Site-Specific Phosphorylation of RTK KIT Kinase Insert Domain: Interactome Landscape Perspectives
Abstract
1. Introduction
2. Results
2.1. KIT KID In Silico Phosphorylation
2.1.1. Modelling of the Phosphorylated KIT KID
2.1.2. General Characterisation of KIDs MD Simulations
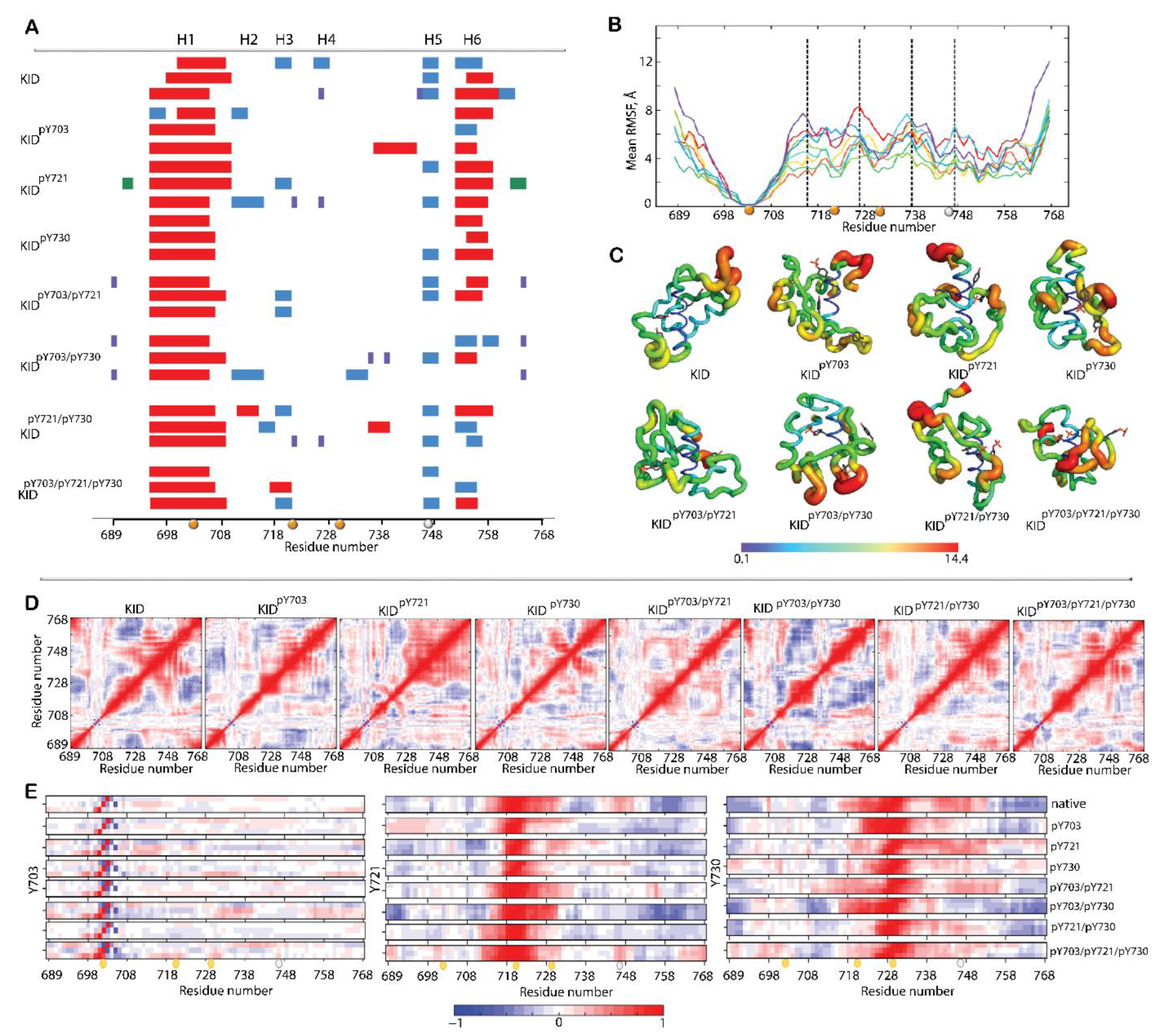
2.1.3. Structural and Dynamical Features of p-KIDs
2.1.4. Shape of p-KIDs and Their Stabilisation by H-Bonds
2.1.5. P-KIDs Solvent Accessibility
2.2. RTK KIT Signalling Protein PI3K
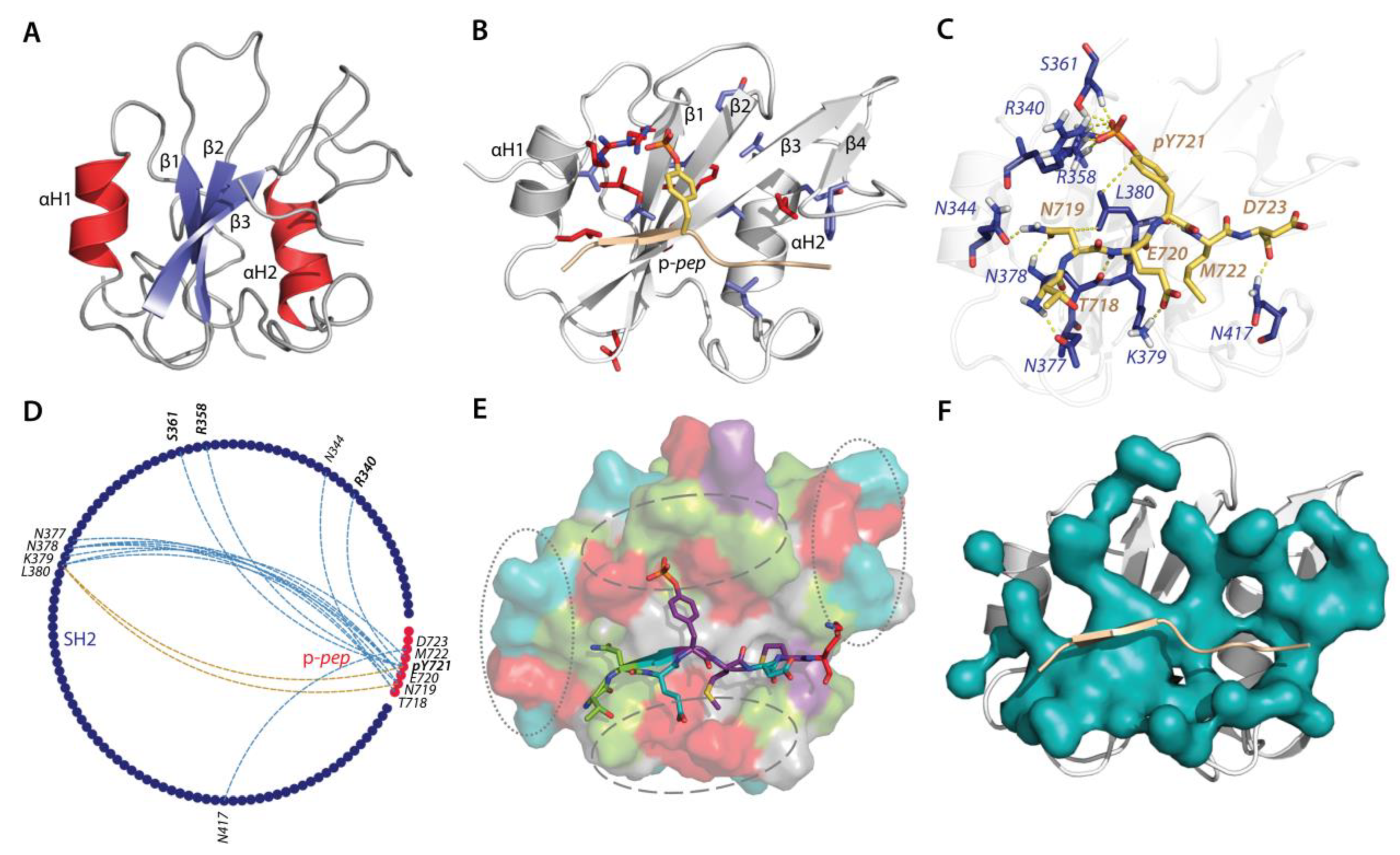
2.2.1. Available Crystallographic Structures Related to KID Binding: Analyses and Hypothesis
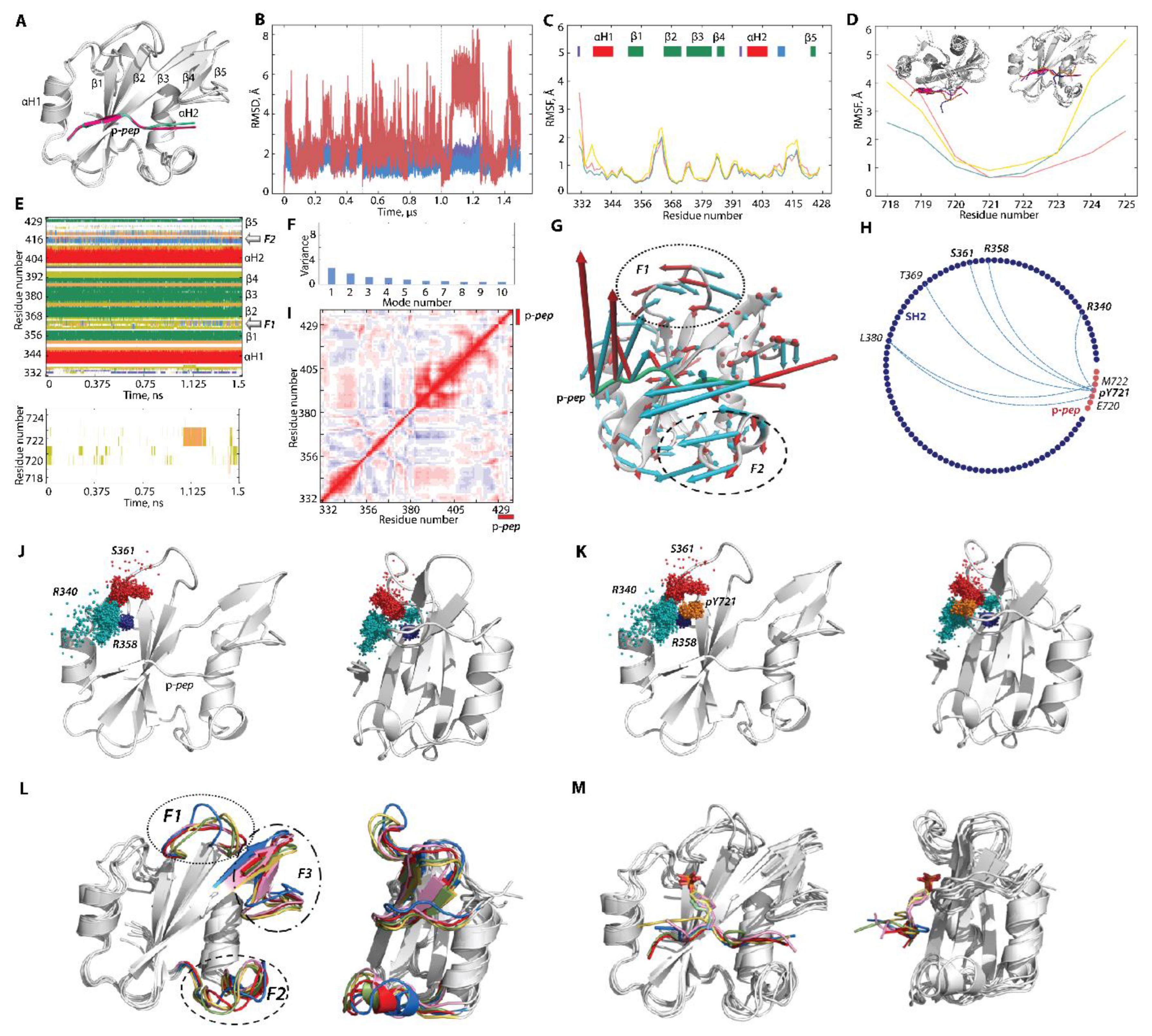
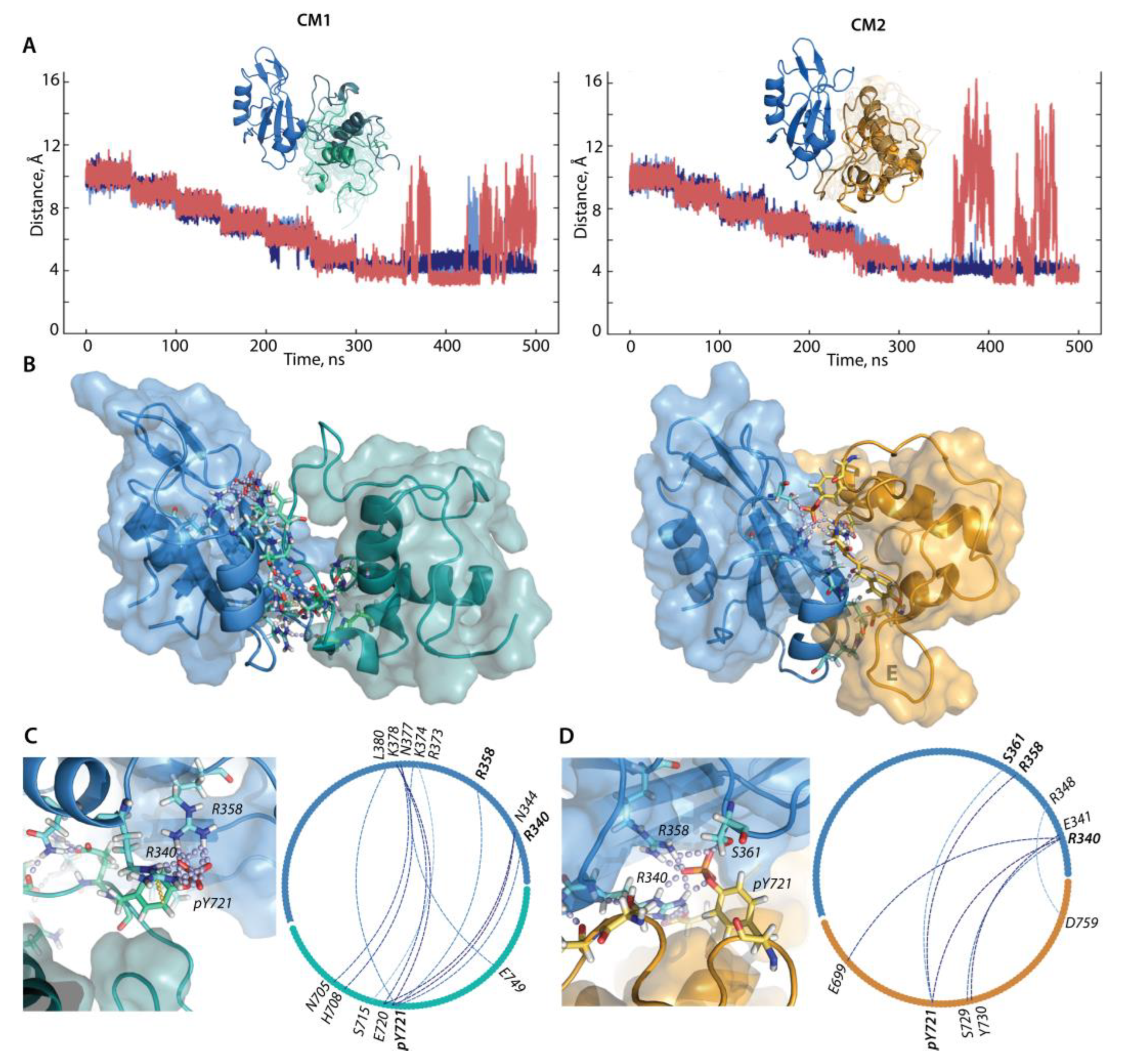
2.2.2. Molecular Dynamics Simulations of p-pep/SH2 Molecular Complex
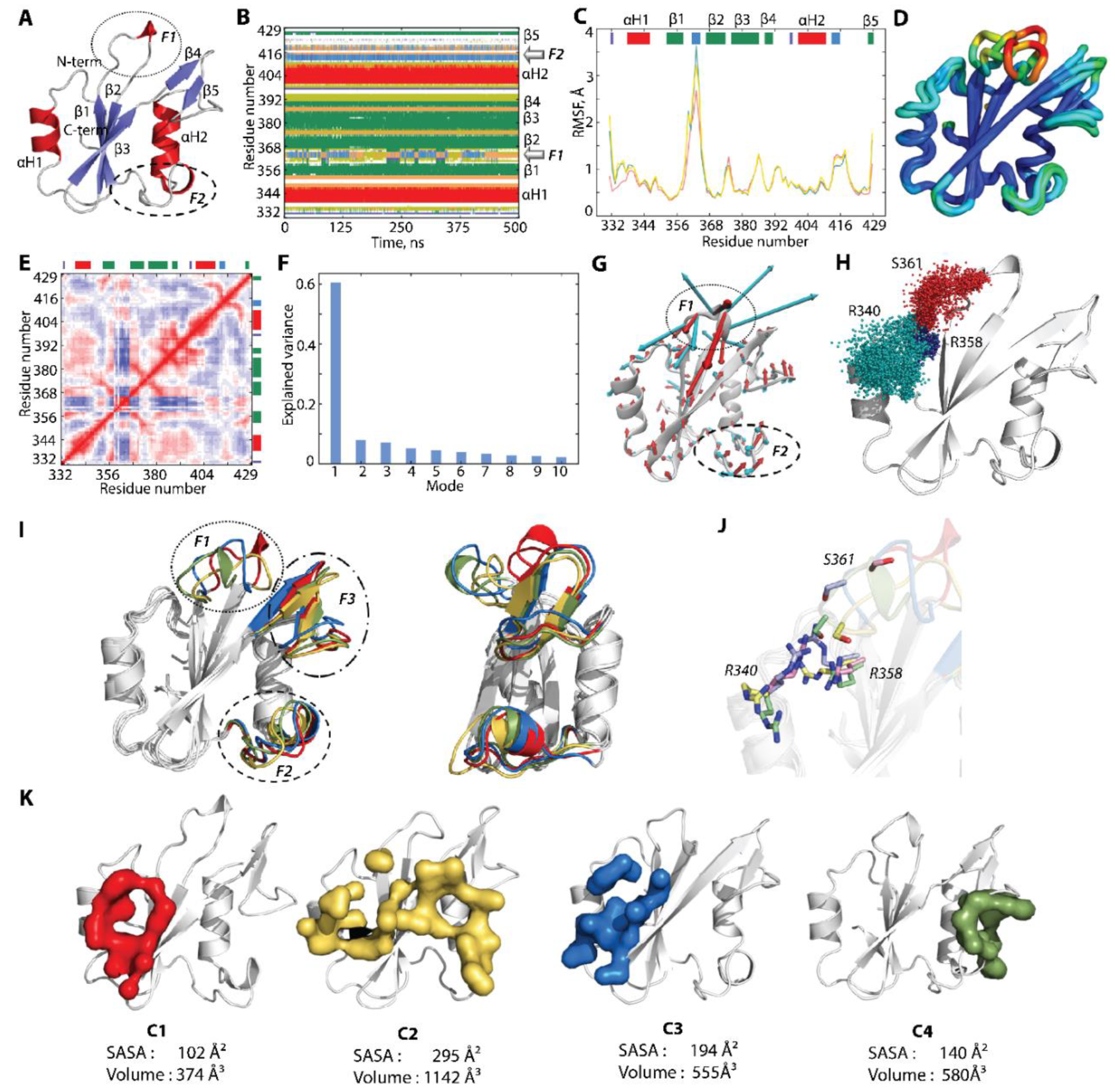
2.2.3. Characterisation of the Free-Ligand SH2 Domain of PI3K
2.3. Protein–Protein Docking: Recognition and Binding of KIT KID and the PI3K SH2 Domain
2.3.1. Can the Crystallographic Structure of the Complex Formed by p-pep of KID and the PI3K SH2 Domain Be Reproduced by Docking?
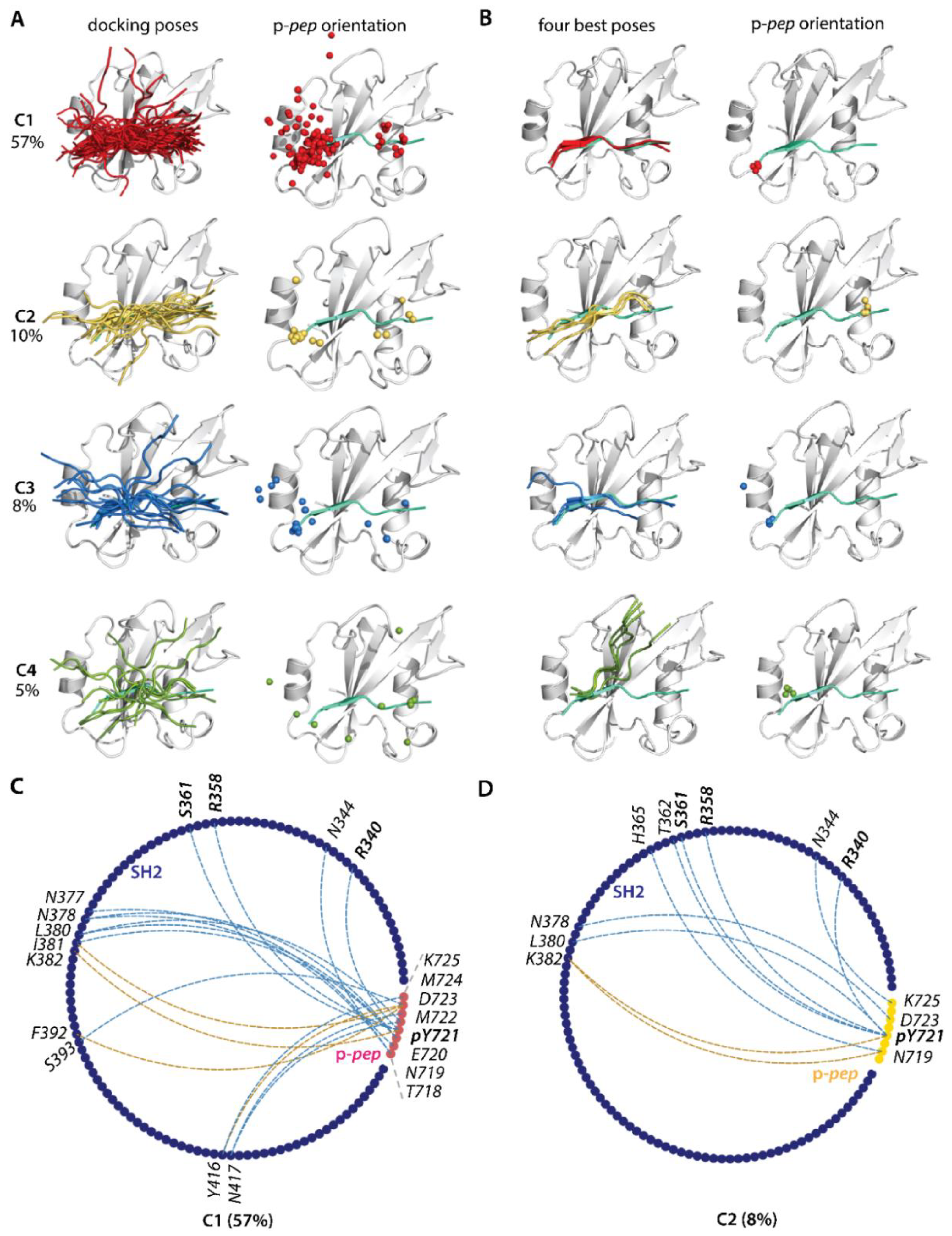
2.3.2. Docking of KID into the PI3K SH2 Domain
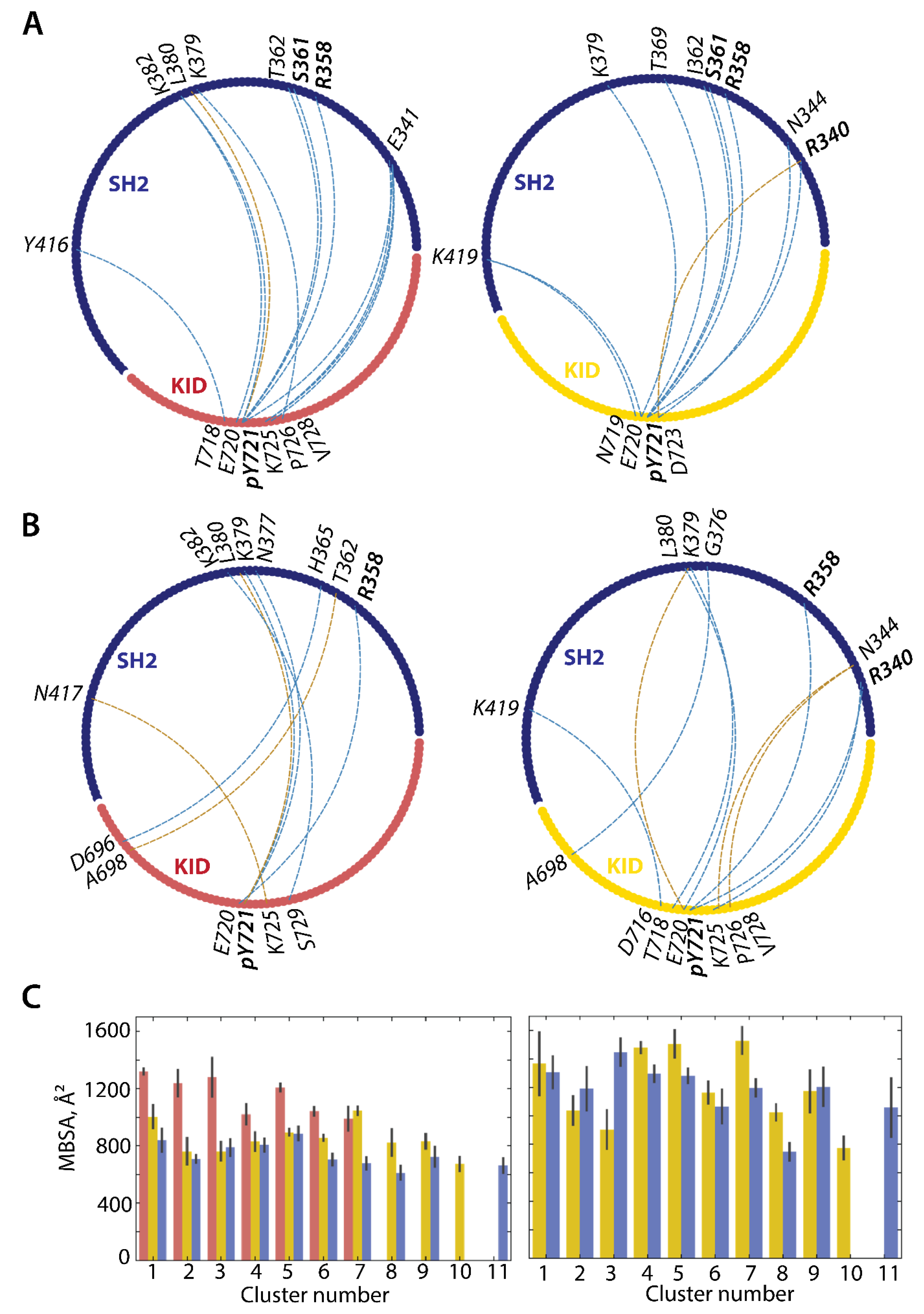
2.3.3. Intuitive User-Guided Modelling of Molecular Complex KIDpY721/SH2
3. Discussion
4. Materials and Methods
4.1. 3D Modelling
4.2. Molecular Dynamic Simulation
4.3. Data Analysis
4.4. Visualisation and Figure Preparation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2000, 103, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J. Receptor tyrosine kinases: Legacy of the first two decades. Cold Spring Harb. Perspect. Biol. 2014, 6, a008912. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradović, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets—The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005, 272, 5129–5148. [Google Scholar] [CrossRef]
- Bondos, S.E.; Dunker, A.K.; Uversky, V.N. Intrinsically disordered proteins play diverse roles in cell signaling. Cell Commun. Signal. CCS 2022, 20, 20. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Ledoux, J.; Tchertanov, L. Receptor Tyrosine Kinase KIT: A New Look for an Old Receptor. In Bioinformatics and Biomedical Engineering, Proceedings of the 9th International Work-Conference, IWBBIO 2022, Maspalomas, Gran Canaria, Spain, 27–30 June 2022; Springer International Publishing: Cham, Switzerland, 2022; pp. 133–137. [Google Scholar]
