Abstract
Fractionation of the stem bark of Enantia chlorantha Oliv yields three alkaloids, palmatine (1), jatrorrhizine (2), columbamine (3), and β-Sitosterol (4). In this investigation, density functional theory (DFT) calculations were carried out to evaluate the electronic structure and properties of 1–4 by DFT-B3LYP/6-31G level of theory using Gaussian 09 software. The highest occupied molecular orbital (HOMO), lowest unoccupied molecular orbital (LUMO), HOMO-LUMO energy difference (band gap), hardness (η), softness (S), dipole moment (μ), electronegativity (χ), hydrophobicity (logP), topological surface area (TPSA), and energy gap (Eg) were calculated. The in vitro cytotoxicity of the compounds was investigated against MCF-7 and HCT116 cancer cell lines using Wi-38 cells as a control. The compounds inhibited the proliferation of the MCF-7 and HCT116 cell lines and induced apoptosis via upregulation of caspase-3, Bax, PARP cleavage, and downregulation of Bcl-2. DFT analyses revealed that compounds 1 and 3 have smaller energy gaps, 0.072 and 0.071eV, respectively, with the highest dipole moments; hence, these compounds are more chemically reactive and exhibit better modulation of caspase-3 enzyme and inhibitory activities of the MCF-3 and HCT116 cell lines. The antimicrobial and antiparasitic evaluation of 1–4 showed moderate efficacy against the bacterial strains and moderate antiparasitic activity against Cichlidogyrus tilapia.
1. Introduction
Cancer has been identified as the major cause of mortality and morbidity worldwide. The American Cancer Society anticipated that there would be 1.9 million new instances of cancer in 2022, resulting in 609,360 deaths in the USA [1]. Most diagnoses are prostate, lung, colorectal, breast, and liver cancer. The World Health Organization (WHO) reported that in 2020, there were 2.3 million women diagnosed with breast cancer and 685,000 deaths globally [2]. Another report also shows 106,180 colon and 108,480 skin cancers, resulting in 11,990 deaths [1]. Surgery, radiation, and chemotherapy are major treatment options for cancers. However, the toxicity problems of radiation and chemotherapy restrict their use and effectiveness due to the associated adverse side effects and reduced quality of life [3]. The development of resistance of certain cancers to chemotherapy, high cost, and limited spectrum of activity of currently used anticancer agents are reasons for the urgent search for new and efficient anticancer agents. Natural products have increasingly become a major source for discovering bioactive and new chemical entities to treat various disease conditions and have also provided starting materials for synthesizing potent and novel pharmacological compounds [4,5,6,7].
Natural products (NPs) are responsible for nearly half of all pharmaceuticals in clinical use. Fifty percent of the small and novel medicinal compounds developed between 1981 and 2006 were of NP origin or their derivatives [8]. And half of the twenty authorized small molecules (novel chemical substances) in 2010 were derived from natural products. The presence of unique and chemically diverse metabolites in plants and microbes enhances the utilization of these products in managing several disease conditions such as cancer, malaria, diabetes, arteriosclerosis, inflammatory diseases, etc. Plants’ secondary metabolites, such as saponins [9], alkaloids [10,11], flavonoids [12,13], diterpenes [14], and sesquiterpenes lactones [15,16], have been implicated as anticancer agents.
In our search for potent phytochemical agents with biological activities against different diseases, we investigated compounds 1–4 (three alkaloids and a sterol; Figure 1) previously isolated by one of the authors [10] from the stem bark of Enantia chlorantha Oliv (Annonaceae), a plant used in Nigerian folk medicine for treating various diseases. This current study evaluated the in vitro anticancer activity of 1–4 against breast cancer (MCF-7) and colon cancer (HCT116), cellular tracing, and molecular docking studies against caspase-3 enzyme targets to predict their possible mechanism of action. Also, the antimicrobial and antiparasitic activities of 1–4 against some pathogenic organisms were evaluated, as well as the electronic properties of the compounds through DFT calculations.
Figure 1.
Chemical structures of compounds 1–4.
2. Results
2.1. Inhibition of Cell Proliferation and Viability of MCF-7 and HCT-116 Cells
All the tested compounds suppressed the growth of MCF-7 and HCT-116 cancer cells in a dose-dependent manner (Figure 2), with a significant drop in cell viability and an increase in cytotoxicity (Figure 3, Figure 4, Figure 5 and Figure 6). The selectivity indexes of the test compounds and the positive control doxorubicin are compared in Figure 7.
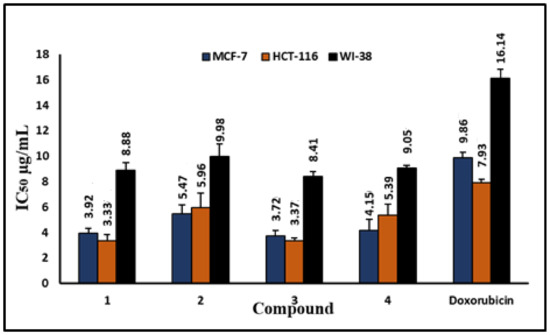
Figure 2.
The IC50 values of compounds 1–4 and doxorubicin against MCF-7, HCT-116, and WI-38 (control) cell lines. The IC50 values are bar graphs, with error bars showing the mean ± SEM.
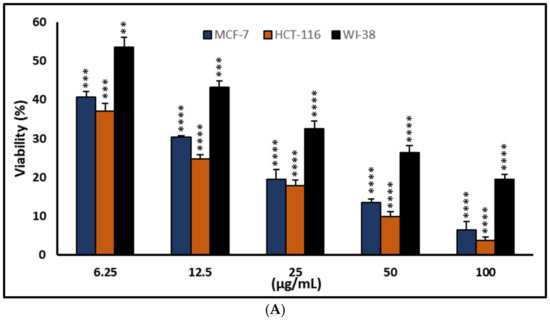
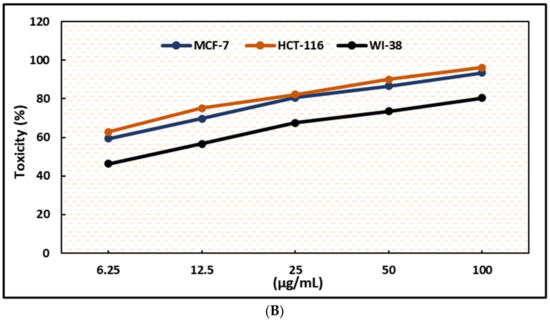
Figure 3.
Effects of compound (1) on MCF-7, HCT-116, and WI-38 cell viability (A) and toxicity (B) after 48 h. MTT was used to assess the proportion of viable cells and their toxicity after cells were incubated at the specified concentrations. Means of triplicate determination, with 7–8 wells per concentration, were used for the plot. The statistical differences between experimental and control cultures are shown as bar graphs with error bars (A) reflecting the means ± SEM; ** p < 0.01, *** p < 0.001, and **** p < 0.0001 vs. +ve doxorubicin-treated cells.
