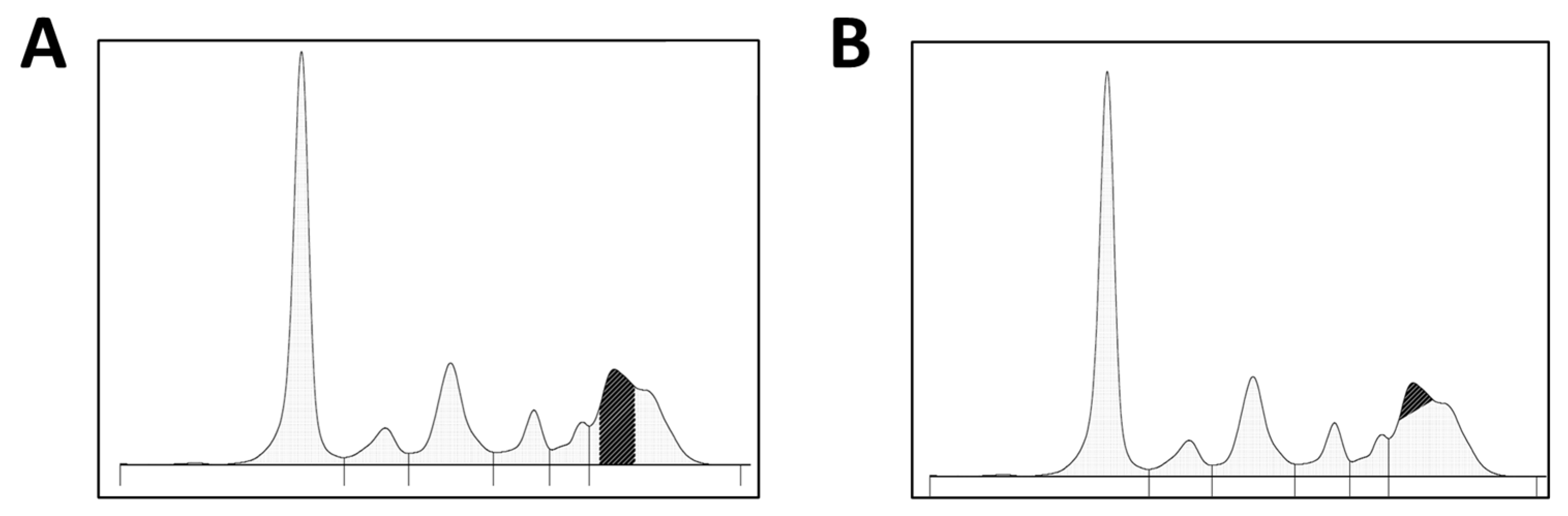
2.2. Data Collection and M-Protein Quantification
2.3. Bias Evaluation and Data Analyses
3. Results
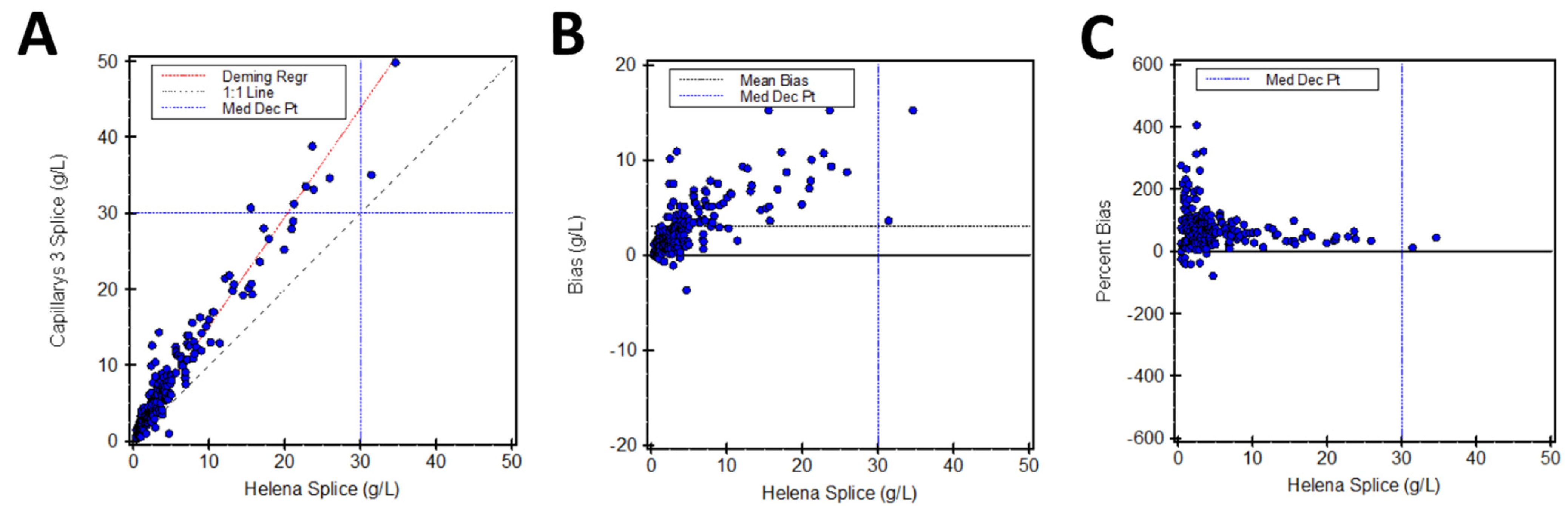
3.1. SPIFE 3000 Splice vs. Capillarys 3 Splice
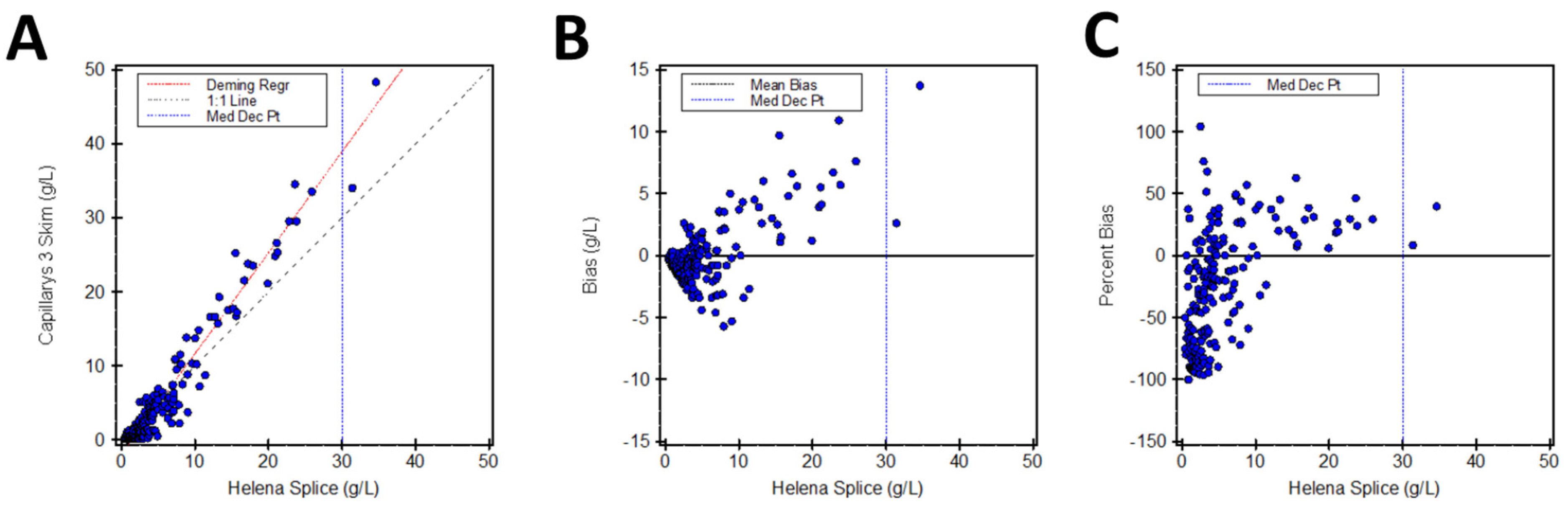
3.2. SPIFE 3000 Splice vs. Capillarys Skim
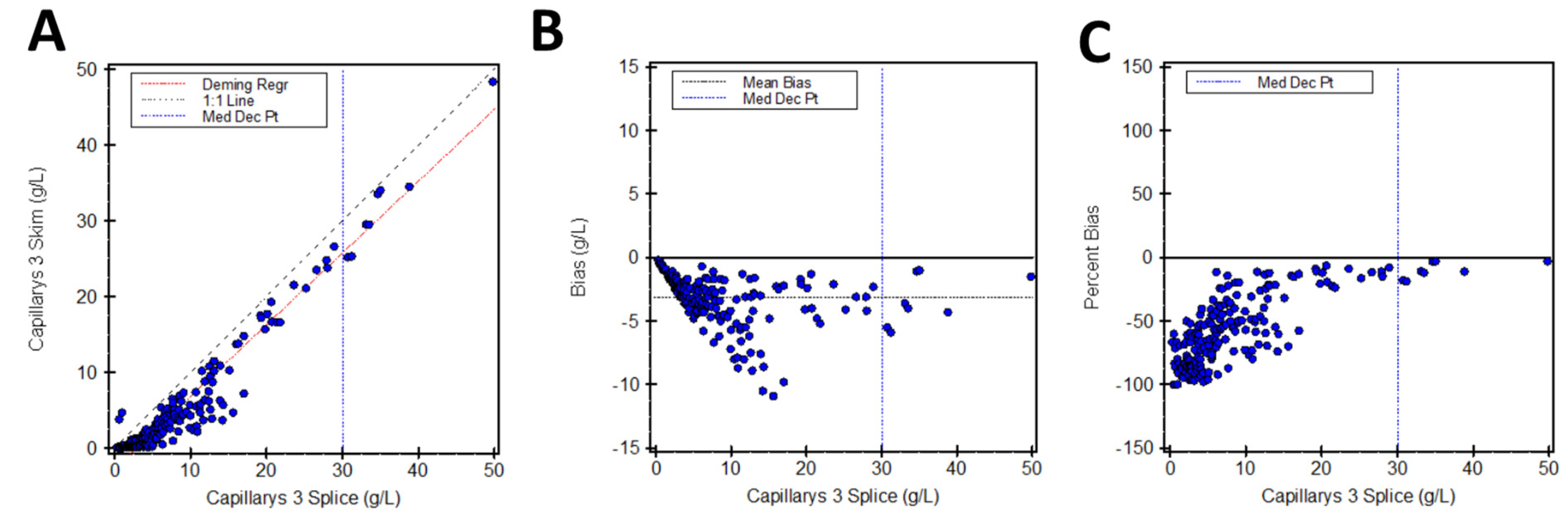
3.3. Capillarys Splice vs. Capillarys Skim
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, S.K.; Callander, N.S.; Hillengass, J.; Liedtke, M.; Baljevic, M.; Campagnaro, E.; Castillo, J.J.; Chandler, J.C.; Cornell, R.F.; Costello, C.; et al. NCCN guidelines insights: Multiple myeloma, version 1.2020. J. Natl. Compr. Cancer Netw. 2019, 17, 1154–1165. [Google Scholar] [CrossRef] [PubMed]
- Caers, J.; Garderet, L.; Kortüm, K.M.; O’Dwyer, M.E.; van de Donk, N.W.C.J.; Binder, M.; Dold, S.M.; Gay, F.; Corre, J.; Beguin, Y.; et al. European myeloma network recommendations on tools for the diagnosis and monitoring of multiple myeloma: What to use and when. Haematologica 2018, 103, 1772–1784. [Google Scholar] [CrossRef] [PubMed]
- Dimopoulos, M.; Kyle, R.; Fermand, J.-P.; Rajkumar, S.V.; San Miguel, J.; Chanan-Khan, A.; Ludwig, H.; Joshua, D.; Mehta, J.; Gertz, M.; et al. Consensus recommendations for Standard Investigative Workup: Report of the International Myeloma Workshop Consensus Panel 3. Blood 2011, 117, 4701–4705. [Google Scholar] [CrossRef] [PubMed]