- Ledoux, J.; Trouvé, A.; Tchertanov, L. The Inherent Coupling of Intrinsically Disordered Regions in the Multidomain Receptor Tyrosine Kinase KIT. Int. J. Mol. Sci. 2022, 23, 1589. [Google Scholar] [CrossRef]
- DiNitto, J.P.; Deshmukh, G.D.; Zhang, Y.; Jacques, S.L.; Coli, R.; Worrall, J.W.; Diehl, W.; English, J.M.; Wu, J.C. Function of activation loop tyrosine phosphorylation in the mechanism of c-Kit auto-activation and its implication in sunitinib resistance. J. Biochem. 2010, 147, 601–609. [Google Scholar] [CrossRef]
- Binns, K.L.; Taylor, P.P.; Sicheri, F.; Pawson, T.; Holland, S.J. Phosphorylation of tyrosine residues in the kinase domain and juxtamembrane region regulates the biological and catalytic activities of eph receptors. Mol. Cell. Biol. 2000, 20, 4791–4805. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.M.; Ilangumaran, S.; La Rose, J.; Chakrabartty, A.; Rottapel, R. Autoinhibition of the kit receptor tyrosine kinase by the cytosolic juxtamembrane region. Mol. Cell. Biol. 2003, 23, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Locascio, L.E.; Donoghue, D.J. KIDs rule: Regulatory phosphorylation of RTKs. Trends Biochem. Sci. 2013, 38, 75–84. [Google Scholar] [CrossRef]
- Amit, I.; Wides, R.; Yarden, Y. Evolvable signaling networks of receptor tyrosine kinases: Relevance of robustness to malignancy and to cancer therapy. Mol. Syst. Biol. 2007, 3, 151. [Google Scholar] [CrossRef]
- Edling, C.E.; Hallberg, B. c-Kit--a hematopoietic cell essential receptor tyrosine kinase. Int. J. Biochem. Cell Biol. 2007, 39, 1995–1998. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, J.D. The Ras-MAPK Signal Transduction Pathway. Sci. Signal. 2010, 3, tr1. [Google Scholar] [CrossRef]
- Shapiro, P. Ras-MAP kinase signaling pathways and control of cell proliferation: Relevance to cancer therapy. Crit. Rev. Clin. Lab. Sci. 2002, 39, 285–330. [Google Scholar] [CrossRef]
- Yee, N.S.; Hsiau, C.W.; Serve, H.; Vosseller, K.; Besmer, P. Mechanism of down-regulation of c-kit receptor. Roles of receptor tyrosine kinase, phosphatidylinositol 3′-kinase, and protein kinase C. J. Biol. Chem. 1994, 269, 31991–31998. [Google Scholar] [CrossRef]