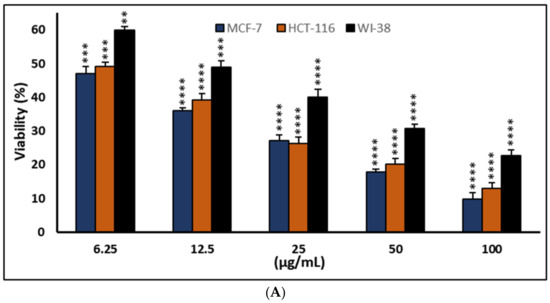
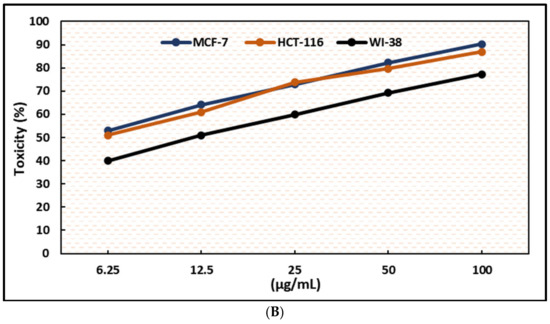
Figure 4.
Effects of compound (2) on MCF-7, HCT-116, and WI-38 cell viability (A) and toxicity (B) post 48 h. MTT was used to assess the proportion of viable cells and toxicity after cells were incubated at the specified concentrations. The values used for plotting are the means of triplicates, with 7–8 wells per concentration examined. Statistical differences between experimental and control cultures are shown as bar graphs (A) with error bars and represented as the means ± SEM; ** p < 0.01, *** p < 0.001, and **** p< 0.0001 vs. +ve doxorubicin-treated cells.
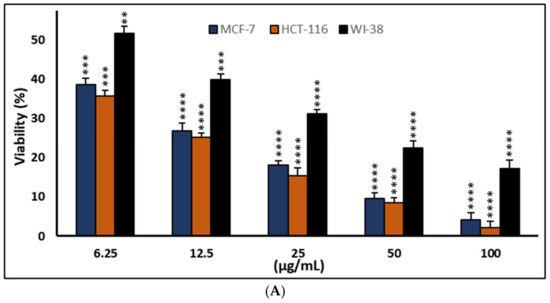
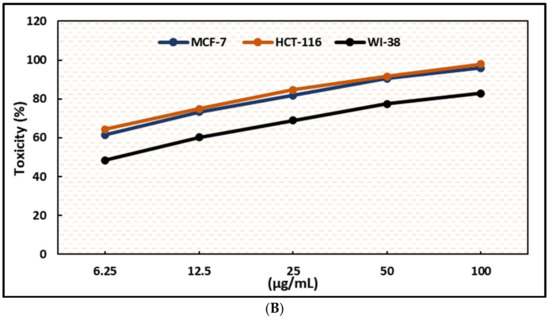
Figure 5.
Effects of compound (3) on MCF-7, HCT-116, and WI-38 cell viability (A) and toxicity (B) after 48 h. MTT was used to assess the proportion of viable cells and their toxicity after cells were incubated at the specified concentrations. The values used for plotting are the means of triplicates, with 7–8 wells per concentration examined. Statistical differences between experimental and control cultures are shown as bar graphs with error bars and mean ± SEM; ** p < 0.01, *** p < 0.001, and **** p < 0.0001 vs. +ve doxorubicin-treated cells.
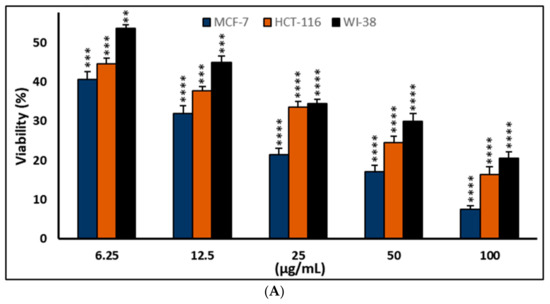
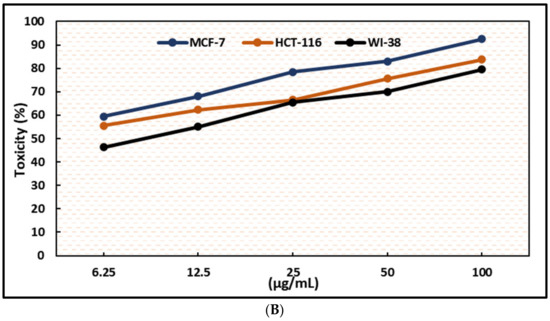
Figure 6.
Effects of compound (4) on MCF-7, HCT-116, and WI-38 cell viability (A) and toxicity (B) post 48 h. MTT was used to assess the proportion of viable cells and their toxicity after cells were incubated at the specified concentrations. The values used for plotting are the means of triplicates, with 7–8 wells examined for each concentration. Statistical differences between experimental and control cultures are shown as bar graphs with error bars denoting mean ± SEM; ** p < 0.01, *** p < 0.001, and **** p < 0.0001 vs. +ve doxorubicin-treated cells.
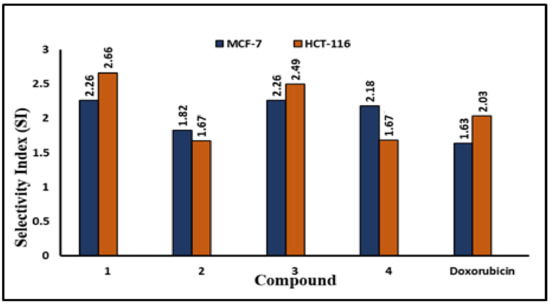
Figure 7.
Comparative SI for the examined compounds 1–4 and doxorubicin on MCF-7 and HCT-116 cancer cells. The SI values are shown as bar graphs.
2.2. Induction of Apoptosis via Bax, Caspase-3, and Cleaved PARP Expression Increase, and Bcl-2 Expression Decrease in Cancer Cells
The induction of apoptosis by compounds 1–4 in MCF-7 and HCT-116 cancer cell lines was carried out by upregulation of pro-apoptotic proteins Bax, caspase-3, and cleaved PARP and downregulation of anti-apoptotic protein Bcl-2 (Figure 8 and Figure 9). The possible schematic of the mechanism of cancer cells apoptotic induction by the tested compounds is shown in Figure 10, which suggest up-regulation of caspase-3 and Bax, with cleavage of PARP and downregulation of Bcl-2, all known to be inducers of apoptosis.
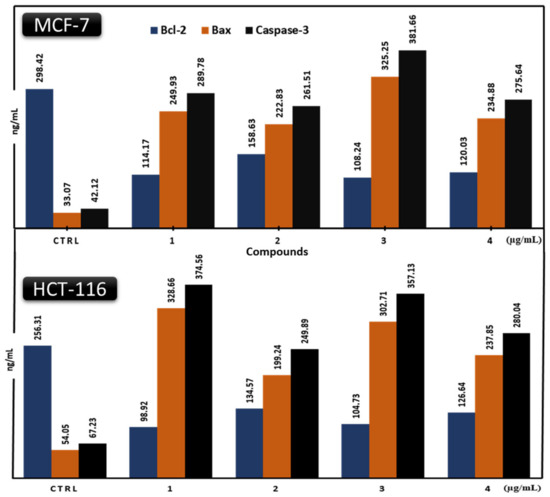
Figure 8.
Comparative modulation of Bcl-2, Bax, and caspase-3 protein expressions in MCF-7 and HCT-116 cancer cells treated with compounds 1–4 (µg/mL) and harvested 48 h after treatments. Bcl-2 values were multiplied by 50 to be shared within Bax and caspase-3 bars on the same chart.
