Fundamental Mechanisms in Membrane Receptology: Old Paradigms, New Concepts and Perspectives
Abstract
1. Introduction
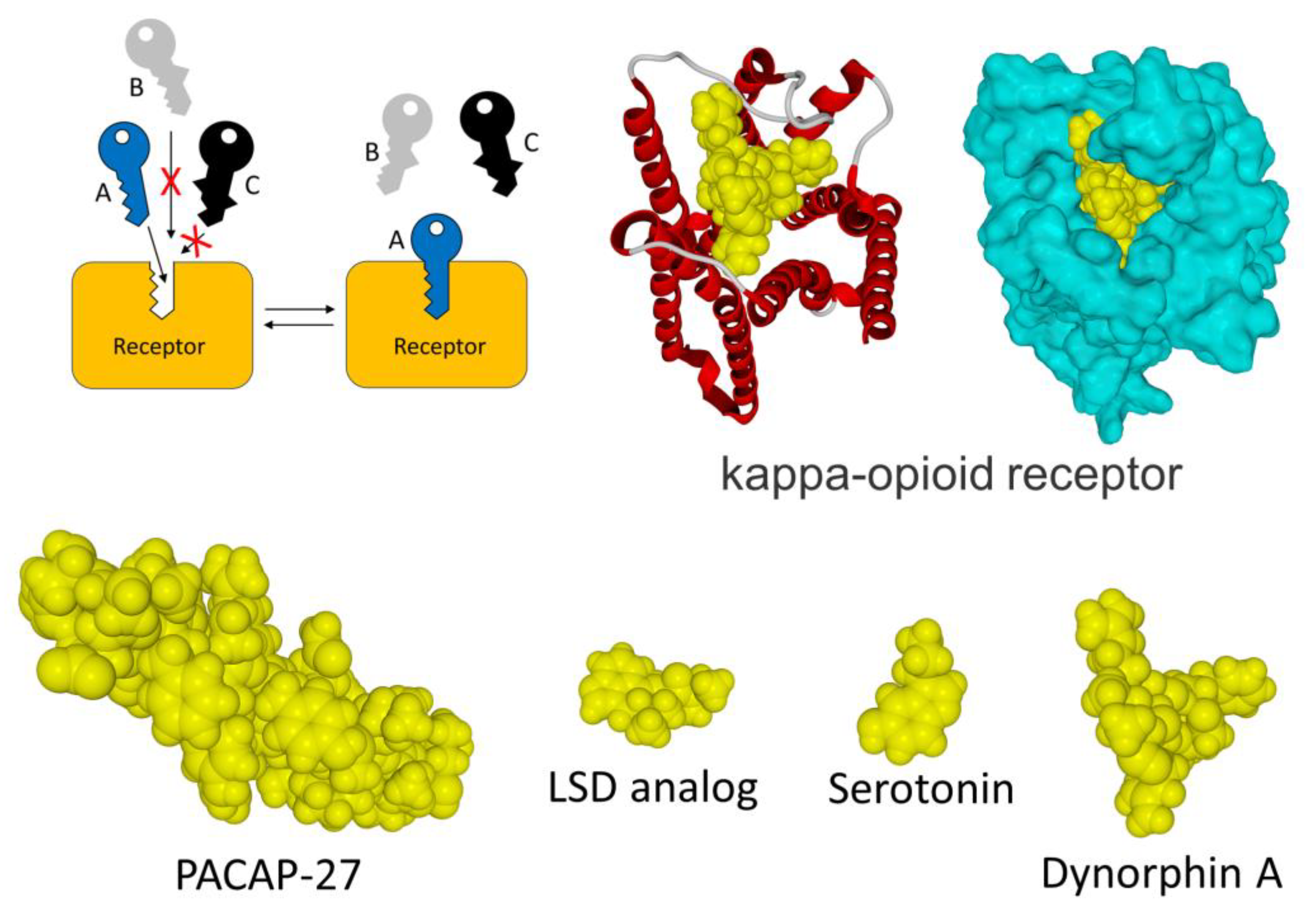
2. The Old Paradigm
3. Limitations of the Old Paradigm
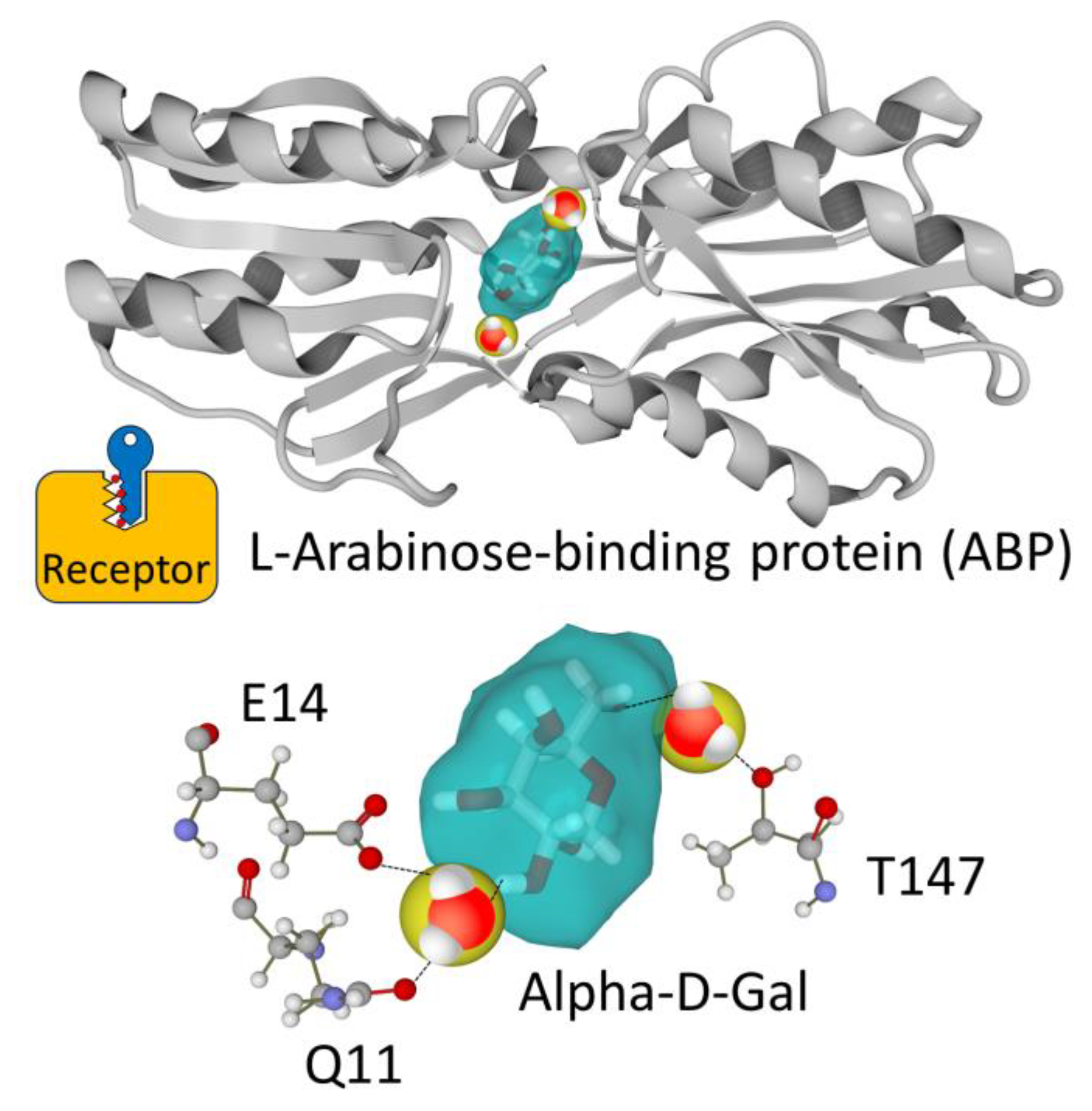
3.1. Water Molecules Bound to Membrane Receptors
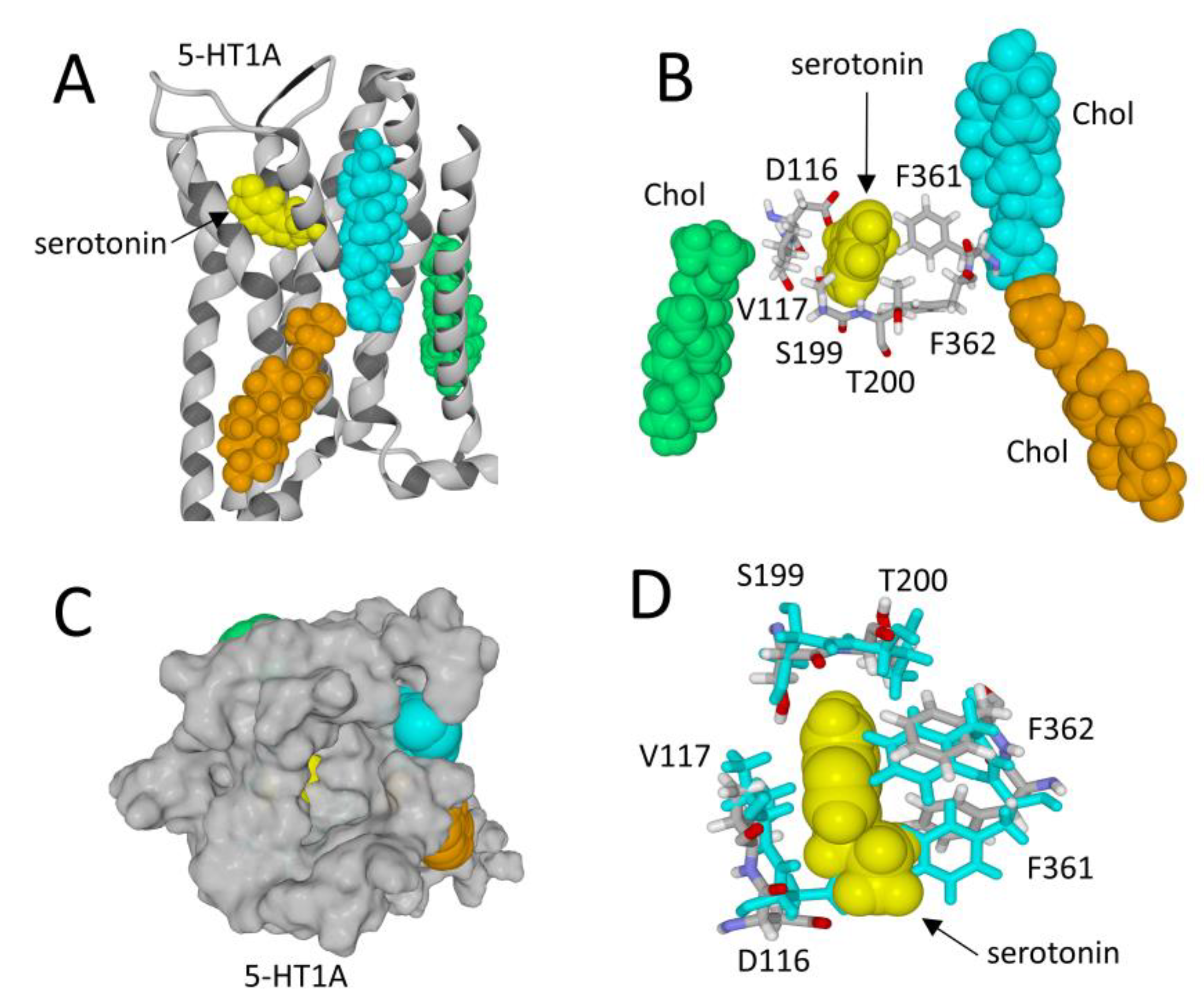
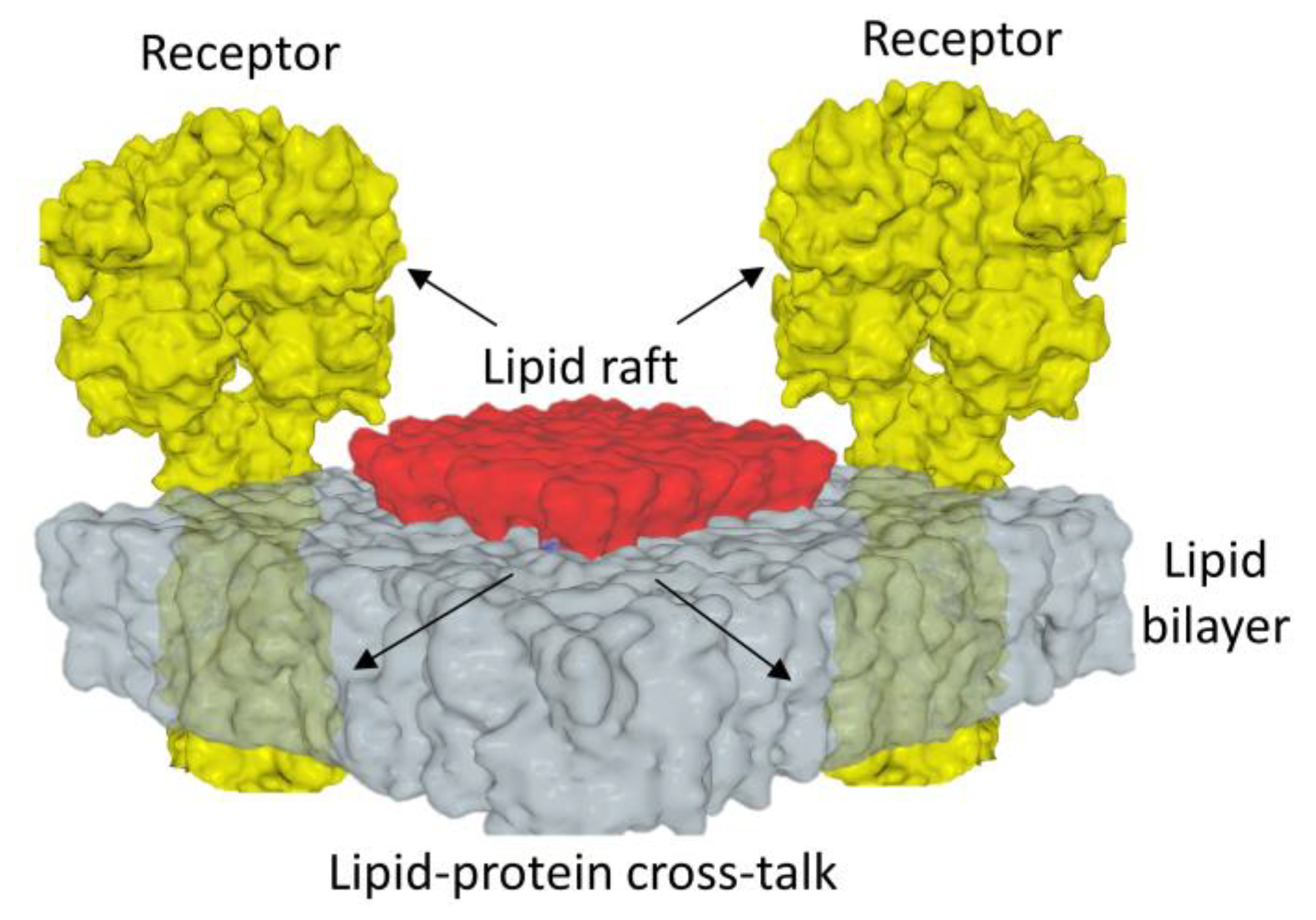
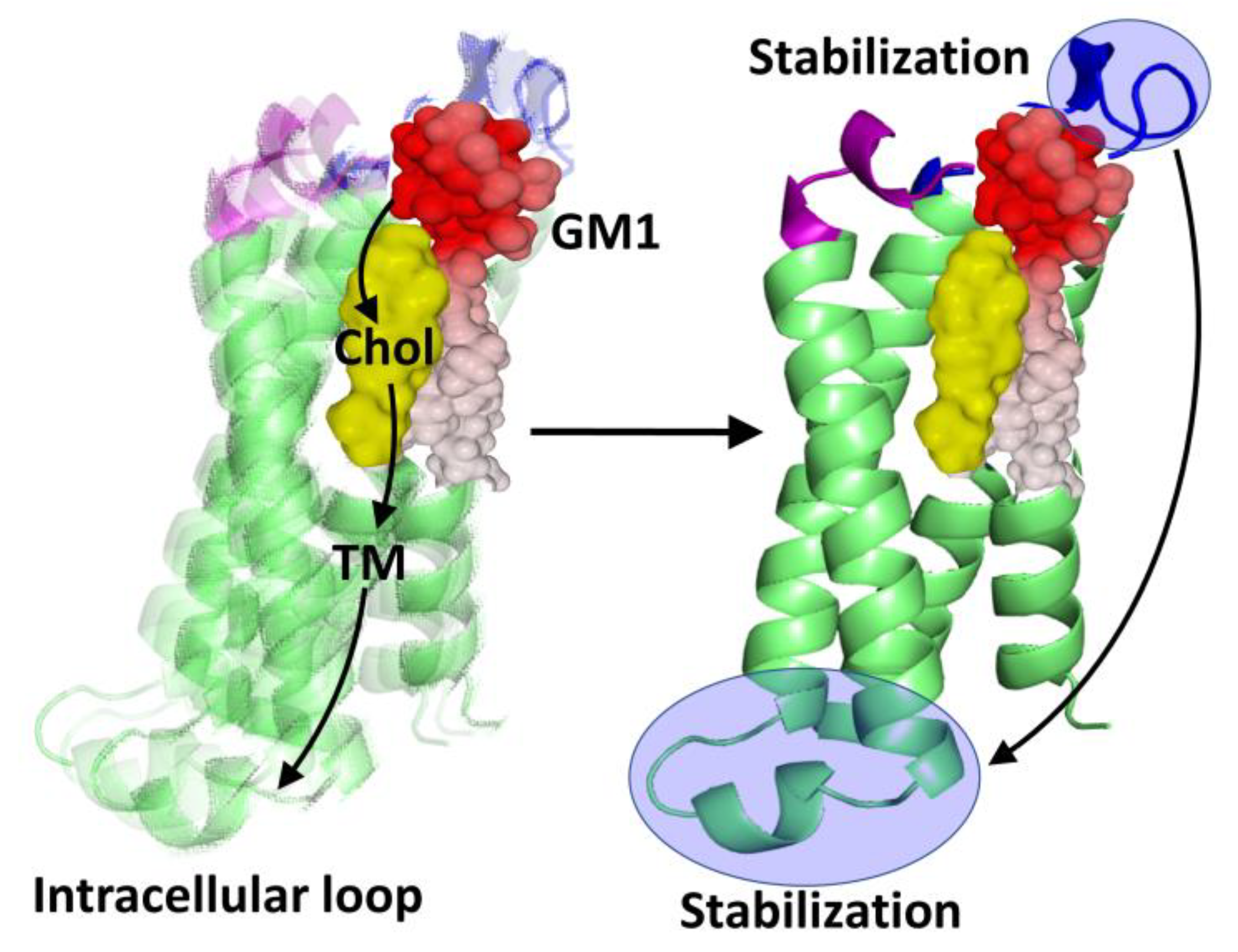
3.2. Membrane Receptors Are Surrounded by Lipids
3.3. Intrinsic Disorder in Receptology: The Puzzling Case of Glucagon
3.4. Conformational Rearrangements following Ligand Binding
