Unit-Emitting Carbon Dots for Cell Imaging and Lipid Droplet Quantification
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Apparatus and Measurements
2.3. C-Dot Fabrication and Separation
2.4. Cell Culture
2.5. Cytotoxicity Assay
2.6. Subcellular Localization
2.7. Cell Stimulation
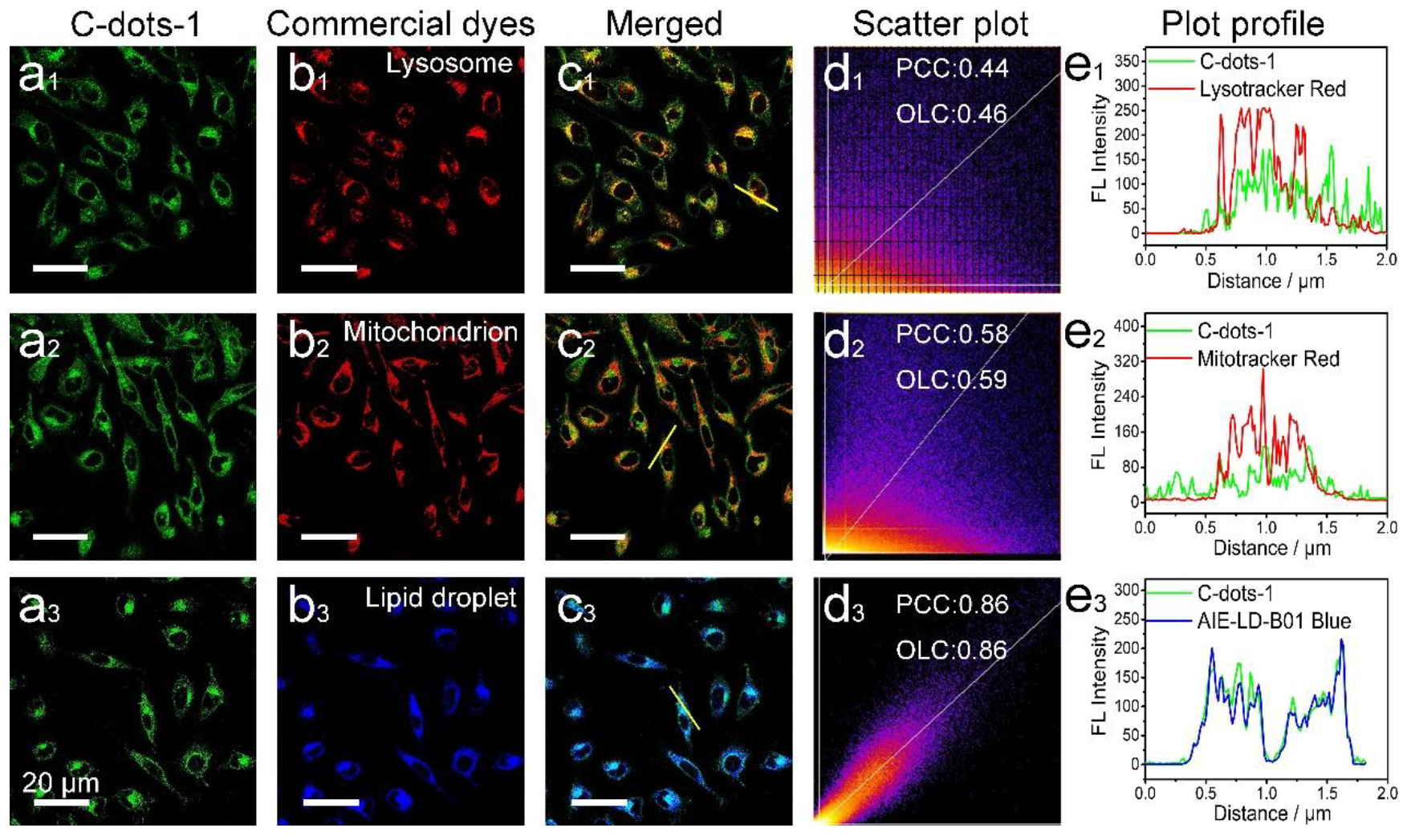
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ðorđević, L.; Arcudi, F.; Cacioppo, M.; Prato, M. A multifunctional chemical toolbox to engineer carbon dots for biomedical and energy applications. Nat. Nanotechnol. 2022, 17, 112–130. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Wang, B.; Qu, X.; Li, S.; Huang, J.; Li, J.; Lu, S.; Zhou, N. State-of-the-art of biomass-derived carbon dots: Preparation, properties, and applications. Chin. Chem. Lett. 2024, 25, 108423. [Google Scholar] [CrossRef]
- Wang, Y.; Xia, Y. Optical, electrochemical and catalytic methods for in-vitro diagnosis using carbonaceous nanoparticles: A review. Microchim. Acta 2019, 186, 50. [Google Scholar] [CrossRef] [PubMed]
- Khan, W.U.; Wang, D.; Wang, Y. Highly green emissive nitrogen-doped carbon dots with excellent thermal stability for bioimaging and solid-state LED. Inorg. Chem. 2018, 57, 15229–15239. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Dash, J.K.; Kumar, P.; Patel, R. Carbon-based quantum dots for photovoltaic devices: A review. ACS Appl. Electron. Mater. 2022, 4, 27–58. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.; Guo, G.; Xia, Y. Liver injury traceability: Spatiotemporally monitoring oxidative stress processes by unit-emitting carbon dots. Anal. Chem. 2023, 95, 2765–2773. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Fan, J.; Mu, H.; Chen, L.; Yang, Y.; Yu, S. Nucleus-targeting orange-emissive carbon dots delivery adriamycin for enhanced anti-liver cancer therapy. Chin. Chem. Lett. 2024, 35, 108947. [Google Scholar] [CrossRef]
- Cheng, B.; Cao, L.; Li, C.; Huo, F.Y.; Meng, Q.F.; Tong, G.; Wu, X.; Bu, L.L.; Rao, L.; Wang, S. Fluorine-doped carbon quantum dots with deep-red emission for hypochlorite determination and cancer cell imaging. Chin. Chem. Lett. 2024, 35, 108969. [Google Scholar] [CrossRef]
- Xin, N.; Gao, D.; Su, B.; Zhou, T.; Zhu, Y.; Wu, C.; Wei, D.; Sun, J.; Fan, H. Orange-emissive carbon dots with high photostability for mitochondrial dynamics tracking in living cells. ACS Sens. 2023, 8, 1161–1172. [Google Scholar] [CrossRef]
- Li, R.S.; Gao, P.F.; Zhang, H.Z.; Zheng, L.L.; Li, C.M.; Wang, J.; Li, Y.F.; Liu, F.; Li, N.; Huang, C.Z. Chiral nanoprobes for targeting and long-term imaging of the Golgi apparatus. Chem. Sci. 2017, 8, 6829–6835. [Google Scholar] [CrossRef]