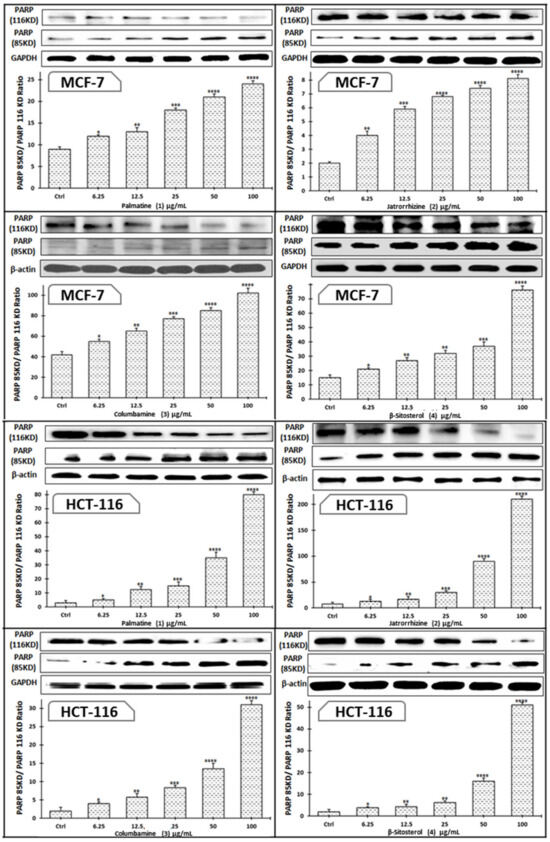
Figure 9.
Effects of compounds 1–4 on the protein expression of cleaved PARP in MCF-7 and HCT-116 cells treated with each drug dose and collected 48 h later. Bars represent the means ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001 vs. +ve doxorubicin-treated cells.
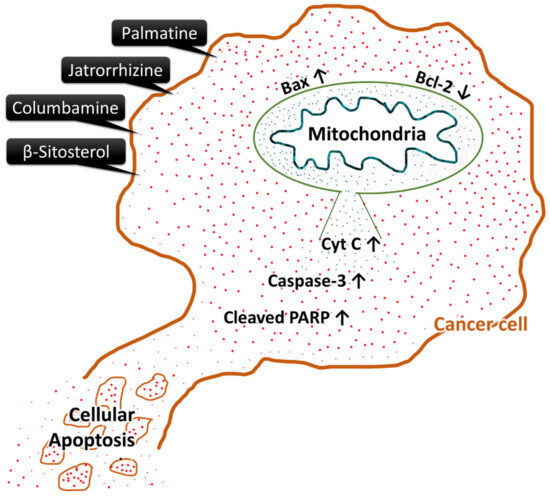
Figure 10.
Schematic of the studied compounds’ possible mechanism of cancer cell apoptosis. This carton is based on the currently available data throughout the present study.
2.3. Molecular Docking of Compounds (1–4)
The current investigation explored the in silico molecular pathways by which compounds 1–4 (Figure 1) triggered cell death in human breast cancer MCF-7 and HCT116 cell lines, concentrating on their apoptotic induction through activation of caspase-3. Molecular docking studies were performed to predict possible binding poses of the compounds in the active site of caspase-3 (Figure 11, Figure 12, Figure 13, Figure 14 and Figure 15).
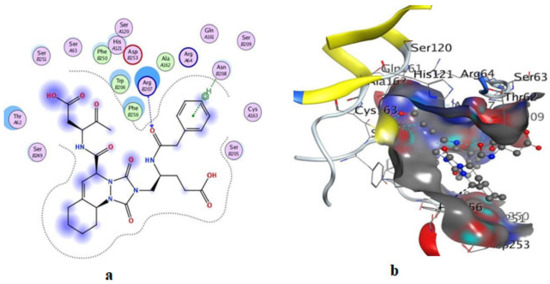
Figure 11.
Illustrations of the interaction of B92 inside the 3KJF active site in 2D (a) and 3D (b).
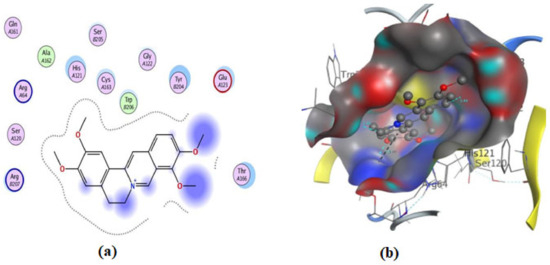
Figure 12.
Illustrations of the interaction of compound 1 inside the 3KJF active site in 2D (a) and 3D (b).
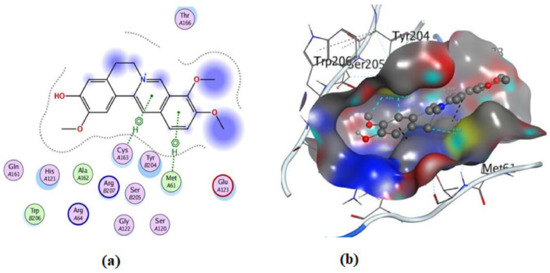
Figure 13.
Illustrations of the interaction of compound 2 inside the 3KJF active site in 2D (a) and 3D (b).
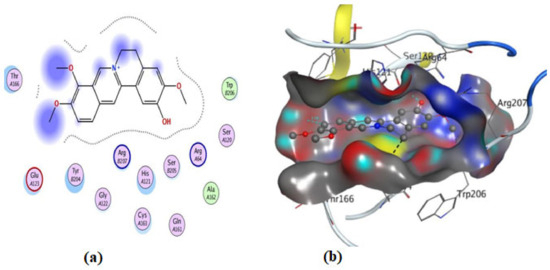
Figure 14.
Illustrations of the interaction of compound 3 inside the 3KJF active site in 2D (a) and 3D (b).
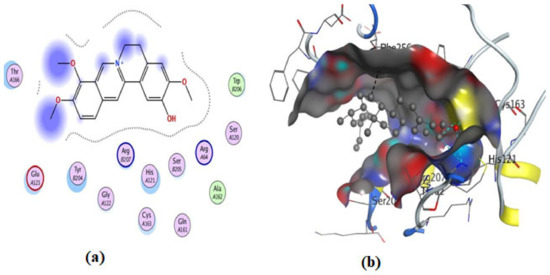
Figure 15.
Illustrations of the interaction of compound 4 inside the 3KJF active site in 2D (a) and 3D (b).
Compounds 1–4 (Figure 16) show a concentration-dependent inhibition of the parasite strains tested. A. hydrophila exhibited the highest sensitivity to all the test compounds compared to others. Palmatine was more potent against all the bacteria strains tested.
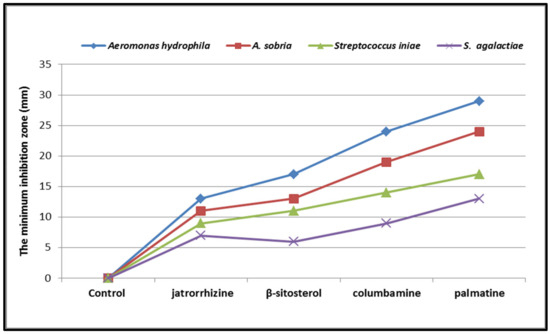
Figure 16.
Antibacterial activity (zones of inhibition, mm) of the test compounds against various bacterial strains. The data are shown as mean ± SEM.
2.4. Antiparasitic Activity
The results of the antiparasitic screening of compounds 1–4 against C. tilapiae is shown in Figure 17. The results revealed that the test compounds (1–4) exhibited time-dependent lethal effects against the test organism.