- Inizan, F.; Hanna, M.; Stolyarchuk, M.; Chauvot de Beauchêne, I.; Tchertanov, L. The First 3D Model of the Full-Length KIT Cytoplasmic Domain Reveals a New Look for an Old Receptor. Sci. Rep. 2020, 10, 5401. [Google Scholar] [CrossRef]
- Blume-Jensen, P.; Wernstedt, C.; Heldin, C.H.; Rönnstrand, L. Identification of the major phosphorylation sites for protein kinase C in kit/stem cell factor receptor in vitro and in intact cells. J. Biol. Chem. 1995, 270, 14192–14200. [Google Scholar] [CrossRef]
- Lennartsson, J.; Jelacic, T.; Linnekin, D.; Shivakrupa, R. Normal and oncogenic forms of the receptor tyrosine kinase kit. Stem Cells 2005, 23, 16–43. [Google Scholar] [CrossRef] [PubMed]
- Hleap, J.S.; Blouin, C. The Semantics of the Modular Architecture of Protein Structures. Curr. Protein Pept. Sci. 2016, 17, 62–71. [Google Scholar] [CrossRef] [PubMed]
- del Sol, A.; Araúzo-Bravo, M.J.; Amoros, D.; Nussinov, R. Modular architecture of protein structures and allosteric communications: Potential implications for signaling proteins and regulatory linkages. Genome Biol. 2007, 8, R92. [Google Scholar] [CrossRef] [PubMed]
- Volinsky, N.; Kholodenko, B.N. Complexity of receptor tyrosine kinase signal processing. Cold Spring Harb. Perspect. Biol. 2013, 5, a009043. [Google Scholar] [CrossRef]
- Bah, A.; Forman-Kay, J.D. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J. Biol. Chem. 2016, 291, 6696–6705. [Google Scholar] [CrossRef]
- Collins, M.O.; Yu, L.; Campuzano, I.; Grant, S.G.; Choudhary, J.S. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol. Cell. Proteom. MCP 2008, 7, 1331–1348. [Google Scholar] [CrossRef] [PubMed]
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O’Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Udgaonkar, J.B. Binding-induced folding under unfolding conditions: Switching between induced fit and conformational selection mechanisms. J. Biol. Chem. 2019, 294, 16942–16952. [Google Scholar] [CrossRef]
- Waudby, C.A.; Alvarez-Teijeiro, S.; Josue Ruiz, E.; Suppinger, S.; Pinotsis, N.; Brown, P.R.; Behrens, A.; Christodoulou, J.; Mylona, A. An intrinsic temporal order of c-JUN N-terminal phosphorylation regulates its activity by orchestrating co-factor recruitment. Nat. Commun. 2022, 13, 6133. [Google Scholar] [CrossRef]