4. New Concepts
4.1. Quantum Mechanims: The Swipe Card Model of Olfaction
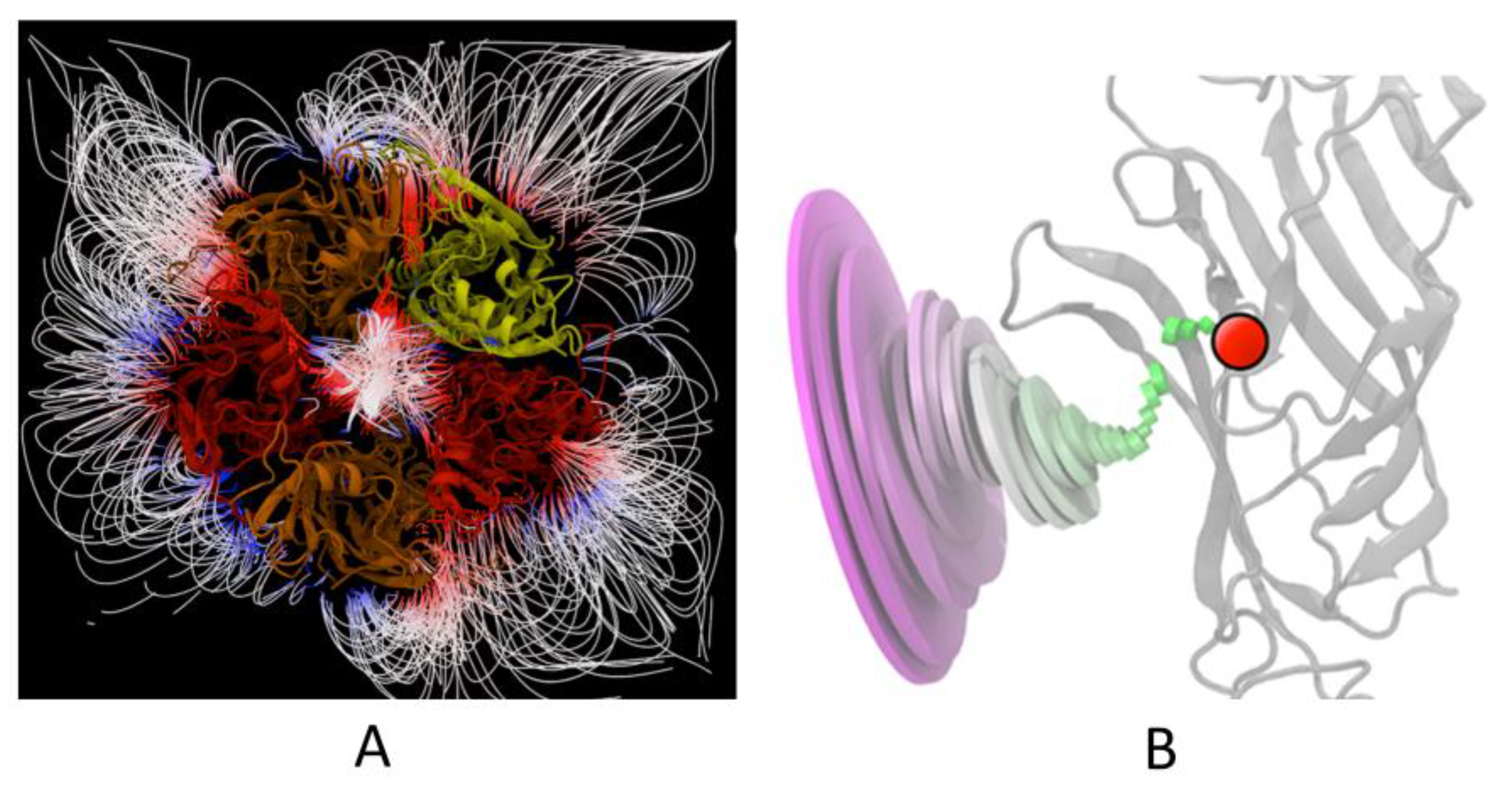
4.2. Electrostatic Surface Potential
5. Summary and Perspectives
- (i)
- Taking water molecules into account should be a systematic reflex for every biologist [13]. The notions of bound water, molecular disorder, hydration/dehydration energy and entropy are likely to modify our vision of ligand–receptor interactions. In addition, the impact of water molecules on the structure—or lack of stable structure—of receptors and ligands is critical.
- (ii)
- The role of membrane lipids, chiefly cholesterol and raft gangliosides [23], remains largely underestimated in receptology. Significantly, numerous studies show that it is possible to annihilate the function of a receptor by modulating the membrane levels of cholesterol and gangliosides that are associated with the receptor [88,89,90]. These lipids exert a chaperone activity on the receptors, such that in their absence these receptors are no longer functional because they are incapable of recognizing their ligand.
- (iii)
- The consideration of molecular disorder and structuring phenomena resulting from the formation of a ligand–receptor complex was approached using the example of glucagon. However, it is likely that such mechanisms could be operative for many other ligand–receptor pairs [51]. But to undertake research in this context, we must realize that about 50% of proteins of the human proteome are either totally or partially disordered [47]. Furthermore, the case of glucagon demonstrates that even short peptides (with less than 30 amino acid residues) can exhibit such characteristics, which takes us even further away from the original key and lock model.