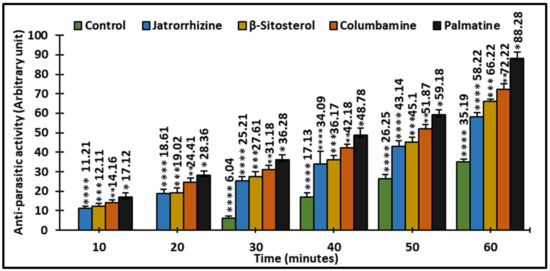
Figure 17.
Antiparasitic activity of currently investigated compounds on time after treatment (min) to death of C. tilapiae (%). Statistical differences from the control are shown as bar graphs with error bars representing the mean ± SEM; * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001 vs. control.
2.5. DFT Calculations
2.5.1. Molecular Electrostatic Potential (MEP)
The results of the DFT calculations of compounds 1–4 are shown in Figure 18. The figure reveals the MEPs generated from the optimized structures at the B3LYP/6–31G (d,p) level of theory for the investigated compounds, 1–4.
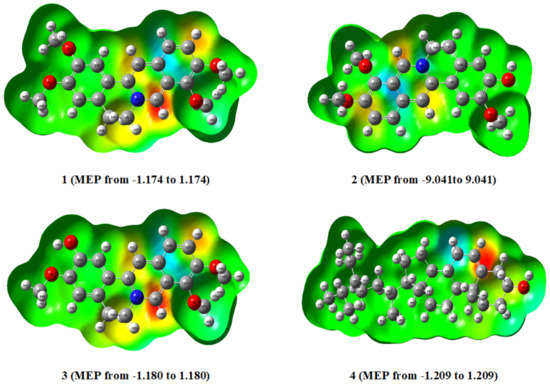
Figure 18.
The MEP surfaces of compounds (1–4). Red indicates negative potential, whereas blue indicates positive potential.
2.5.2. In Silico Study of Physicochemical and ADME Characteristics
Computational testing was used to assess the physicochemical and ADME properties of the test compounds. Compounds 1–4 (Figure 1) have acceptable ADME values and significant drug-like qualities and are likely to be orally active, according to Lipinski’s rule of five and Veber’s criterion (Table S2). It has been shown that palmatine, jatrorrhizine, columbamine, and β-sitosterol exhibit rotational bonds arranged between (C4–C6), suggesting molecular stability toward their biological targets. The topological polar surface area (TPSA) ranges from 20.23 Å2 to 51.80Å2 (Table S2). Based on pharmacokinetic and medicinal chemistry features, compounds 1–3 show high gastric absorption and the potential to pass the blood–brain barrier (BBB) (Table 1). A putative non-substrate P-glycoprotein (Pgp) is an effluent transport system, driving medications and other chemicals to leave the cell. Pgp is one of the factors leading to multidrug resistance in cancer chemotherapy and drug pharmacokinetics [17].

Table 1.
Pharmacokinetic and medicinal chemistry characteristics of compounds (1–4).
Consequently, the test compounds were investigated using the Swiss ADME website. Except for compound 4, it was revealed that most hits are not P-gp protein substrates, hence their potential as antiproliferative agents. The study showed that β-sitosterol (4) has a very low probability of escaping the cell, resulting in a significant impact (as shown in Table 1). Bioavailability, a plasma quantity indicator, is the most critical factor influencing absorption. Notable is that compounds 1–4 have a bioavailability of 0.55, which is quite significant. The compounds have synthetic accessibility scores ranging from 3.05 to 6.30, suggesting they can be synthesized in large amounts (Table 1). Similarly, Table 2 shows the dipole moments of all the compounds investigated, revealing that compounds 1 and 3 have larger dipole moments than 4.

Table 2.
DFT parameters calculated for the compounds (1–4).
3. Discussion
In this study, the MTT assay was used to evaluate the cytotoxicity of the title compounds (1–4) against the two mammalian cancer cell lines, MCF-7 (human breast cancer cell line) and HCT116 (colorectal cancer cell line) as well as the normal cell line (WI-38).
Several investigations have shown the cytotoxicity of 1–4 isolated from different plants against the MCF-7 cell line [4,18,19] and HCT116 [20,21,22]. This study investigated the molecular mechanism of action of palmatine (1), jatrorrhizine (2), columbamine (3), and β-sitosterol (4) through caspase-3-induced apoptosis in both MCF-7 and HCT116 cell lines, which have not been reported previously in the literature.
As shown in Figure 6, the test compounds suppressed the growth of MCF-7 and HCT-116 cancer cells in a dose-dependent manner, with a significant drop in cell viability and an increase in cytotoxicity (Figure 2, Figure 3, Figure 4 and Figure 5), with p < 0.0001. The IC50 values range between 3.33 and 5.96 µg/mL, with palmatine (1) showing significant inhibition of human colon cancer (HCT116) cells at IC50 = 3. 33 µg/mL and a selectivity index (SI) of 2.66 and jatrorrhizine (2) showing the highest IC50 = 5.96 µg/mL against HCT-116 cells (Figure 6 and Figure 7). The IC50 ranges for all compounds against MCF-7 cancer cells were 3.72–5.47 μg/mL. Compound 3 (columbamine) exhibited the highest antiproliferative activity against the MCF-7 cell line at an IC50 = 3.37 µg/mL (p < 0.0001), while jatrorrhizine (2) was the least effective at the same concentration (Figure 6). Doxorubicin was used as a standard antiproliferative agent (positive control). The IC50 values of all compounds and doxorubicin against WI-38 (human diploid cell line) ranged from 8.41—16.14 µg/mL (Figure 6). Therefore, the cytotoxicity of the compounds against MCF-7 and HCT116 cell lines was significantly higher compared with doxorubicin, which showed an IC50 values range of 7.93–9.86 µg/mL against the two cell lines (Figure 6).
The MCF-7 and HCT-116 cell lines were induced to undergo apoptosis by producing specific regulatory proteins in response to the presence of each drug (Figure 8 and Figure 9). Each drug boosted the pro-apoptotic proteins Bax and caspase-3 and cleaved PARP in the two cell lines while decreasing the anti-apoptotic protein Bcl-2. These values are significant (p < 0.0001) and compared favorably to doxorubicin. The Bax ranges for all compounds against MCF-7 and HCT-116 cancer cells were 222.83–325.25 ng/mL and 199.24–328.66 ng/mL, respectively (Figure 8), with the results showing that the lowest was jatrorrhizine (2) and the highest was palmatine (1) in the HCT-116 cancer cell line. The Bcl-2 values for all compounds range from 1.97 to 3.17 ng/mL. In contrast, against the HCT-116 cancer cell line, palmatine (1) showed the lowest Bcl-2 value, while jatrorrhizine (2) had the highest against the MCF-7 cancer cell line. The Bcl-2 ranges for all compounds on MCF-7 and HCT-116 cancer cells were 2.16–3.17 ng/mL and 1.97–2.69 ng/mL, respectively. The caspase-3 values for all compounds were within the 249.89–381.66 ng/mL range. The lowest was for jatrorrhizine (2) in the HCT-116 cancer cell line, and the highest was for columbamine (3) in the MCF-7 cancer cell line. The caspase-3 ranges for all compounds on MCF-7 and HCT-116 cancer cells were 261.51–381.66 ng/mL and 249.89–374.56 ng/mL, respectively (Figure 8). These findings demonstrated that each compound significantly induced apoptosis and antiproliferative activities in MCF-7 and HCT-116 cancer cells through the mitochondrial mechanism, as proposed in Figure 10. Extracts of plants and compounds have been reported to increase the expression of apoptosis-inducing proteins in treated cells, leading to inhibition of the proliferation of cancer cells [23].