- Wu, L.; Li, X.; Ling, Y.; Huang, C.; Jia, N. Morpholine derivative-functionalized carbon dots-based fluorescent probe for highly selective lysosomal imaging in living cells. ACS Appl. Mater. Interfaces 2017, 9, 28222–28232. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Zhang, Y.; Wu, K.; Min, H.; Wei, D.; Sun, J.; Yang, H.; Fan, H. One-step synthesis of ultrabright amphiphilic carbon dots for rapid and precise tracking lipid droplets dynamics in biosystems. Biosens. Bioelectron. 2022, 200, 113928. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Bhattacharya, A.; Hazra, N.; Gayen, K.; Sen, P.; Banerjee, A. Yellow-emitting carbon dots for selective fluorescence imaging of lipid droplets in living cells. Langmuir 2022, 38, 8829–8836. [Google Scholar] [CrossRef] [PubMed]
- She, C.; Wang, Z.; Zeng, J.; Wu, F. G Orange/red dual-emissive boron- and nitrogen-codoped carbon dots for wash-free and selective staining of lipid droplets in live cells. Carbon 2022, 191, 636–645. [Google Scholar] [CrossRef]
- Ohsaki, Y.; Suzuki, M.; Fujimoto, T. Open questions in lipid droplet biology. Chem. Biol. 2014, 21, 86e96. [Google Scholar] [CrossRef] [PubMed]
- Thiam, A.R.; Dugail, I. Lipid droplet-membrane contact sites-from protein binding to function. J. Cell Sci. 2019, 132, jcs230169. [Google Scholar] [CrossRef]
- Welte, M.A.; Gould, A.P. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 2017, 1862, 1260–1272. [Google Scholar] [CrossRef]
- Liang, T.; Wen, D.; Chen, G.; Chan, A.; Chen, Z.; Li, H.; Wang, Z.; Han, X.; Jiang, L.; Zhu, J.J.; et al. Adipocyte-derived anticancer lipid droplets. Adv. Mater. 2021, 33, 2100629. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Ma, C.; Yang, W.; Yin, W.; Wei, J.; Li, N. Facile construction of boranyl complexes with aggregation-induced emission characteristics and their specific lipid droplet imaging applications. Chem. Commun. 2019, 55, 8494–8497. [Google Scholar] [CrossRef]
- Xu, S.; Zhang, X.; Liu, P. Lipid Droplet Proteins and Metabolic Diseases. Biochim. Biophys. Acta, Mol. Basis Dis. 2018, 1864, 1968–1983. [Google Scholar] [CrossRef]
- Dalhaimer, P. Lipid droplets in disease. Cell 2019, 8, 974. [Google Scholar] [CrossRef] [PubMed]
- Sayed, S.M.; Li, X.F.; Jia, H.R.; Durrani, S.F.; Wu, G.; Lu, X. A dibenzothiophene core-based small-molecule AIE probe for wash-free and selective staining of lipid droplets in live mammalian and fungal cells. Sensor. Actuator. B Chem. 2021, 343, 130128. [Google Scholar] [CrossRef]
- Chen, J.; Liu, W.; Fang, X.; Qiao, Q.; Xu, Z. BODIPY 493 acts as a bright buffering fluorogenic probe for super-resolution imaging of lipid droplet dynamics. Chin. Chem. Lett. 2022, 33, 5042–5046. [Google Scholar] [CrossRef]