- Genzen, J.R.; Murray, D.L.; Abel, G.; Meng, Q.H.; Baltaro, R.J.; Rhoads, D.D.; Delgado, J.C.; Souers, R.J.; Bashleben, C.; Keren, D.F.; et al. Screening and diagnosis of monoclonal gammopathies: An International Survey of Laboratory Practice. Arch. Pathol. Lab. Med. 2017, 142, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.-V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Avet-Loiseau, H.; Oliva, S.; Lokhorst, H.M.; Goldschmidt, H.; Rosinol, L.; Richardson, P.; Caltagirone, S.; Lahuerta, J.J.; Facon, T.; et al. Revised international staging system for multiple myeloma: A report from International Myeloma Working Group. J. Clin. Oncol. 2015, 33, 2863–2869. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Paiva, B.; Anderson, K.C.; Durie, B.; Landgren, O.; Moreau, P.; Munshi, N.; Lonial, S.; Bladé, J.; Mateos, M.-V.; et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016, 17, e328–e346. [Google Scholar] [CrossRef] [PubMed]
- Willrich, M.A.V.; Katzmann, J.A. Laboratory testing requirements for diagnosis and follow-up of multiple myeloma and related plasma cell dyscrasias. Clin. Chem. Lab. Med. (CCLM) 2016, 54, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Willrich, M.A.; Long, T.A.; Bashleben, C.; Fink, S.L.; Rudolf, J.W.; Peterson, D.; Wener, M.H.; Baltaro, R.J.; Genzen, J.R.; Ansari, M.Q.; et al. Performance of perpendicular drop versus tangent skimming gating of M-protein in proficiency testing challenges. Clin. Chem. Lab. Med. (CCLM) 2020, 59, e19–e22. [Google Scholar] [CrossRef] [PubMed]
