Mass Spectrometry and 3D Modeling Indicate the SBK2 Kinase Phosphorylates Splicing Factor SRSF7 to Regulate Cardiac Development
Abstract
1. Introduction
2. Results
2.1. SBK2 Overexpression Impacts Ventricular Structure and Function
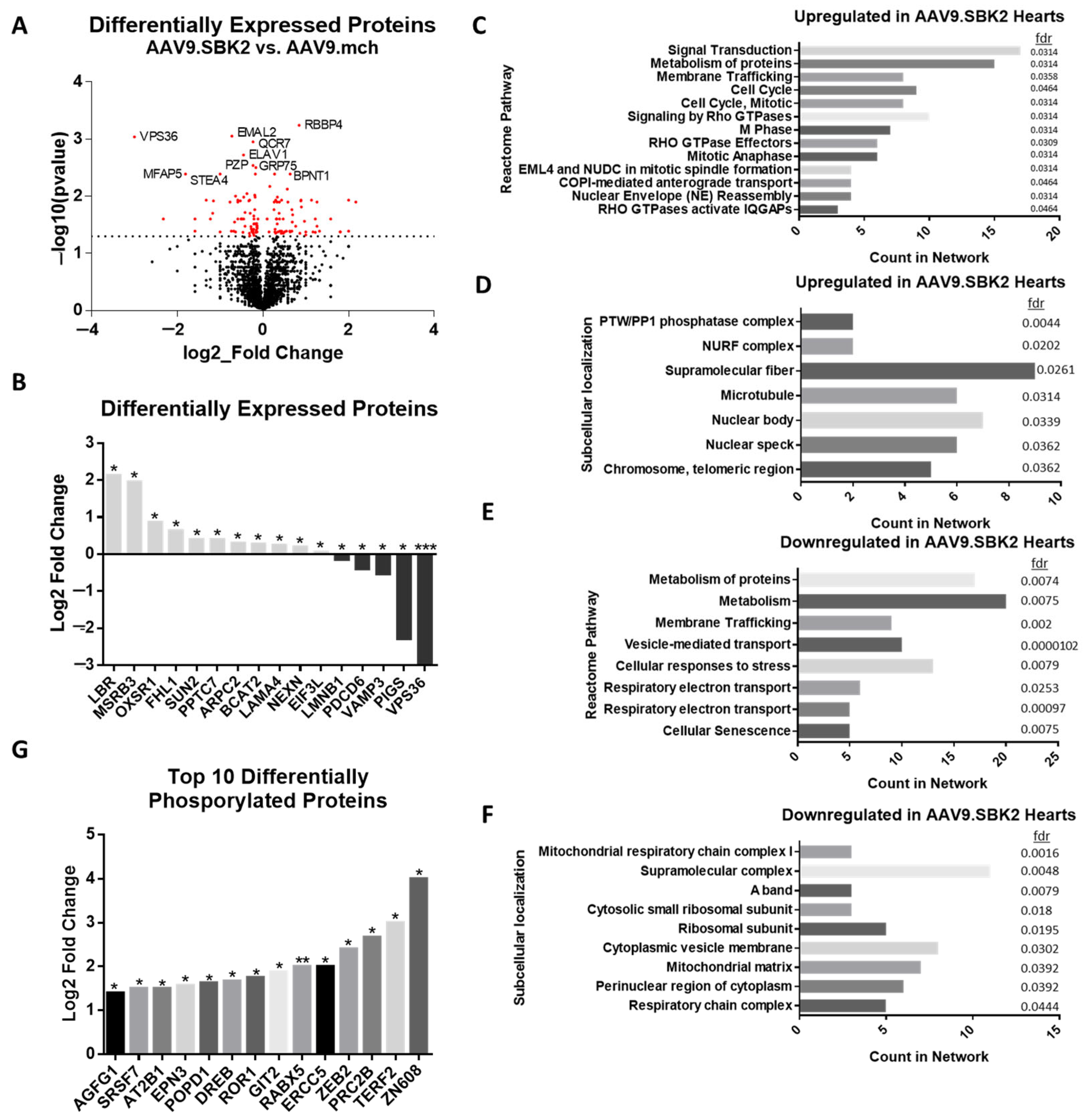
2.2. Mass Spectrometry Reveals Proteome Changes Resulting from SBK2 Overexpression in 2-Week-Old Ventricles
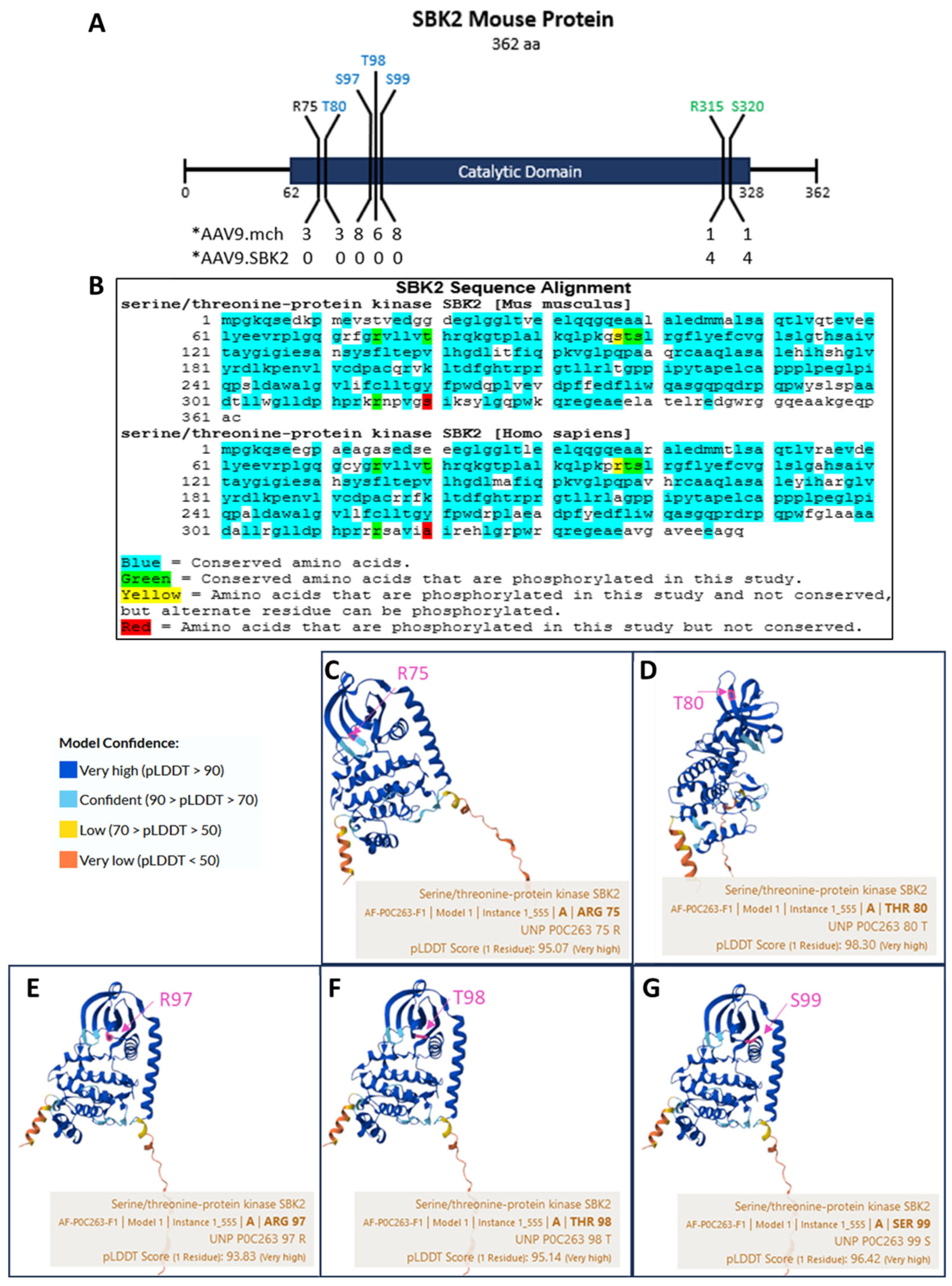
2.3. Phosphorylation of SBK2
2.4. SBK2 R315 Phosphorylation Occurs on a Conserved Amino Acid of a Turn Motif
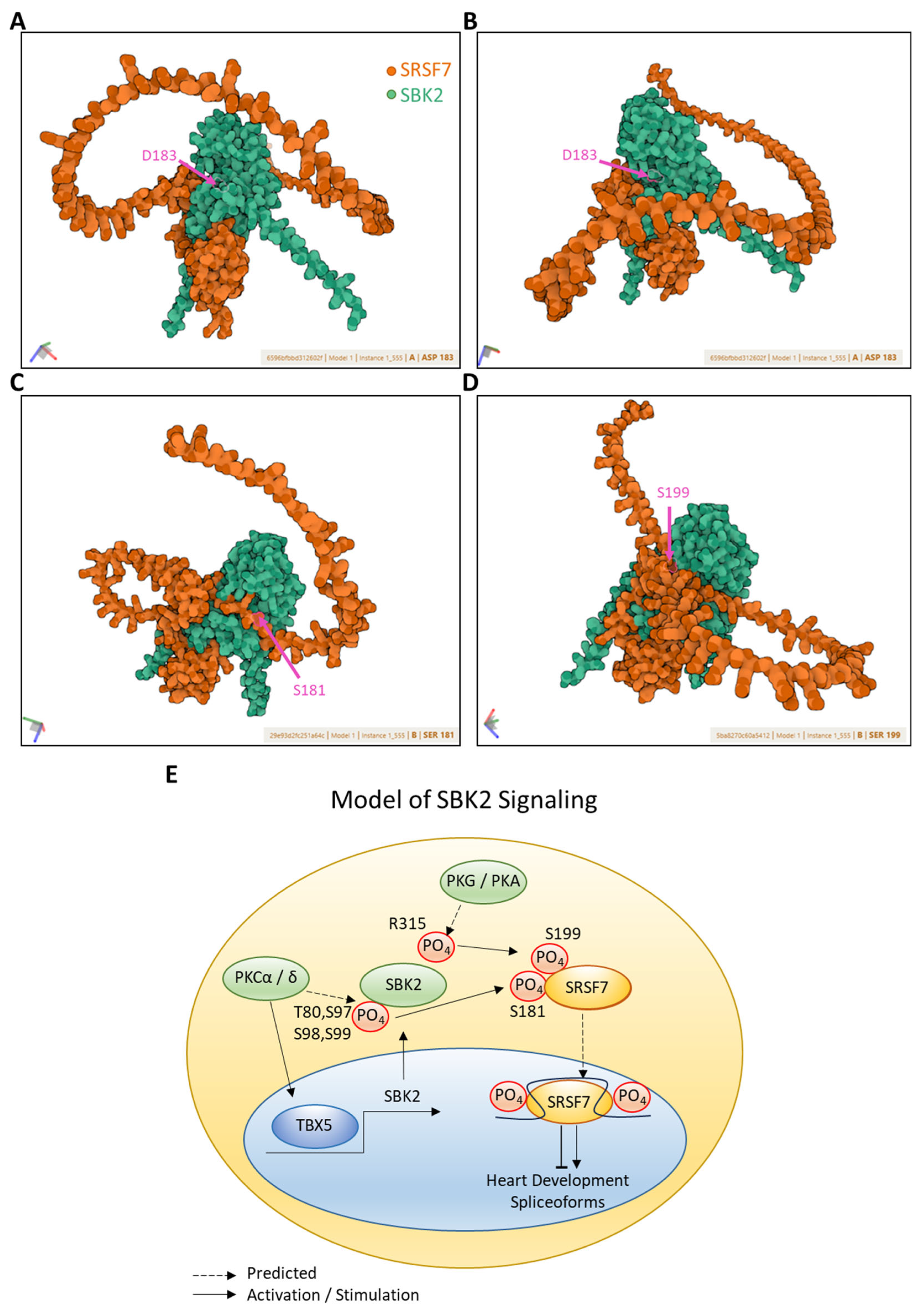
2.5. Modeling of SBK2 Binding to Phospho-Target Proteins
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RT-qPCR
4.3. Western Blotting
4.4. AAV Generation and Injection
4.5. Echocardiography
4.6. Mass Spectrometry
4.6.1. Protein Isolation for Mass Spectrometry Analysis
4.6.2. Protein Identification by Mass Spectrometry Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV9.SBK2 | AAV9.cTNT.SBK2.P2A.mcherry |
| AAV9.mch | AAV9.cTNT.P2A.mcherry |
| CRYAB | Crystallin Alpha B |
| DRC | Desmin-Related Cardiomyopathy |
| R120G | Crystallin Alpha B R120G mutation |
| RABX5/RABGEF1 | Rab5 GDP/GTP exchange factor |
| SBK2 | SH3 Domain Binding Kinase Family Member 2 |
| SRSF7 | Serine/arginine-rich splicing factor 7 |
References
- van Gorp, P.R.; Zhang, J.; Liu, J.; Tsonaka, R.; Mei, H.; Dekker, S.O.; Bart, C.I.; De Coster, T.; Post, H.; Heck, A.J.; et al. Sbk2, a newly discovered atrium-enriched regulator of sarcomere integrity. Circ. Res. 2022, 131, 24–41. [Google Scholar] [CrossRef]