On the other hand, an in silico investigation was conducted to establish the molecular pathways by which the test compounds (1–4) induced cell death in the MCF-7 and HCT116 cell lines, concentrating on their apoptotic induction through activation of the caspase-3 protein. The docking studies were targeted at the active site of the caspase-3 protein to reveal the binding poses of the compounds (Supplementary Materials, Table S1). The docking process was first verified by re-docking the co-crystallized ligand (B92) in the binding pocket of the enzyme with an energy score (S) = −6.38 kcal/mol and RMSD of 2.69, which produced five H-bonds with Arg 207, Cys 163, and Thr 62 (Table S1 and Figure 11). Furthermore, the amino acid residue Phe 256 formed an arene–H interaction with the CH2-cinnoline ring proton, and the residue Asn 208 showed H-arene contact with the phenyl ring (Figure 11). The ligand was removed from the complex, and then docking of compounds (1–4) was performed; the results obtained are recorded in Table S1. As shown in Figure 11, Figure 12, Figure 13, Figure 14 and Figure 15, compounds (1–4) possess a good binding affinity to caspase-3 active sites, with different numbers of H-bonds and H-pi bond interactions. Compound 1 has a docking score energy (S) = −5.71 kcal/mol with three established hydrogen bonds with Ala 254, Arg 207, and Gly 122. Also, Arg 207 revealed H-arene interaction with the phenyl moiety of the isoquinoline ring (Figure 12). Compound 2 exhibited five hydrogen bonds with Cys 163, Met 61, Glu 123, and Gly 122; conversely, His 121-residue showed two H-arene contacts with the phenyl moiety of isoquinoline and dihydro isoquinoline rings (Figure 13). Similarly, compound 3 produced three H-bonds with Ser 120, Glu 123, and Gly 122 and showed H-arene interaction with Tyr 204 residue (Figure 14). Finally, compound 4 had a −6.19 Kcal/mol docking score comparable to that of B92 (−6.38 Kcal/mol), the natural ligand, and revealed four H-bonds, including Cys163 and His 121 amino acids; in addition, the proton of -CH2-cyclopentane ring in compound 4 was oriented in close contact with Phe 256, forming an H–arene interaction (Figure 15).
Molecular electrostatic potential (MEP) is essential for understanding a molecule’s relative polarity and reactivity. MEP is also beneficial for predicting and analyzing molecular interactions like hydrogen bonding, enzyme–substrate interactions, and drug receptors. According to the MEP analysis (Figure 18), compound 4 has a negative electron density around the =CH- proton moiety of the phenanthrene ring. In contrast, compounds (1–3) have negative potential around the N-CH moiety of the isoquinoline ring, which is an attractive site for positively charged species. These areas are thus associated with electrophilic reactivity. On the other hand, a positive potential is found in the protons of some aromatic rings in all derivatives (1–3) connected to nucleophilic reactivity, as illustrated in Figure 19. The dipole moments of all the compounds investigated are shown in Table 2. The results revealed that compounds 1 and 3 have larger dipole moments. Consequently, 1–3 may permit more interaction with high dipole moment species, especially in biological systems [24]. The DFT calculations reveal that compounds 1 and 3 have smaller energy gaps, 0.072 and 0.071eV, respectively (Table S1 and Figure 19), with the highest dipole moments and, as such, are more chemically reactive and exhibited better modulation of caspase-3 enzyme and inhibitory activities of the MCF-3 and HCT116 cell lines (Figure 6).
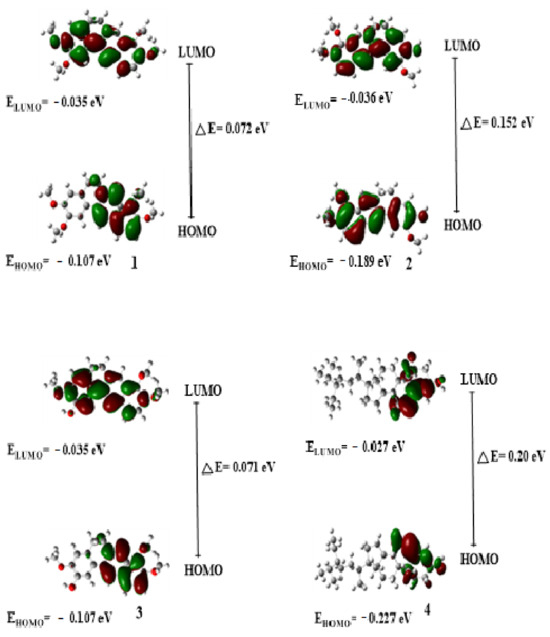
Figure 19.
The HOMO, LUMO, and △E energies of the studied compounds (1–4).
Frontier molecular orbitals (FMOs) are an effective method for analyzing the molecular interactions inside a molecule. The highest occupied molecular orbitals (HOMOs) function as electron donors, whereas the lowest unoccupied molecular orbitals (LUMOs) function as electron acceptors. The lower the value of E, the more polarized and active the molecule is, and the lower its kinetic bioactivity [25]. On the other hand, Figure 19 shows all isolated compounds’ HOMO-LUMO orbitals and energy values. Compounds 1 and 3 have smaller energy gaps, 0.072 and 0.071eV, respectively, compared with other compounds, as shown in Table 2. It is, therefore, expected that 1 and 3 may exhibit better biological activity. This is also confirmed from the in vitro studies wherein the compounds showed IC50 values of 3.9259 and 3.7202 µg/mL against the MCF-7 cells, 3.3366 and 3.3713 µg/mL, respectively, against the HCT116 cells. The molecular descriptors obtained in this study include the highest occupied molecular orbital (HOMO), lowest unoccupied molecular orbital (LUMO), HOMO-LUMO energy difference (band gap), softness, chemical hardness, dipole moment, hydrophobicity (log P), and topological polar surface area (TPSA) as shown in Table 1 and Table S1. The HOMO and LUMO are essential in binding the compounds to the target sites of the biomolecule. As a result, the extent of the nonbonding interactions (hydrogen bonding and hydrophilic interactions) between the drug molecules and receptors depends on the magnitude of these parameters. The energy difference between the LUMO and HOMO (band gap) is shown in Table 1. The lower the energy gap, the higher the propensity of the compound to donate electrons to neighboring molecules. This parameter is important in protein–ligand interactions between the bioactivity of the studied compounds. The energy difference (band gap) is ranked in the following order: 2 > 3 > 1 > 4. Similarly, the lipophilicity of the compounds (Table S1), calculated as LogP, i.e., the ability of a compound to penetrate biological membranes, shows that, except for compound 4, the others have very good lipophilicity values and hence good drug-likeness properties. The ranking of LogP values of compounds 2 and 3 (1.78) < 1 (2.01) < 4 (6.73). The Moriguchi octanol–water partition coefficient stipulates that it should be less than 4.15 [26].