- (iv)
- If we combine this notion of IDPs with the chaperone activity of membrane lipids, we understand the failure of the AlphaFold program for predicting the structure of membrane proteins [41]. My personal experience leads me to believe that too many colleagues have a lot of confidence in the protein structures proposed by AlphaFold, whose self-appreciation is probably slightly exaggerated [91]. In fact, although it is usually very good in the prediction of globular proteins, this is unfortunately not the case for membrane proteins. Specific examples of AlphaFold’s low reliability in the case of membrane proteins have recently been published [41,42]. We should therefore not overestimate the capabilities of this program.
- (v)
- Conformational waves induced by ligands on membrane receptors are often interpreted as isolated phenomena totally disconnected from membrane lipids [92]. This is clearly not the case. The unique configuration of lipid rafts, functionally associating cholesterol molecules and gangliosides as well as tail-to-tail cholesterol dimers, underlines how these microdomains are adapted to the transmission of conformational information across the plasma membrane [64]. The establishment of these mechanisms required a long co-evolution of receptors and lipids, with cholesterol replacing bacterial hopanoids and gangliosides replacing ancestral glycosylated lipids [93,94,95,96]. Concomitantly, bacterial receptors gradually evolved into synaptic receptors, modulating hopanoid recognition patterns to make them even more efficient for raft cholesterol [97].
- (vi)
- If there is one area of receptology that is still very underrated, it is that involving quantum mechanisms [72]. However, the swipe card model of odorant recognition [73] is particularly attractive, and it should inspire vocations among our young researchers. This open field of research is an opportunity which should stimulate the imagination of our students. Incidentally, this should start with a questioning of professors who neglect this promising new dimension of biology. Indeed, quantum biology is not sufficiently taught in university biology courses.
- (vii)
- Finally, I consider the electrostatic surface potential as the most intuitive of fundamental notions [12]. This is perhaps the most important and, in any case, the most accessible parameter for understanding molecular interactions, which is still the basis of receptology.
6. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rang, H. The receptor concept: Pharmacology’s big idea. Br. J. Pharmacol. 2006, 147, S9–S16. [Google Scholar] [CrossRef]
- Urban, J.D.; Clarke, W.P.; Von Zastrow, M.; Nichols, D.E.; Kobilka, B.; Weinstein, H.; Javitch, J.A.; Roth, B.L.; Christopoulos, A.; Sexton, P.M. Functional selectivity and classical concepts of quantitative pharmacology. J. Pharmacol. Exp. Ther. 2007, 320, 1–13. [Google Scholar] [CrossRef]
- Stone, M.J.; Hayward, J.A.; Huang, C.; E. Huma, Z.; Sanchez, J. Mechanisms of Regulation of the Chemokine-Receptor Network. Int. J. Mol. Sci. 2017, 18, 342. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Yahi, N. Brain Lipids in Synaptic Function and Neurological Disease: Clues to Innovative Therapeutic Strategies for Brain Disorders; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Ben-Naim, A. The lock and key model for Molecular Recognition. Is it time for a paradigm shift? arXiv preprint 2018, arXiv:1806.03499. [Google Scholar]
- Wang, Y.; Zhuang, Y.; DiBerto, J.F.; Zhou, X.E.; Schmitz, G.P.; Yuan, Q.; Jain, M.K.; Liu, W.; Melcher, K.; Jiang, Y.; et al. Structures of the entire human opioid receptor family. Cell 2023, 186, 413–427.e417. [Google Scholar] [CrossRef]
- Gianni, S.; Dogan, J.; Jemth, P. Distinguishing induced fit from conformational selection. Biophys. Chem. 2014, 189, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Yu, E.W.; Koshland, D.E., Jr. Propagating conformational changes over long (and short) distances in proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 9517–9520. [Google Scholar] [CrossRef]
- Vogt, A.D.; Di Cera, E. Conformational selection is a dominant mechanism of ligand binding. Biochemistry 2013, 52, 5723–5729. [Google Scholar] [CrossRef] [PubMed]
- Changeux, J.-P.; Edelstein, S. Conformational selection or induced fit? 50 years of debate resolved. F1000 Biol. Rep. 2011, 3, 19. [Google Scholar] [CrossRef]
- Wlodarski, T.; Zagrovic, B. Conformational selection and induced fit mechanism underlie specificity in noncovalent interactions with ubiquitin. Proc. Natl. Acad. Sci. USA 2009, 106, 19346–19351. [Google Scholar] [CrossRef]
- Fantini, J.; Azzaz, F.; Chahinian, H.; Yahi, N. Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era. Viruses 2023, 15, 284. [Google Scholar] [CrossRef]
- Chaplin, M. Do we underestimate the importance of water in cell biology? Nat. Rev. Mol. Cell Biol. 2006, 7, 861–866. [Google Scholar] [CrossRef]
- Ringe, D. What makes a binding site a binding site? Curr. Opin. Struct. Biol. 1995, 5, 825–829. [Google Scholar] [CrossRef]
- Di Scala, C.; Fantini, J.; Yahi, N.; Barrantes, F.J.; Chahinian, H. Anandamide revisited: How cholesterol and ceramides control receptor-dependent and receptor-independent signal transmission pathways of a lipid neurotransmitter. Biomolecules 2018, 8, 31. [Google Scholar] [CrossRef]
- Breiten, B.; Lockett, M.R.; Sherman, W.; Fujita, S.; Al-Sayah, M.; Lange, H.; Bowers, C.M.; Heroux, A.; Krilov, G.; Whitesides, G.M. Water networks contribute to enthalpy/entropy compensation in protein–ligand binding. J. Am. Chem. Soc. 2013, 135, 15579–15584. [Google Scholar] [CrossRef] [PubMed]