- Keren, D.F.; Schroeder, L. Challenges of measuring monoclonal proteins in serum. Clin. Chem. Lab. Med. (CCLM) 2016, 54, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.J.; Taher, J.; Kulasingam, V.; Chan, P.C. To skim or splice? comparing the quantification of M-proteins using two peak-integration protocols across multiple electrophoresis platforms. Clin. Biochem. 2022, 102, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Schild, C.; Wermuth, B.; Trapp-Chiappini, D.; Egger, F.; Nuoffer, J.-M. Reliability of M protein quantification: Comparison of two peak integration methods on capillarys 2. Clin. Chem. Lab. Med. 2008, 46, 876–877. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bergón, E.; Miranda, I.; Miravalles, E. Linearity and detection limit in the measurement of serum M-protein with the capillary zone electrophoresis system Capillarys®. Clin. Chem. Lab. Med. (CCLM) 2005, 43, 721–723. [Google Scholar] [CrossRef] [PubMed]
- McCudden, C.R.; Mathews, S.P.; Hainsworth, S.A.; Chapman, J.F.; Hammett-Stabler, C.A.; Willis, M.S.; Grenache, D.G. Performance comparison of capillary and agarose gel electrophoresis for the identification and characterization of monoclonal immunoglobulins. Am. J. Clin. Pathol. 2008, 129, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Katzmann, J.A.; Clark, R.; Sanders, E.; Landers, J.P.; Kyle, R.A. Prospective study of serum protein capillary zone electrophoresis and immunotyping off monoclonal proteins by immunosubtraction. Am. J. Clin. Pathol. 1998, 110, 503–509. [Google Scholar] [CrossRef] [PubMed][Green Version]