The antiparasitic and antimicrobial screening of the test compounds (1–4) are shown in Figure 16 and Figure 17. The compounds caused a lethal effect on the C. tilapia parasite. The parasites initially showed turbulent movements, contracting with violent twitching. This was followed by the parasites separating from the host. The parasites continued to contract, although at a reduced pace, until death. Parasite mortality was concentration- and time-dependent. The percentage of killed parasites in the control group rose from around 6 after 30 min to 35 after 60 min. After treatment with palmatine, about 17% of the parasites died after 10 min (p < 0.001), while at 60 min, the percentage of parasite death was more than 88 % (Figure 17). Palmatine exhibited the highest antiparasitic activity against C. tilapiae (88.28%) compared with the other compounds. In a similar vein, the compounds showed moderate activity against the different strains of pathogenic organisms (Aeromonas hydrophilia, Aeromonas sobria, Streptococcus agalactiae, and Streptococcus iniae) tested (Figure 16). Amongst the four compounds, palmatine exhibited better activity against Aeromonas hydrophilia with a zone of inhibition > 29mm. However, a study reported that palmatine was not effective against Aeromonas hydrophilia, an opportunistic pathogen of aquaculture, showing an MIC value of 256 µg/mL compared with the standard antibiotic Enrofloxacin with an MIC value of 4 µg/mL [27].
4. Materials and Methods
The compounds (1–4) used in this experiment were isolated from the stem bark of Enantia chlorantha, as previously reported [10]. Antiproliferative screening assay was performed on two cancer cell lines (HCT-116 and MCF-7) using MTT (Sigma-Aldrich, Saint Louis, MO, USA) assay and WI-38 human fibroblast cell line to evaluate cytotoxicity.
4.1. Anticancer Assay
4.1.1. Cell Cultures
The cancer cell lines, human colon cancer cells (HCT116) and breast cancer (MCF-7), in addition to normal human lung fibroblast (WI-38) were obtained from VACSERA Co., Cairo, Egypt. All cell lines were grown in an acceptable medium (HCT116: McCoy’s 5a; MCF-7: DMEM-high glucose; WI-38: EMEM) supplemented with 10% FCS, 2 mM of glutamine, 100 units of penicillin and streptomycin, and 5% CO2. The culture media were changed every three to four days. At 85–90 percent confluence, the cultures were passaged using suitable procedures for downstream application using 0.25 percent trypsin/EDTA solution [17].
4.1.2. MTT Assay for Cytotoxicity Evaluation
A cell proliferation kit was used to measure the cytotoxicity of 1–4 against MCF-7, HCT116, and WI-38. Accordingly, 5000 cells per well were grown for 24 h in 96-well culture plates. Individual cells were provided at different conc. of 1–4 (0, 6.25, 12.5, and 25 µg/mL) for 48 h at 37 °C and 5% humidity in a CO2 incubator, and 100 g/mL doxorubicin was employed as a positive control. All cells were treated for 2 h with 10 L of MTT powder at a concentration of 0.5 mg/mL until violet crystals of varying colors developed, indicating metabolic activity. The insoluble crystals of formazan were dissolved in phosphate-buffered saline. The colorimetric absorbance was measured at 620 nm and 570 nm using a SynergyTM 2 Multi-Mode Microplate Reader (BioTek Inc., Winooski, VT, USA). The cell viability was computed using the formula in [28].
The IC50 was computed using the IC50 calculator [19]. The treatments and analytical procedures were carried out at least three times.
4.1.3. Determination of Selectivity Index (SI)
The SI evaluates the extract’s cytotoxic selectivity toward cancer cells compared with normal control cells [18]. The SI is equal to the half-maximal (IC50) concentration of the extract in the normal control cells against that of the test samples (cancer cell lines) [19].
4.1.4. Determination of Caspase-3 Activity
The level of caspase-3 activity was determined using the Invitrogen Caspase-3 assay (active) Human kit (Thermofisher Scientific Co., Waltham, MA, USA). Following a quick PBS wash, the cancer cells were collected and lysed by adding an extraction solution containing 1 mM of PMSF (stock is 0.3 M in DMSO) and a combination of protease inhibitors (Sigma-Aldrich Cat. # P-2714, St. Louis, MO, USA). After that, 500 µL per 5 mL of cell extraction buffer–protease inhibitors (1 mL per 1 × 107 cells) was added. The lysate was diluted just before the experiment. Using a microplate reader AC3000 (Azure Biosystems, Inc, Dublin, CA, USA), the OD of every well was calculated within 30 min set at 450 nm.
4.1.5. Determination of Bax and Bcl-2
Cancer cells were cultured in DMEM supplemented with 5% FBS at 37 °C and treated with the test compounds (1–4) using lysed cell extract. The cell lysate was then diluted with the standard buffer and analyzed with a Bax ELISA (EIA-4487) kit (DRG Instruments, Marburg, Germany) for human active Bax and Bcl-2 and a Zymed Bcl-2 ELISA Kit (Thermofisher Scientific Co., Waltham, MA, USA). Bax and Bcl-2 levels were determined in a suspension of cancer cells.
4.1.6. Western Blotting
The medium was discarded 24 h after MCF-7 and HCT-116 and every compound was administered to cells. The cells were washed with cold PBS (10 mmol/L, pH 7.4) and then incubated in an ice-cold lysis buffer [50 mmol/L Tris–HCl, 150 mmol/L NaCl, 1 mmol/L EGTA, 1 mmol/L EDTA, 20 mmol/L NaF, 100 mmol/L Na3VO4, 0.5% NP40, 1% Triton X-100, and 1 mmol/L PMSF (pH 7.4)] with a recently introduced inhibitor mixture (Inhibitor Cocktail Set III, Calbiochem, La Jolla, CA) thirty minutes above the ice [22]. The cells were scraped, and the cell solution was run through a 21.5-gauge needle in a microfuge tube many times to separate any cell aggregates [23]. The whole cell lysate (supernatant) was kept at −80 °C for subsequent analysis after centrifugation at 14,000 g for 25 min at 4 °C [22]. PARP (catalog# 32563 & 9542), GAPDH (catalog# 2118), β-actin (catalog# 4970), anti-rabbit HRP (catalog# 7074), and anti-mouse HRP secondary (catalog# 7076) antibodies, all diluted 1:1000, were acquired from Cell Signaling Technology (Beverly, MA, USA), while GAPDH (catalog# sc-47724) and β-actin (catalog# sc-8432) antibodies, both diluted 1:1000, were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz Co., Santa Cruz, CA, USA). Mini-protean precast Tris-Glycine gels were manufactured by BioRad (Hercules, CA, USA) and the ECL detection system was manufactured by GE Healthcare (Buckinghamshire, UK). Gentian violet solution 2% w/v was purchased from Ricca Chemical Company (Arlington, TX, USA) and precast Tris-Glycine gels were purchased from Invitrogen Novex (Corning, NY, USA). Approximately 25–40 g of protein was separated on 8–12% polyacrylamide gels and put into a nitrocellulose membrane. Blots were blocked using blocking buffer [7% nonfat dry milk/1% Tween 20; in 20 mmol/L TBS (pH 7.6)] for 1 h at room temperature, followed by incubation with a suitable monoclonal or polyclonal primary antibody in blocking buffer for 2 h at room temperature or overnight at 4 °C, and then incubated with either an ant-rabbit HRP-conjugated antibody (1:1000) or anti-mouse HRP-conjugated antibody (1:1000). The blots were treated to enhanced chemiluminescence (Thermo Scientific Pierce, Rockford, IL, USA) and autoradiography was performed utilizing imaging equipment from BioRad version 1.3.2.0 (Hercules, CA, USA). The digitalized scientific software Quantity One (BioRad) version 1.3.2.0 (Hercules, CA, USA) was utilized to analyze the Western blot to determine the densitometric measurements of the bands. The treatment procedure was repeated thrice, and each protein expression was tested with similar outcomes [18,22].