- Salazar, C.; Höfer, T. Kinetic models of phosphorylation cycles: A systematic approach using the rapid-equilibrium approximation for protein–protein interactions. Biosystems 2006, 83, 195–206. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Nolte, R.T.; Eck, M.J.; Schlessinger, J.; Shoelson, S.E.; Harrison, S.C. Crystal structure of the PI 3-kinase p85 amino-terminal SH2 domain and its phosphopeptide complexes. Nat. Struct. Biol. 1996, 3, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Ledoux, J.; Trouvé, A.; Tchertanov, L. Folding and Intrinsic Disorder of the Receptor Tyrosine Kinase KIT Insert Domain Seen by Conventional Molecular Dynamics Simulations. Int. J. Mol. Sci. 2021, 22, 7375. [Google Scholar] [CrossRef] [PubMed]
- Lennartsson, J.; Rönnstrand, L. Stem cell factor receptor/c-Kit: From basic science to clinical implications. Physiol. Rev. 2012, 92, 1619–1649. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J.; Lemmon, M.A. SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE Signal Transduct. Knowl. Environ. 2003, 2003, Re12. [Google Scholar] [CrossRef]
- Songyang, Z.; Shoelson, S.E.; Chaudhuri, M.; Gish, G.; Pawson, T.; Haser, W.G.; King, F.; Roberts, T.; Ratnofsky, S.; Lechleider, R.J.; et al. SH2 domains recognize specific phosphopeptide sequences. Cell 1993, 72, 767–778. [Google Scholar] [CrossRef]
- Backer, J.M.; Myers, M.G., Jr.; Shoelson, S.E.; Chin, D.J.; Sun, X.J.; Miralpeix, M.; Hu, P.; Margolis, B.; Skolnik, E.Y.; Schlessinger, J.; et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992, 11, 3469–3479. [Google Scholar] [CrossRef]
- Wagner, M.J.; Stacey, M.M.; Liu, B.A.; Pawson, T. Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb. Perspect. Biol. 2013, 5, a008987. [Google Scholar] [CrossRef]
- Le Guilloux, V.; Schmidtke, P.; Tuffery, P. Fpocket: An open source platform for ligand pocket detection. BMC Bioinform. 2009, 10, 168. [Google Scholar] [CrossRef]