- Quiocho, F.A.; Wilson, D.K.; Vyas, N.K. Substrate specificity and affinity of a protein modulated by bound water molecules. Nature 1989, 340, 404–407. [Google Scholar] [CrossRef]
- Renard, K.; Byrne, B. Insights into the role of membrane lipids in the structure, function and regulation of integral membrane proteins. Int. J. Mol. Sci. 2021, 22, 9026. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Barrantes, F.J. Sphingolipid/cholesterol regulation of neurotransmitter receptor conformation and function. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2009, 1788, 2345–2361. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Garmy, N.; Mahfoud, R.; Yahi, N. Lipid rafts: Structure, function and role in HIV, Alzheimer’s and prion diseases. Expert Rev. Mol. Med. 2002, 4, 1–22. [Google Scholar] [CrossRef]
- Simons, K.; Ikonen, E. Functional rafts in cell membranes. Nature 1997, 387, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Simons, K.; Toomre, D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000, 1, 31–39. [Google Scholar] [CrossRef]
- Samhan-Arias, A.K.; Poejo, J.; Marques-da-Silva, D.; Martínez-Costa, O.H.; Gutierrez-Merino, C. Are There Lipid Membrane-Domain Subtypes in Neurons with Different Roles in Calcium Signaling? Molecules 2023, 28, 7909. [Google Scholar] [CrossRef]
- Lee, A.G. How lipids and proteins interact in a membrane: A molecular approach. Mol. BioSystems 2005, 1, 203–212. [Google Scholar] [CrossRef]
- Levental, I.; Lyman, E. Regulation of membrane protein structure and function by their lipid nano-environment. Nat. Rev. Mol. Cell Biol. 2023, 24, 107–122. [Google Scholar] [CrossRef]
- Koehl, A.; Hu, H.; Feng, D.; Sun, B.; Zhang, Y.; Robertson, M.J.; Chu, M.; Kobilka, T.S.; Laeremans, T.; Steyaert, J.; et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 2019, 566, 79–84. [Google Scholar] [CrossRef]
- Matveeva, M.; Lefebvre, M.; Chahinian, H.; Yahi, N.; Fantini, J. Host membranes as drivers of virus evolution. Viruses 2023, 15, 1854. [Google Scholar] [CrossRef] [PubMed]
- Paila, Y.D.; Tiwari, S.; Sengupta, D.; Chattopadhyay, A. Molecular modeling of the human serotonin 1A receptor: Role of membrane cholesterol in ligand binding of the receptor. Molecular BioSystems 2021, 7, 224–234. [Google Scholar]
- Sarkar, P.; Chattopadhyay, A. Cholesterol interaction motifs in G protein-coupled receptors: Slippery hot spots? Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1481. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Epand, R.M.; Barrantes, F.J. Cholesterol-recognition motifs in membrane proteins. Direct Mech. Cholest. Modul. Protein Funct. 2019, 1135, 3–25. [Google Scholar]
- Di Scala, C.; Baier, C.J.; Evans, L.S.; Williamson, P.T.; Fantini, J.; Barrantes, F.J. Relevance of CARC and CRAC cholesterol-recognition motifs in the nicotinic acetylcholine receptor and other membrane-bound receptors. Curr. Top. Membr. 2017, 80, 3–23. [Google Scholar]
- Fantini, J.; Barrantes, F.J. How cholesterol interacts with membrane proteins: An exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 2013, 4, 31. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Di Scala, C.; Baier, C.J.; Barrantes, F.J. Molecular mechanisms of protein-cholesterol interactions in plasma membranes: Functional distinction between topological (tilted) and consensus (CARC/CRAC) domains. Chem. Phys. Lipids 2016, 199, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Di Scala, C.; Evans, L.S.; Williamson, P.T.; Barrantes, F.J. A mirror code for protein-cholesterol interactions in the two leaflets of biological membranes. Sci. Rep. 2016, 6, 21907. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J. Lipid rafts and human diseases: Why we need to target gangliosides. FEBS Open Bio 2023, 13, 1636–1650. [Google Scholar] [CrossRef] [PubMed]
- Azzaz, F.; Yahi, N.; Di Scala, C.; Chahinian, H.; Fantini, J. Ganglioside binding domains in proteins: Physiological and pathological mechanisms. Adv. Protein Chem. Struct. Biol. 2022, 128, 289–324. [Google Scholar] [PubMed]
- Xu, P.; Huang, S.; Zhang, H.; Mao, C.; Zhou, X.E.; Cheng, X.; Simon, I.A.; Shen, D.D.; Yen, H.Y.; Robinson, C.V.; et al. Structural insights into the lipid and ligand regulation of serotonin receptors. Nature 2021, 592, 469–473. [Google Scholar] [CrossRef]
- Gimpl, G.; Burger, K.; Fahrenholz, F. Cholesterol as modulator of receptor function. Biochemistry 1997, 36, 10959–10974. [Google Scholar] [CrossRef]
- Opekarova, M.; Tanner, W. Specific lipid requirements of membrane proteins—A putative bottleneck in heterologous expression. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2003, 1610, 11–22. [Google Scholar] [CrossRef]
- Han, R.; Yoon, H.; Kim, G.; Lee, H.; Lee, Y. Revolutionizing Medicinal Chemistry: The Application of Artificial Intelligence (AI) in Early Drug Discovery. Pharmaceuticals 2023, 16, 1259. [Google Scholar] [CrossRef]
- Azzaz, F.; Fantini, J. The epigenetic dimension of protein structure. Biomol. Concepts 2022, 13, 55–60. [Google Scholar] [CrossRef]
- Azzaz, F.; Yahi, N.; Chahinian, H.; Fantini, J. The Epigenetic Dimension of Protein Structure Is an Intrinsic Weakness of the AlphaFold Program. Biomolecules 2022, 12, 1527. [Google Scholar] [CrossRef] [PubMed]
- Tourlet, S.; Radjasandirane, R.; Diharce, J.; de Brevern, A.G. AlphaFold2 Update and Perspectives. BioMedInformatics 2023, 3, 378–390. [Google Scholar] [CrossRef]