4.2. Antibacterial Activity
Compounds 1–4 were evaluated for antibacterial activity against Aeromonas hydrophilia, Aeromonas sobria, Streptococcus agalactiae, and Streptococcus iniae acquired from the Department of Microbiology, Science Faculty, Al-Azhar University, Egypt. The antibacterial potency of the test compounds was carried out using the cup plate diffusion method as described in [24].
4.3. Antiparasitic Activity
Contaminated Nile tilapia (Oreochromis niloticus) with parasite Cichlidogyrus tilapia, acquired in May 2022 from Abbassa, Sharkia Governorate, Egypt, was used for this assay. Briefly, the gill arches of the fish were removed into Petri plates containing distilled water. Cut portions of gill filaments carrying parasites were inserted in 12-well plates with 10 mL of distilled water and 10 parasites in each well. The compounds (1–4) were applied against C. Tilapia in three replicates at time zero (0 h). Each treatment contained wells with deionized water without the test chemicals that served as a control. Using a dissecting microscope, parasites were inspected every 10 min and mortalities were reported. Parasites were reported dead if there was no response to touch or reaction when moved to clean wells containing distilled water. Treatment is deemed successful if 100% parasite death is achieved within twenty-four hours [25]. The antiparasitic effectiveness of each treatment and control group was determined by the following:
where AE represents the antiparasitic effectiveness, B represents the mean number of survivals in the control group, and T represents the mean survival in the treatment group [26].
AE = [B − T] × 100%/B
4.4. Molecular Docking Study
The 2015.10 edition of the Molecular Operating Environment (MOE) 2022.02 Chemical Computing Group ULC, Montreal, Canada, was used to compute the docking study of the investigated compounds. ChemDraw18.0 (PerkinEimer Informatics Inc., MA, US) was used to draw the structures of the compounds (1–4), which were then saved as MDL MOL files. Before molecular docking, the following procedures were carried out: a) conversion of ligands’ 2D structures to their 3D forms; b) polar hydrogen atoms were added or removed; and c) the compound’s energy was minimized with the MMFF94x force field until an RMSD (root-mean-square deviation) of atomic position gradient of 0.01 Kcal mol−1 Å−1 was reached and saved as MOE.
The B92 crystal structure with the caspase-3 active site (PDB: ID 3KJF) was retrieved from the protein data bank. The 3D protonation method, in which hydrogen atoms were added to the usual enzyme structure, was used to prepare it for docking. The compounds (1–4) were docked into caspase-3 through MOE-Dock using the triangle matcher placement approach, the London dG scoring function, and force field refinement on the top 5 postures per compound. B92 was redocked with the 3KJF active site to confirm the docking method.
4.5. DFT Calculations
The structures of compounds 1–4 were drawn in Gauss view 6.0 (Semichem Inc., Shawnee, KS, USA). The DFT methods were used to optimize the ground state of all candidate compounds (1–4) using the Gaussian 09 package of Frisch et al. (2019) as described in [27]. In this study, Becke’s 3-parameter formula Hybrid Lee–Yang–Parr exchange-correlation (B3LYP) functions with a 6–31G (d,p) basis set was used for gas-phase compound optimization [28]. Using the same level of theory, frequency calculations were undertaken to ensure that all structures found were minimal local energy structures. Finally, the optimized structures at the B3LYP/6–31G (d,p) level of theory were utilized to calculate the molecular electrostatic potential (MEP) of all compounds investigated.
In Silico Study of Physicochemical and ADME Characteristics
The structures of compounds (1–4) were converted to SMILE names using the Swiss ADME website (http:/swissadme.ch/index.php, accessed 7 October 2022) to predict physical and chemical properties, pharmacokinetics, and medicinal chemistry parameters. Computational testing was used to evaluate the physicochemical and ADME characteristics of the intended substances.
4.6. Statistical Analysis
All experiments were conducted in triplicate, and the results were analyzed using Microsoft Excel (version 2010, Microsoft Inc., US) and reported as mean ± standard error of the mean (SEM). Statistical significance was determined by one-way ANOVA for multiple groups, followed by Tukey’s test for multiple comparisons. p ˂ 0.05 was considered significant.
5. Conclusions
This study demonstrated the in vitro antiproliferative potentials of palmatine, columbamine, jatrorrhizine, and β-sitosterol (1–4) against MCF-7 and HCT116 cancer cells. The compounds also showed significant antibacterial efficacy against selected Gram-positive and Gram-negative pathogenic bacteria strains and antiparasitic activity against Cichlidogyrus tilapiae, a known parasite of fishes. In silico docking of the compounds against the caspase-3 enzyme receptor site demonstrated that the compounds’ anticancer activities might be related to the modulation of apoptosis. The compounds also showed significant interactions with different amino acids and exhibited high binding affinity to the caspase-3 receptor. DFT analysis revealed that compounds 1 and 3 had the highest dipole moment and lowest energy gap compared with other compounds; hence, they are less stable and chemically more reactive. Based on Lipinski’s rule, in silico investigation revealed that the compounds have ADME values in the acceptable range and many drug-like features and could serve as pharmaceutical leads.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ddc3010017/s1. Table S1: The data of compounds (1–4) and B92 pending docking in caspase-3 (PDB ID: 3KJF) active spots and Table S2: Physicochemical properties of compounds (1–4) (TPSA, Lipinski’s rule of five and Veber’s rule).
Author Contributions
Conceptualization, V.O.I., A.F., A.A.Z. and M.A.E.B.; methodology, M.A.E.B., M.A.E.A.E.-A., I.R. and I.M.M.G.; formal analysis, H.A.E.-N., I.R. and I.M.M.G.; investigation, E.S.A.-A., S.Y., I.R. and M.A.S.A.; data curation, M.A.M.E.-T. and M.A.E.B.; writing—original draft preparation, V.O.I., M.A.E.B., I.R. and R.M.S.; writing—review and editing, all authors; supervision, A.F. All authors have read and agreed to the published version of the manuscript.