- van Zundert, G.C.P.; Rodrigues, J.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Takemura, K.; Kitao, A. More efficient screening of protein-protein complex model structures for reducing the number of candidates. Biophys. Phys. 2019, 16, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Guharoy, M.; Robert, C.H.; Chakrabarti, P.; Janin, J. Reassessing buried surface areas in protein-protein complexes. Protein Sci. A Publ. Protein Soc. 2013, 22, 1453–1457. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.A. Buried and Accessible Surface Area Control Intrinsic Protein Flexibility. J. Mol. Biol. 2013, 425, 3250–3263. [Google Scholar] [CrossRef] [PubMed]
- Stolyarchuk, M.; Ledoux, J.; Maignant, E.; Trouvé, A.; Tchertanov, L. Identification of the Primary Factors Determining the Specificity of Human VKORC1 Recognition by Thioredoxin-Fold Proteins. Int. J. Mol. Sci. 2021, 22, 802. [Google Scholar] [CrossRef]
- Miao, Y.; Feher, V.A.; McCammon, J.A. Gaussian Accelerated Molecular Dynamics: Unconstrained Enhanced Sampling and Free Energy Calculation. J. Chem. Theory Comput. 2015, 11, 3584–3595. [Google Scholar] [CrossRef]
- Miao, Y.; McCammon, J.A. Gaussian Accelerated Molecular Dynamics: Theory, Implementation, and Applications. Annu. Rep. Comput. Chem. 2017, 13, 231–278. [Google Scholar] [CrossRef]
- Lim, W.A. The modular logic of signaling proteins: Building allosteric switches from simple binding domains. Curr. Opin. Struct. Biol. 2002, 12, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Buck, E.; Iyengar, R. Organization and Functions of Interacting Domains for Signaling by Protein-Protein Interactions. Sci. STKE 2003, 2003, re14. [Google Scholar] [CrossRef]
- Dueber, J.E.; Yeh, B.J.; Bhattacharyya, R.P.; Lim, W.A. Rewiring cell signaling: The logic and plasticity of eukaryotic protein circuitry. Curr. Opin. Struct. Biol. 2004, 14, 690–699. [Google Scholar] [CrossRef]
- Dueber, J.E.; Yeh, B.J.; Chak, K.; Lim, W.A. Reprogramming control of an allosteric signaling switch through modular recombination. Science 2003, 301, 1904–1908. [Google Scholar] [CrossRef]
- Berlow, R.B.; Dyson, H.J.; Wright, P.E. Expanding the Paradigm: Intrinsically Disordered Proteins and Allosteric Regulation. J. Mol. Biol. 2018, 430, 2309–2320. [Google Scholar] [CrossRef]
- Tompa, P.; Davey, N.E.; Gibson, T.J.; Babu, M.M. A million peptide motifs for the molecular biologist. Mol. Cell 2014, 55, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. A Publ. Protein Soc. 2013, 22, 693–724. [Google Scholar] [CrossRef] [PubMed]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Demarest, S.J.; Martinez-Yamout, M.; Chung, J.; Chen, H.; Xu, W.; Dyson, H.J.; Evans, R.M.; Wright, P.E. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 2002, 415, 549–553. [Google Scholar] [CrossRef]