- Boulos, I.; Jabbour, J.; Khoury, S.; Mikhael, N.; Tishkova, V.; Candoni, N.; Ghadieh, H.E.; Veesler, S.; Bassim, Y.; Azar, S.; et al. Exploring the World of Membrane Proteins: Techniques and Methods for Understanding Structure, Function, and Dynamics. Molecules 2023, 28, 7176. [Google Scholar] [CrossRef]
- Mani, H.; Chang, C.-C.; Hsu, H.-J.; Yang, C.-H.; Yen, J.-H.; Liou, J.-W. Comparison, Analysis, and Molecular Dynamics Simulations of Structures of a Viral Protein Modeled Using Various Computational Tools. Bioengineering 2023, 10, 1004. [Google Scholar] [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins and their environment: Effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009, 28, 305–325. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. The mysterious unfoldome: Structureless, underappreciated, yet vital part of any given proteome. J. Biomed. Biotechnol. 2010, 2010, 568068. [Google Scholar] [CrossRef] [PubMed]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015, 282, 1182–1189. [Google Scholar] [CrossRef]
- Uversky, V.N. Dancing Protein Clouds: The Strange Biology and Chaotic Physics of Intrinsically Disordered Proteins. J. Biol. Chem. 2016, 291, 6681–6688. [Google Scholar] [CrossRef]
- Pullen, R.A.; Jenkins, J.A.; Tickle, I.J.; Wood, S.P.; Blundell, T.L. The relation of polypeptide hormone structure and flexibility to receptor binding: The relevance of X-ray studies on insulins, glucagon and human placental lactogen. Mol. Cell. Biochem. 1975, 8, 5–20. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol. 2015, 16, 18–29. [Google Scholar] [CrossRef]
- Boesch, C.; Bundi, A.; Oppliger, M.; Wüthrich, K. 1H nuclear-magnetic-resonance studies of the molecular conformation of monomeric glucagon in aqueous solution. Eur. J. Biochem. 1978, 91, 209–214. [Google Scholar] [CrossRef]
- Zhang, H.; Qiao, A.; Yang, L.; Van Eps, N.; Frederiksen, K.S.; Yang, D.; Dai, A.; Cai, X.; Zhang, H.; Yi, C.; et al. Structure of the glucagon receptor in complex with a glucagon analogue. Nature 2018, 553, 106–110. [Google Scholar] [CrossRef]
- Chou, P.Y.; Fasman, G.D. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry 1974, 13, 211–222. [Google Scholar] [CrossRef]
- Sasaki, K.; Dockerill, S.; Adamiak, D.A.; Tickle, I.J.; Blundell, T. X-ray analysis of glucagon and its relationship to receptor binding. Nature 1975, 257, 751–757. [Google Scholar] [CrossRef]
- Shoemaker, B.A.; Portman, J.J.; Wolynes, P.G. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc. Natl. Acad. Sci. USA 2000, 97, 8868–8873. [Google Scholar] [CrossRef]
- Sugase, K.; Dyson, H.J.; Wright, P.E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 2007, 447, 1021–1025. [Google Scholar] [CrossRef]
- Endres, N.F.; Das, R.; Smith, A.W.; Arkhipov, A.; Kovacs, E.; Huang, Y.; Pelton, J.G.; Shan, Y.; Shaw, D.E.; Wemmer, D.E. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 2013, 152, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Ottemann, K.M.; Xiao, W.; Shin, Y.-K.; Koshland, D.E., Jr. A piston model for transmembrane signaling of the aspartate receptor. Science 1999, 285, 1751–1754. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xiao, X.; Lee, B.H.; Vu, S.; Yang, W.; Yarov-Yarovoy, V.; Zheng, J. The conformational wave in capsaicin activation of transient receptor potential vanilloid 1 ion channel. Nat. Commun. 2018, 9, 2879. [Google Scholar] [CrossRef] [PubMed]
- Cochary, E.F.; Bizzozero, O.A.; Sapirstein, V.S.; Nolan, C.E.; Fischer, I. Presence of the plasma membrane proteolipid (plasmolipin) in myelin. J. Neurochem. 1990, 55, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Shulgin, A.; Lebedev, T.; Prassolov, V.; Spirin, P. Plasmolipin and its role in cell processes. Mol. Biol. 2021, 55, 773–785. [Google Scholar] [CrossRef]
- Bosse, F.; Hasse, B.; Pippirs, U.; Greiner-Petter, R.; Müller, H.W. Proteolipid plasmolipin: Localization in polarized cells, regulated expression and lipid raft association in CNS and PNS myelin. J. Neurochem. 2003, 86, 508–518. [Google Scholar] [CrossRef]
- Azzaz, F.; Mazzarino, M.; Chahinian, H.; Yahi, N.; Scala, C.D.; Fantini, J. Structure of the Myelin Sheath Proteolipid Plasmolipin (PLLP) in a Ganglioside-Containing Lipid Raft. FBL 2023, 28, 157. [Google Scholar] [CrossRef]
- Harris, J.S.; Epps, D.E.; Davio, S.R.; Kezdy, F.J. Evidence for transbilayer, tail-to-tail cholesterol dimers in dipalmitoylglycerophosphocholine liposomes. Biochemistry 1995, 34, 3851–3857. [Google Scholar] [CrossRef]
- Naglekar, A.; Chattopadhyay, A.; Sengupta, D. Palmitoylation of the Glucagon-like Peptide-1 Receptor Modulates Cholesterol Interactions at the Receptor-Lipid Microenvironment. J. Phys. Chem. B 2023, 127, 11000–11010. [Google Scholar] [CrossRef]
- Khelashvili, G.; Menon, A.K. Phospholipid Scrambling by G Protein-Coupled Receptors. Annu. Rev. Biophys. 2022, 51, 39–61. [Google Scholar] [CrossRef]
- Turin, L. A Spectroscopic Mechanism for Primary Olfactory Reception. Chem. Senses 1996, 21, 773–791. [Google Scholar] [CrossRef] [PubMed]
- Bittner, E.R.; Madalan, A.; Czader, A.; Roman, G. Quantum origins of molecular recognition and olfaction in Drosophila. J. Chem. Phys. 2012, 137, 22a551. [Google Scholar] [CrossRef] [PubMed]
- Franco, M.I.; Turin, L.; Mershin, A.; Skoulakis, E.M. Molecular vibration-sensing component in Drosophila melanogaster olfaction. Proc. Natl. Acad. Sci. USA 2011, 108, 3797–3802. [Google Scholar] [CrossRef] [PubMed]
- Brookes, J.C.; Hartoutsiou, F.; Horsfield, A.P.; Stoneham, A.M. Could Humans Recognize Odor by Phonon Assisted Tunneling? Phys. Rev. Lett. 2007, 98, 038101. [Google Scholar] [CrossRef] [PubMed]