Funding
The Deanship of Scientific Research at Umm Al-Qura University supported this work by grant code (22UQU4290565DSR117).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
The authors thank the National Center for Natural Products Research (NCNPR), University of Mississippi, Oxford, MS, USA, and the University of Benin, Benin City, Nigeria, for providing laboratory space and some equipment for this work.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.M.; Kehm, R.D.; Terry, M.B. Global breast cancer incidence and mortality trends by region, age-groups, and fertility patterns. eClinicalMedicine 2021, 38, 100985. [Google Scholar] [CrossRef]
- Fridlender, M.; Kapulnik, Y.; Koltai, H. Plant derived substances with anticancer activity: From folklore to practice. Front. Plant Sci. 2015, 6, 799. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Bashar, M.A.E.; Rady, I.; El-Naggar, H.A.; Abd El-Maoula, L.M.; Mehany, A.B.M. Antiproliferative Activity, Proapoptotic Effect, and Cell Cycle Arrest in Human Cancer Cells of Some Marine Natural Product Extract. Oxid. Med. Cell. Longev. 2020, 25, 7948705. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, H.A.; Bashar, M.A.E.; Rady, I.; El-Wetidy, M.S.; Suleiman, W.B.; Al-Otibi, F.O.; Al-Rashed, S.A.; Abd El-Maoula, L.M.; Salem, E.S.S.; Attia, E.M.; et al. Two Red Sea Sponge Extracts (Negombata magnifica and Callyspongia siphonella) Induced Anticancer and Antimicrobial Activity. Appl. Sci. 2022, 12, 1400. [Google Scholar] [CrossRef]
- Harvey, A.L. Natural products in drug discovery. Drug Discov. Today 2008, 13, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Supuran, C.T. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Last 25 Years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef]
- Yassin, A.M.; El-Deeb, N.M.; Metwaly, A.M.; El Fawal, G.F.; Radwan, M.M.; Hafez, E.E. Induction of Apoptosis in Human Cancer Cells Through Extrinsic and Intrinsic Pathways by Balanites aegyptiaca Furostanol Saponins and Saponin-Coated SilverNanoparticles. Appl. Biochem. Biotechnol. 2017, 182, 1675–1693. [Google Scholar] [CrossRef]
- Imieje, V.O.; Zaki, A.A.; Fasinu, P.S.; Ali, Z.; Khan, I.A.; Tekwani, B.; Khan, S.I.; Nosa, E.O.; Falodun, A. Antiprotozoal and Cytotoxicity Studies of Fractions and Compounds from Enantia chlorantha. Trop. J. Nat. Prod. Res. 2017, 1, 89–94. [Google Scholar] [CrossRef][Green Version]
- Metwaly, A.M.; Ghoneim, M.M.; Musa, A. Two new antileishmanial diketopiperazine alkaloids from the endophytic fungus Trichosporum sp. Derpharmachemica 2015, 11, 322. [Google Scholar]
- Jalmakhanbetova, R.I.; Suleimen, Y.M.; Oyama, M.; Elkaeed, E.B.; Eissa, I.H.; Suleimen, R.N.; Metwaly, A.M.; Ishmuratova, M.Y. Isolation and In Silico Anti-COVID-19 Main Protease (Mpro) Activities of Flavonoids and a Sesquiterpene Lactone from Artemisia sublessingiana. J. Chem. 2021, 2021, 5547013. [Google Scholar] [CrossRef]
- Roy, A.; Khan, A.; Ahmad, I.; Alghamdi, S.; Rajab, B.S.; Babalghith, A.O.; Alshahrani, M.Y.; Islam, S.; Islam, M.R. Flavonoids a Bioactive Compound from Medicinal Plants and Its Therapeutic Applications. Biomed. Res. Int. 2022, 2022, 5445291. [Google Scholar] [CrossRef] [PubMed]
- Almutairi, S.; Edrada-Ebel, R.; Fearnley, J.; Igoli, J.O.; Alotaibi, W.; Clements, C.J.; Gray, A.I.; Watson, D.G. Isolation of diterpenes and flavonoids from a new type of propolis from Saudi Arabia. Phytochem. Lett. 2014, 10, 160–163. [Google Scholar] [CrossRef]
- Dhyani, P.; Sati, P.; Sharma, E.; Attri, D.C.; Bahukhandi, A.; Tynybekov, B.; Szopa, A.; Sharifi-Rad, J.; Calina, D.; Suleria, H.A.; et al. Sesquiterpenoid lactones as potential anticancer agents: An update on molecular mechanisms and recent studies. Cancer Cell Int. 2022, 22, 305. [Google Scholar] [CrossRef]
- Zhang, S.; Won, Y.K.; Ong, C.N.; Shen, H.M. Anti-cancer potential of sesquiterpene lactones: Bioactivity and molecular mechanisms. Curr. Med. Chem. Anti-Cancer Agents. 2005, 5, 239–249. [Google Scholar] [CrossRef]
- Prachayasittikul, V.; Worachartcheewan, A.; Shoombuatong, W.; Prachayasittikul, V.; Nantasenamat, C. Classification of P-glycoprotein-interacting compounds using machine learning methods. EXCLI J. 2015, 14, 958–970. [Google Scholar]
- Salam, H.S.; Tawfik, M.M.; Elnagar, M.R.; Mohammed, M.A.; Zarka, M.A.; Awad, N.S. Potential Apoptotic Activities of Hylocereus undatus Peel and Pulp Extracts in MCF-7 and Caco-2 Cancer Cell Lines. Plants 2022, 11, 2192. [Google Scholar] [CrossRef]
- Prayong, P.; Barusrux, S.; Weerapreeyakul, N. Cytotoxic activity screening of some indigenous Thai plants. Fitoterapia 2008, 79, 598–601. [Google Scholar] [CrossRef]
- Bézivin, C.; Tomasi, S.; Lohézic-Le Dévéhat, F.; Boustie, J. Cytotoxic activity of some lichen extracts on murine and human cancer cell lines. Phytomed. Int. J. Phytother. Phytopharm. 2003, 10, 499–503. [Google Scholar] [CrossRef]
- Sanna, V.; Singh, C.K.; Jashari, R.; Adhami, V.M.; Chamcheu, J.C.; Rady, I.; Sechi, M.; Mukhtar, H.; Siddiqui, I.A. Targeted nanoparticles encapsulating (−)-epigallocatechin-3-gallate for prostate cancer prevention and therapy. Sci. Rep. 2017, 7, 41573. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Siddiqui, I.A.; Nihal, M.; Pilla, S.; Rosenthal, K.; Mukhtar, H.; Gong, S. Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer. Biomaterials 2013, 34, 5244–5253. [Google Scholar] [CrossRef] [PubMed]
- Grabarska, A.; Wróblewska-Łuczka, P.; Kukula-Koch, W.; Łuszczki, J.J.; Kalpoutzakis, E.; Adamczuk, G.; Skaltsounis, A.L.; Stepulak, A. Palmatine, a Bioactive Protoberberine Alkaloid Isolated from Berberis cretica, Inhibits the Growth of Human Estrogen Receptor-Positive Breast Cancer Cells and Acts Synergistically and Additively with Doxorubicin. Molecules 2021, 26, 6253. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.S.; Borland, M.; Brain, C.; Chen, C.H.; Cheng, H.; Chopra, R.; Chung, K.; Groarke, J.; He, G.; Hou, Y.; et al. 4-(Pyrazol-4-yl)-pyrimidines as selective inhibitors of cyclin-dependent kinase 4/6. J. Med. Chem. 2010, 53, 7938–7957. [Google Scholar] [CrossRef] [PubMed]
- Klingmüller, U.; Schilling, M.; Depner, S.; D’Alessandro, L.A. Biological foundations of signal transduction, systems biology and aberrations in disease. In Computational Systems Biology: From Molecular Mechanisms to Disease; Elsevier Inc.: Amsterdam, The Netherlands, 2013; pp. 45–64. [Google Scholar]
- Zhang, X.P.; Li, W.X.; Ai, T.S.; Zou, H.; Wu, S.G.; Wang, G.T. The efficacy of four common anthelmintic drugs and traditional Chinese medicinal plant extracts to control Dactylogyrus vastator (Monogenea). Aquaculture 2014, 420, 302–307. [Google Scholar] [CrossRef]
- Dong, J.; Yan, T.; Yang, Q.; Song, Y.; Cheng, B.; Zhou, S.; Liu, Y.; Ai, X. Palmatine Inhibits the Pathogenicity of Aeromonas hydrophila by Reducing Aerolysin Expression. Foods 2022, 11, 3250. [Google Scholar] [CrossRef] [PubMed]
- Ruta, L.L.; Farcasanu, I.C.; Bacalum, M.; Răileanu, M.; Rostas, A.M.; Daniliuc, C.; Chifiriuc, M.C.; Măruțescu, L.; Popa, M.; Badea, M.; et al. Biological activity of triazolopyrimidine copper (II) complexes modulated by an auxiliary NN-chelating heterocycle ligands. Molecules 2021, 26, 6772. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).