- Waters, L.; Yue, B.; Veverka, V.; Renshaw, P.; Bramham, J.; Matsuda, S.; Frenkiel, T.; Kelly, G.; Muskett, F.; Carr, M.; et al. Structural Diversity in p160/CREB-binding Protein Coactivator Complexes. J. Biol. Chem. 2006, 281, 14787–14795. [Google Scholar] [CrossRef]
- Hilser, V.J.; Thompson, E.B. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 8311–8315. [Google Scholar] [CrossRef]
- Motlagh, H.N.; Wrabl, J.O.; Li, J.; Hilser, V.J. The ensemble nature of allostery. Nature 2014, 508, 331–339. [Google Scholar] [CrossRef]
- Dunker, A.K.; Babu, M.M.; Barbar, E.; Blackledge, M.; Bondos, S.E.; Dosztányi, Z.; Dyson, H.J.; Forman-Kay, J.; Fuxreiter, M.; Gsponer, J.; et al. What’s in a name? Why these proteins are intrinsically disordered: Why these proteins are intrinsically disordered. Intrinsically Disord. Proteins 2013, 1, e24157. [Google Scholar] [CrossRef]
- Panda, A.; Tuller, T. Exploring Potential Signals of Selection for Disordered Residues in Prokaryotic and Eukaryotic Proteins. Genom. Proteom. Bioinform. 2020, 18, 549–564. [Google Scholar] [CrossRef]
- Wandless, T.J. SH2 domains: A question of independence. Curr. Biol. CB 1996, 6, 125–127. [Google Scholar] [CrossRef]
- Mittag, T.; Orlicky, S.; Choy, W.-Y.; Tang, X.; Lin, H.; Sicheri, F.; Kay, L.E.; Tyers, M.; Forman-Kay, J.D. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. USA 2008, 105, 17772–17777. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.H.; Weis, W.I. The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin. Cell 2001, 105, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Lev, S.; Givol, D.; Yarden, Y. Interkinase domain of kit contains the binding site for phosphatidylinositol 3′ kinase. Proc. Natl. Acad. Sci. USA 1992, 89, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Vajravelu, B.N.; Hong, K.U.; Al-Maqtari, T.; Cao, P.; Keith, M.C.L.; Wysoczynski, M.; Zhao, J.; Moore Iv, J.B.; Bolli, R. C-Kit Promotes Growth and Migration of Human Cardiac Progenitor Cells via the PI3K-AKT and MEK-ERK Pathways. PLoS ONE 2015, 10, e0140798. [Google Scholar] [CrossRef]