- Brookes, J.C. Quantum effects in biology: Golden rule in enzymes, olfaction, photosynthesis and magnetodetection. Proc. R. Soc. A Math. Phys. Eng. Sci. 2017, 473, 20160822. [Google Scholar] [CrossRef] [PubMed]
- Brookes, J.C.; Horsfield, A.P.; Stoneham, A.M. The Swipe Card Model of Odorant Recognition. Sensors 2012, 12, 15709–15749. [Google Scholar] [CrossRef] [PubMed]
- Weiner, P.K.; Langridge, R.; Blaney, J.M.; Schaefer, R.; Kollman, P.A. Electrostatic potential molecular surfaces. Proc. Natl. Acad. Sci. USA 1982, 79, 3754–3758. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. Phospholipid Synthesis and Transport in Mammalian Cells. Traffic 2015, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.; London, E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998, 14, 111–136. [Google Scholar] [CrossRef] [PubMed]
- Ikonen, E. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 2001, 13, 470–477. [Google Scholar] [CrossRef]
- Gousset, K.; Wolkers, W.F.; Tsvetkova, N.M.; Oliver, A.E.; Field, C.L.; Walker, N.J.; Crowe, J.H.; Tablin, F. Evidence for a physiological role for membrane rafts in human platelets. J. Cell. Physiol. 2002, 190, 117–128. [Google Scholar] [CrossRef]
- Barnett-Norris, J.; Lynch, D.; Reggio, P.H. Lipids, lipid rafts and caveolae: Their importance for GPCR signaling and their centrality to the endocannabinoid system. Life Sci. 2005, 77, 1625–1639. [Google Scholar] [CrossRef]
- Kawarabayashi, T.; Nakamura, T.; Sato, K.; Seino, Y.; Ichii, S.; Nakahata, N.; Takatama, M.; Westaway, D.; George-Hyslop, P.S.; Shoji, M. Lipid rafts act as a common platform for amyloid-β oligomer-induced Alzheimer’s disease pathology. J. Alzheimer’s Dis. 2022, 87, 1189–1203. [Google Scholar] [CrossRef]
- Sviridov, D.; Mukhamedova, N.; Miller, Y.I. Lipid rafts as a therapeutic target: Thematic review series: Biology of lipid rafts. J. Lipid Res. 2020, 61, 687–695. [Google Scholar] [CrossRef]
- Schengrund, C.-L. Lipid rafts: Keys to neurodegeneration. Brain Res. Bull. 2010, 82, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Mañes, S.; del Real, G.; Martínez-a, C. Pathogens: Raft hijackers. Nat. Rev. Immunol. 2003, 3, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Chahinian, H.; Yahi, N. Convergent Evolution Dynamics of SARS-CoV-2 and HIV Surface Envelope Glycoproteins Driven by Host Cell Surface Receptors and Lipid Rafts: Lessons for the Future. Int. J. Mol. Sci. 2023, 24, 1923. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Azzaz, F.; Chahinian, H. Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating Covid-19 outbreaks. J. Infect. 2021, 83, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, A.; Paila, Y.D.; Shrivastava, S.; Tiwari, S.; Singh, P.; Fantini, J. Sphingolipid-binding domain in the serotonin(1A) receptor. Adv. Exp. Med. Biol. 2012, 749, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, T.S.; Lightstone, F.C. An electrostatic funnel in the GABA-binding pathway. PLoS Comput. Biol. 2016, 12, e1004831. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Pucadyil, T.J.; Paila, Y.D.; Ganguly, S.; Chattopadhyay, A. Chronic cholesterol depletion using statin impairs the function and dynamics of human serotonin(1A) receptors. Biochemistry 2010, 49, 5426–5435. [Google Scholar] [CrossRef] [PubMed]
- Burger, K.; Gimpl, G.; Fahrenholz, F. Regulation of receptor function by cholesterol. Cell. Mol. Life Sci. CMLS 2000, 57, 1577–1592. [Google Scholar] [CrossRef]
- Miljan, E.A.; Bremer, E.G. Regulation of growth factor receptors by gangliosides. Sci. STKE Signal Transduct. Knowl. Environ. 2002, 2002, re15. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Grosman, C.; Zhou, M.; Auerbach, A. Mapping the conformational wave of acetylcholine receptor channel gating. Nature 2000, 403, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Bloch, K.E. Speculations on the evolution of sterol structure and function. CRC Crit. Rev. Biochem. 1979, 7, 1–5. [Google Scholar] [CrossRef]
- Yao, W.; Zhang, W.; He, W.; Xiao, W.; Chen, Y.; Zhu, Y.; Zheng, F.; Zhang, C. Lipidomic chemotaxonomy aligned with phylogeny of Halobacteria. Front. Microbiol. 2023, 14, 1297600. [Google Scholar] [CrossRef]
- Ourisson, G.; Rohmer, M.; Poralla, K. Prokaryotic hopanoids and other polyterpenoid sterol surrogates. Annu. Rev. Microbiol. 1987, 41, 301–333. [Google Scholar] [CrossRef]
- Pearson, A. Resolving a piece of the archaeal lipid puzzle. Proc. Natl. Acad. Sci. USA 2019, 116, 22423–22425. [Google Scholar] [CrossRef] [PubMed]
- Barrantes, F.J.; Fantini, J. From hopanoids to cholesterol: Molecular clocks of pentameric ligand-gated ion channels. Prog. Lipid Res. 2016, 63, 1–13. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fantini, J. Fundamental Mechanisms in Membrane Receptology: Old Paradigms, New Concepts and Perspectives. Receptors 2024, 3, 107-121. https://doi.org/10.3390/receptors3010006
Fantini J. Fundamental Mechanisms in Membrane Receptology: Old Paradigms, New Concepts and Perspectives. Receptors. 2024; 3(1):107-121. https://doi.org/10.3390/receptors3010006
Chicago/Turabian StyleFantini, Jacques. 2024. "Fundamental Mechanisms in Membrane Receptology: Old Paradigms, New Concepts and Perspectives" Receptors 3, no. 1: 107-121. https://doi.org/10.3390/receptors3010006
APA StyleFantini, J. (2024). Fundamental Mechanisms in Membrane Receptology: Old Paradigms, New Concepts and Perspectives. Receptors, 3(1), 107-121. https://doi.org/10.3390/receptors3010006