- McLendon, P.M.; Robbins, J. Desmin-related cardiomyopathy: An unfolding story. Am. J. Physiol.-Heart Circ. Physiol. 2011, 301, H1220–H1228. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Osinska, H.; Dorn, G.W.; Nieman, M.; Lorenz, J.N.; Gerdes, A.M.; Witt, S.; Kimball, T.; Gulick, J.; Robbins, J. Mouse model of desmin-related cardiomyopathy. Circulation 2001, 103, 2402–2407. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, L.G.; Dalakas, M.C. Tragedy in a heartbeat: Malfunctioning desmin causes skeletal and cardiac muscle disease. J. Clin. Investig. 2009, 119, 1806–1813. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Osinska, H.; Klevitsky, R.; Gerdes, A.M.; Nieman, M.; Lorenz, J.; Hewett, T.; Robbins, J. Expression of R120G–αB-crystallin causes aberrant desmin and αB-crystallin aggregation and cardiomyopathy in mice. Circ. Res. 2001, 89, 84–91. [Google Scholar] [CrossRef]
- Jordan, E.; Peterson, L.; Ai, T.; Asatryan, B.; Bronicki, L.; Brown, E.; Celeghin, R.; Edwards, M.; Fan, J.; Ingles, J. Evidence-based assessment of genes in dilated cardiomyopathy. Circulation 2021, 144, 7–19. [Google Scholar] [CrossRef]
- Bermudez-Jimenez, F.J.; Protonotarios, A.; García-Hernández, S.; Pérez Asensio, A.; Rampazzo, A.; Zorio, E.; Brodehl, A.; Arias, M.A.; Macías-Ruiz, R.; Fernández-Armenta, J.; et al. Phenotype and clinical outcomes in desmin-related arrhythmogenic cardiomyopathy. Clin. Electrophysiol. 2024, 10, 1178–1190. [Google Scholar] [CrossRef]
- Vicart, P. On noxious desmin: Functional effects of a novel heterozygous desmin insertion mutation on the extrasarcomeric desmin cytoskeleton and mitochondria. Hum. Mol. Genet. 2003, 12, 657–669. [Google Scholar] [CrossRef]
- Maloyan, A.; Sanbe, A.; Osinska, H.; Westfall, M.; Robinson, D.; Imahashi, K.I.; Murphy, E.; Robbins, J. Mitochondrial dysfunction and apoptosis underlie the pathogenic process in α-B-crystallin desmin-related cardiomyopathy. Circulation 2005, 112, 3451–3461. [Google Scholar] [CrossRef]