- Marcus, Y.; Hefter, G. Ion Pairing. Chem. Rev. 2006, 106, 4585–4621. [Google Scholar] [CrossRef]
- Stanfield, R.L.; Wilson, I.A. Protein-peptide interactions. Curr. Opin. Struct. Biol. 1995, 5, 103–113. [Google Scholar] [CrossRef]
- Keskin, O.; Gursoy, A.; Ma, B.; Nussinov, R. Principles of protein-protein interactions: What are the preferred ways for proteins to interact? Chem. Rev. 2008, 108, 1225–1244. [Google Scholar] [CrossRef]
- Keskin, O.; Haliloglu, T.; Ma, B.Y.; Nussinov, R. Protein-protein interactions: Structurally conserved residues at protein-protein interfaces. Biophys. J. 2004, 86, 267A. [Google Scholar]
- Keskin, O.; Ma, B.; Nussinov, R. Hot regions in protein–protein interactions: The organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005, 345, 1281–1294. [Google Scholar] [CrossRef]
- Gianni, S.; Dogan, J.; Jemth, P. Coupled binding and folding of intrinsically disordered proteins: What can we learn from kinetics? Curr. Opin. Struct. Biol. 2016, 36, 18–24. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. Unraveling hot spots in binding interfaces: Progress and challenges. Curr. Opin. Struct. Biol. 2002, 12, 14–20. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The case for open-source software in drug discovery. Drug Discov. Today 2005, 10, 213–217. [Google Scholar] [CrossRef]
- Mol, C.D.; Lim, K.B.; Sridhar, V.; Zou, H.; Chien, E.Y.; Sang, B.C.; Nowakowski, J.; Kassel, D.B.; Cronin, C.N.; McRee, D.E. Structure of a c-kit product complex reveals the basis for kinase transactivation. J. Biol. Chem. 2003, 278, 31461–31464. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinform. 2016, 54, 5.6.1–5.6.37. [Google Scholar] [CrossRef]
- Shen, M.Y.; Sali, A. Statistical potential for assessment and prediction of protein structures. Protein Sci. A Publ. Protein Soc. 2006, 15, 2507–2524. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Case, D.A.; Cheatham, T.E., 3rd; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M., Jr.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef]
- Meagher, K.L.; Redman, L.T.; Carlson, H.A. Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 2003, 24, 1016–1025. [Google Scholar] [CrossRef]