| X Method | Y Method | Regression Equation | Predicted “Y” Concentration at Clinical Cut Point of 30 g/L (95% CI) | % Difference from 30 g/L |
|---|---|---|---|---|
| SPIFE 3000 Splice | Capillarys 3 Splice | y = 1.437x + 0.76 | 43.9 g/L (42.6–45.1) | 46.3% |
| SPIFE 3000 Splice | Capillarys 3 Skim | y = 1.363x − 1.98 | 38.9 g/L (37.8–40.1) | 29.7% |
| Capillarys 3 Splice | Capillarys 3 Skim | y = 0.947x − 2.65 | 25.8 g/L (25.0–26.5) | −14.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larkin, B.; Mahaney, L.; Abegunde, S.; Shea, J.L. Comparison of Two Different Integration Methods for Quantifying Monoclonal Proteins on Agarose Gel and Capillary Zone Electrophoresis Instruments. LabMed 2024, 1, 14-21. https://doi.org/10.3390/labmed1010004
Larkin B, Mahaney L, Abegunde S, Shea JL. Comparison of Two Different Integration Methods for Quantifying Monoclonal Proteins on Agarose Gel and Capillary Zone Electrophoresis Instruments. LabMed. 2024; 1(1):14-21. https://doi.org/10.3390/labmed1010004
Chicago/Turabian StyleLarkin, Brittany, Laura Mahaney, Samuel Abegunde, and Jennifer L. Shea. 2024. "Comparison of Two Different Integration Methods for Quantifying Monoclonal Proteins on Agarose Gel and Capillary Zone Electrophoresis Instruments" LabMed 1, no. 1: 14-21. https://doi.org/10.3390/labmed1010004
APA StyleLarkin, B., Mahaney, L., Abegunde, S., & Shea, J. L. (2024). Comparison of Two Different Integration Methods for Quantifying Monoclonal Proteins on Agarose Gel and Capillary Zone Electrophoresis Instruments. LabMed, 1(1), 14-21. https://doi.org/10.3390/labmed1010004