- Oka, H.; Nakau, K.; Imanishi, R.; Furukawa, T.; Tanabe, Y.; Hirono, K.; Hata, Y.; Nishida, N.; Azuma, H. A case report of a rare heterozygous variant in the desmin gene associated with hypertrophic cardiomyopathy and complete atrioventricular block. CJC Open 2021, 3, 1195–1198. [Google Scholar] [CrossRef]
- Tannous, P.; Zhu, H.; Johnstone, J.L.; Shelton, J.M.; Rajasekaran, N.S.; Benjamin, I.J.; Nguyen, L.; Gerard, R.D.; Levine, B.; Rothermel, B.A.; et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc. Natl. Acad. Sci. USA 2008, 105, 9745–9750. [Google Scholar] [CrossRef]
- Li, J.; Horak, K.M.; Su, H.; Sanbe, A.; Robbins, J.; Wang, X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. J. Clin. Investig. 2011, 121, 3689–3700. [Google Scholar] [CrossRef]
- Zheng, H.; Tang, M.; Zheng, Q.; Kumarapeli, A.R.K.; Horak, K.M.; Tian, Z.; Wang, X. Doxycycline attenuates protein aggregation in cardiomyocytes and improves survival of a mouse model of cardiac proteinopathy. J. Am. Coll. Cardiol. 2010, 56, 1418–1426. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Pan, B.; Xiao, P.; Bouska, M.; Lewno, M.T.; Xing, Y.; Zeng, E.; Liang, H.; Li, F.; Gao, X. Sustained but Decoyed Activation of the JAK1-STAT Pathway by Aberrant Protein Aggregation Exacerbates Proteotoxicity. Circulation 2024, 150, 1302–1305, Erratum in Circulation 2025, 152, e47. [Google Scholar] [CrossRef]
- Yang, L.; Parajuli, N.; Wu, P.; Liu, J.; Wang, X. S14-phosphorylated RPN6 mediates proteasome activation by PKA and alleviates proteinopathy. Circ. Res. 2023, 133, 572–587. [Google Scholar] [CrossRef]
- Lindsey, M.L.; Kassiri, Z.; Virag, J.A.; de Castro Brás, L.E.; Scherrer-Crosbie, M. Guidelines for measuring cardiac physiology in mice. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H733–H752. [Google Scholar] [CrossRef]