- Berendsen, H.J.C.; Postma, J.P.M.; Gunsteren, W.F.v.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Duane, S.; Kennedy, A.D.; Pendleton, B.J.; Roweth, D. Hybrid Monte Carlo. Phys. Lett. B 1987, 195, 216–222. [Google Scholar] [CrossRef]
- Andersen, H.C. Rattle: A “velocity” version of the shake algorithm for molecular dynamics calculations. J. Comput. Phys. 1983, 52, 24–34. [Google Scholar] [CrossRef]
- Roe, D.R.; Cheatham, T.E. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure—Pattern-Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Lyman, E.; Zuckerman, D.M. Ensemble-based convergence analysis of biomolecular trajectories. Biophys. J. 2006, 91, 164–172. [Google Scholar] [CrossRef]
- Bahar, I.; Lezon, T.R.; Bakan, A.; Shrivastava, I.H. Normal Mode Analysis of Biomolecular Structures: Functional Mechanisms of Membrane Proteins. Chem. Rev. 2010, 110, 1463–1497. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein–Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Bakan, A.; Dutta, A.; Mao, W.; Liu, Y.; Chennubhotla, C.; Lezon, T.R.; Bahar, I. Evol and ProDy for bridging protein sequence evolution and structural dynamics. Bioinformatics 2014, 30, 2681–2683. [Google Scholar] [CrossRef] [PubMed]
- Bakan, A.; Meireles, L.M.; Bahar, I. ProDy: Protein dynamics inferred from theory and experiments. Bioinformatics 2011, 27, 1575–1577. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ledoux, J.; Tchertanov, L. Site-Specific Phosphorylation of RTK KIT Kinase Insert Domain: Interactome Landscape Perspectives. Kinases Phosphatases 2023, 1, 39-71. https://doi.org/10.3390/kinasesphosphatases1010005
Ledoux J, Tchertanov L. Site-Specific Phosphorylation of RTK KIT Kinase Insert Domain: Interactome Landscape Perspectives. Kinases and Phosphatases. 2023; 1(1):39-71. https://doi.org/10.3390/kinasesphosphatases1010005
Chicago/Turabian StyleLedoux, Julie, and Luba Tchertanov. 2023. "Site-Specific Phosphorylation of RTK KIT Kinase Insert Domain: Interactome Landscape Perspectives" Kinases and Phosphatases 1, no. 1: 39-71. https://doi.org/10.3390/kinasesphosphatases1010005
APA StyleLedoux, J., & Tchertanov, L. (2023). Site-Specific Phosphorylation of RTK KIT Kinase Insert Domain: Interactome Landscape Perspectives. Kinases and Phosphatases, 1(1), 39-71. https://doi.org/10.3390/kinasesphosphatases1010005