- Hornum, M.; Mulberg, M.W.; Szomek, M.; Reinholdt, P.; Brewer, J.R.; Wüstner, D.; Kongsted, J.; Nielsen, P. Substituted 9-diethylaminobenzo[a]phenoxazin-5-ones (Nile red analogues): Synthesis and photophysical properties. J. Org. Chem. 2021, 86, 1471–1488. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xing, H.; Park, H.; Zhang, R.; Peng, C.; Sung, H.H.Y.; Williams, I.D.; Ma, C.; Wong, K.S.; Li, S.; et al. Deep-red aggregation-induced emission luminogen based on dithiofuvalene-fused benzothiadiazole for lipid droplet-specific imaging. ACS Mater. Lett. 2022, 4, 159–164. [Google Scholar] [CrossRef]
- Wang, C.; Guo, G.; Wang, Y.; Yin, X.; Zong, H.; Xia, Y. Twins of minimalistic carbon dots: Uniform emitting-units and molecular level repeatable photoluminescence. Adv. Opt. Mater. 2022, 10, 2102827. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, J.; Sun, Y.Q.; Liu, M.; Guo, W. Carbon−dipyrromethenes: Bright cationic fluorescent dyes and potential application in revealing cellular trafficking of mitochondrial glutathione conjugates. J. Am. Chem. Soc. 2020, 142, 17069–17078. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.Y.; Raines, R.T. Mechanism of ribonuclease an endocytosis: Analogies to cell-penetrating peptides. Biochemistry 2011, 50, 8374–8382. [Google Scholar] [CrossRef] [PubMed]
- Ujimoto, Y.F.; Noduka, J.O.; Omma, K.J.H.; Amaguchi, S.Y.; Ori, M.M.; Igashi, Y.H.; Akita, M.M.; Inoshita, T.K.; Oda, J.N.; Tabe, H.I.; et al. Long-chain fatty acids induce lipid droplet formation in a cultured human hepatocyte in a manner dependent of acyl-CoA synthetase. Biol. Pharm. Bull. 2006, 29, 2174–2180. [Google Scholar] [CrossRef]
- Wang, K.; Ma, S.; Ma, Y.; Zhao, Y.; Xing, M.; Zhou, L.; Cao, D.; Lin, W. Aurone derivative revealing the metabolism of lipid droplets and monitoring oxidative stress in living cells. Anal. Chem. 2020, 92, 6631–6636. [Google Scholar] [CrossRef]
- Nguyen, T.B.; Olzmann, J.A. Lipid droplets and lipotoxicity during autophagy. Autophagy 2017, 13, 2002–2003. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, A.D.; Siniossoglou, S. Spatial distribution of lipid droplets during starvation: Implications for lipophagy. Commun. Integr. Bio. 2016, 9, e1183854. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Xia, Y. Unit-Emitting Carbon Dots for Cell Imaging and Lipid Droplet Quantification. Targets 2024, 2, 126-136. https://doi.org/10.3390/targets2020008
Xu Y, Xia Y. Unit-Emitting Carbon Dots for Cell Imaging and Lipid Droplet Quantification. Targets. 2024; 2(2):126-136. https://doi.org/10.3390/targets2020008
Chicago/Turabian StyleXu, Yanli, and Yunsheng Xia. 2024. "Unit-Emitting Carbon Dots for Cell Imaging and Lipid Droplet Quantification" Targets 2, no. 2: 126-136. https://doi.org/10.3390/targets2020008
APA StyleXu, Y., & Xia, Y. (2024). Unit-Emitting Carbon Dots for Cell Imaging and Lipid Droplet Quantification. Targets, 2(2), 126-136. https://doi.org/10.3390/targets2020008