- Bish, L.T.; Morine, K.; Sleeper, M.M.; Sanmiguel, J.; Wu, D.; Gao, G.; Wilson, J.M.; Sweeney, H.L. Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum. Gene Ther. 2008, 19, 1359–1368. [Google Scholar] [CrossRef]
- Prasad, K.M.; Xu, Y.; Yang, Z.; Acton, S.T.; French, B.A. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: In vivo gene delivery follows a Poisson distribution. Gene Ther. 2011, 18, 43–52. [Google Scholar] [CrossRef]
- Donnelly, M.L.; Luke, G.; Mehrotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’mechanism indicates not a proteolytic reaction, but a novel translational effect: A putative ribosomal ‘skip’. J. Gen. Virol. 2001, 82, 1013–1025. [Google Scholar] [CrossRef]
- Liu, R. Complex functionality of protein phosphatase 1 isoforms in the heart. Cell. Signal. 2021, 85, 110059. [Google Scholar] [CrossRef]
- Sun, X.; Tang, X.; Qiu, H. Cardiac-Specific Suppression of Valosin-Containing Protein Induces Progressive Heart Failure and Premature Mortality Correlating with Temporal Dysregulations in mTOR Complex 2 and Protein Phosphatase 1. Int. J. Mol. Sci. 2024, 25, 6445. [Google Scholar] [CrossRef]
- Yamano, K.; Wang, C.; Sarraf, S.A.; Münch, C.; Kikuchi, R.; Noda, N.N.; Hizukuri, Y.; Kanemaki, M.T.; Harper, W.; Tanaka, K.; et al. Endosomal Rab cycles regulate Parkin-mediated mitophagy. Elife 2018, 7, e31326. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Sigrist, C.J.; Cerutti, L.; De Castro, E.; Langendijk-Genevaux, P.S.; Bulliard, V.; Bairoch, A.; Hulo, N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010, 38 (Suppl. S1), D161–D166. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.N.; Noble, M.E.; Owen, D.J. Active and inactive protein kinases: Structural basis for regulation. Cell 1996, 85, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Gógl, G.; Kornev, A.P.; Reményi, A.; Taylor, S.S. Disordered protein kinase regions in regulation of kinase domain cores. Trends Biochem. Sci. 2019, 44, 300–311. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Žemgulytė, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef]
- Cáceres, J.F.; Screaton, G.R.; Krainer, A.R. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998, 12, 55–66. [Google Scholar] [CrossRef]
- Kadota, Y.; Jam, F.A.; Yukiue, H.; Terakado, I.; Morimune, T.; Tano, A.; Tanaka, Y.; Akahane, S.; Fukumura, M.; Tooyama, I. Srsf7 establishes the juvenile transcriptome through age-dependent alternative splicing in mice. iScience 2020, 23, 100929. [Google Scholar] [CrossRef]
- Le, M.N.; Nguyen, T.D.; Nguyen, T.A. SRSF7 and SRSF3 depend on RNA sequencing motifs and secondary structures to regulate Microprocessor. Life Sci. Alliance 2023, 6, e202201779. [Google Scholar] [CrossRef] [PubMed]
- Schwich, O.D.; Blümel, N.; Keller, M.; Wegener, M.; Setty, S.T.; Brunstein, M.E.; Poser, I.; Mozos, I.R.D.L.; Suess, B.; Münch, C.; et al. SRSF3 and SRSF7 modulate 3′ UTR length through suppression or activation of proximal polyadenylation sites and regulation of CFIm levels. Genome Biol. 2021, 22, 82. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.E.; Choi, D.H.; Kim, W.H.; Park, Y.S. Up-regulation of PKCα and δ during beating cardiomyocyte differentiation of P19CL6 cells with suppressed apoptotic cell populations. Mol. Cell. Toxicol. 2024, 20, 167–176. [Google Scholar] [CrossRef]
- Steimle, J.D.; Moskowitz, I.P. TBX5: A key regulator of heart development. Curr. Top. Dev. Biol. 2017, 122, 195–221. [Google Scholar] [PubMed]
- Siatra, P.; Vatsellas, G.; Chatzianastasiou, A.; Balafas, E.; Manolakou, T.; Papapetropoulos, A.; Agapaki, A.; Mouchtouri, E.T.; Ruchaya, P.J.; Korovesi, A.G.; et al. Return of the Tbx5; lineage-tracing reveals ventricular cardiomyocyte-like precursors in the injured adult mammalian heart. NPJ Regen. Med. 2023, 8, 13. [Google Scholar] [CrossRef]
- Sweat, M.E.; Cao, Y.; Zhang, X.; Burnicka-Turek, O.; Perez-Cervantes, C.; Kulandaisamy, A.; Lu, F.; Keating, E.M.; Akerberg, B.N.; Ma, Q.; et al. Tbx5 maintains atrial identity in postnatal cardiomyocytes by regulating an atrial-specific enhancer network. Nat. Cardiovasc. Res. 2023, 2, 881–898, Correction in Nat. Cardiovasc. Res. 2023, 2, 1095. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouska, M.; Callegari, E.; Paez, D.; Wang, X. Mass Spectrometry and 3D Modeling Indicate the SBK2 Kinase Phosphorylates Splicing Factor SRSF7 to Regulate Cardiac Development. Kinases Phosphatases 2025, 3, 20. https://doi.org/10.3390/kinasesphosphatases3040020
Bouska M, Callegari E, Paez D, Wang X. Mass Spectrometry and 3D Modeling Indicate the SBK2 Kinase Phosphorylates Splicing Factor SRSF7 to Regulate Cardiac Development. Kinases and Phosphatases. 2025; 3(4):20. https://doi.org/10.3390/kinasesphosphatases3040020
Chicago/Turabian StyleBouska, Mark, Eduardo Callegari, Daniela Paez, and Xuejun Wang. 2025. "Mass Spectrometry and 3D Modeling Indicate the SBK2 Kinase Phosphorylates Splicing Factor SRSF7 to Regulate Cardiac Development" Kinases and Phosphatases 3, no. 4: 20. https://doi.org/10.3390/kinasesphosphatases3040020
APA StyleBouska, M., Callegari, E., Paez, D., & Wang, X. (2025). Mass Spectrometry and 3D Modeling Indicate the SBK2 Kinase Phosphorylates Splicing Factor SRSF7 to Regulate Cardiac Development. Kinases and Phosphatases, 3(4), 20. https://doi.org/10.3390/kinasesphosphatases3040020






