The NR4A Orphan Receptor Modulator C-DIM12 Selectively Alters Inflammatory Mediators in Myeloid Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. MTT Assay
2.3. Resazurin Assay
2.4. RealTime-Glo™ MT Cell Viability Assay
2.5. CellTox™ Green Cytotoxicity Assay
2.6. MultiTox-Fluor Multiplex Cytotoxicity Assay
- Viability substrate: excitation ~400 nm; emission ~505 nm.
- Cytotoxicity substrate: excitation ~485 nm; emission ~520 nm.
2.7. Caspase-Glo3/7 Assay
2.8. Proteome Profiler Human XL Cytokine Array Kit
2.9. MCP-1 and CCL5 ELISA
2.10. CXCL1/GROα ELISA
2.11. QUANTI-Luc™
2.12. Gene Expression Analysis
2.13. Glucose Uptake
2.14. Real-Time Cell Metabolic Analysis
2.15. Statistics
3. Results
Effect of C-DIM12 on Inflammatory Outcome in THP-1 Cells
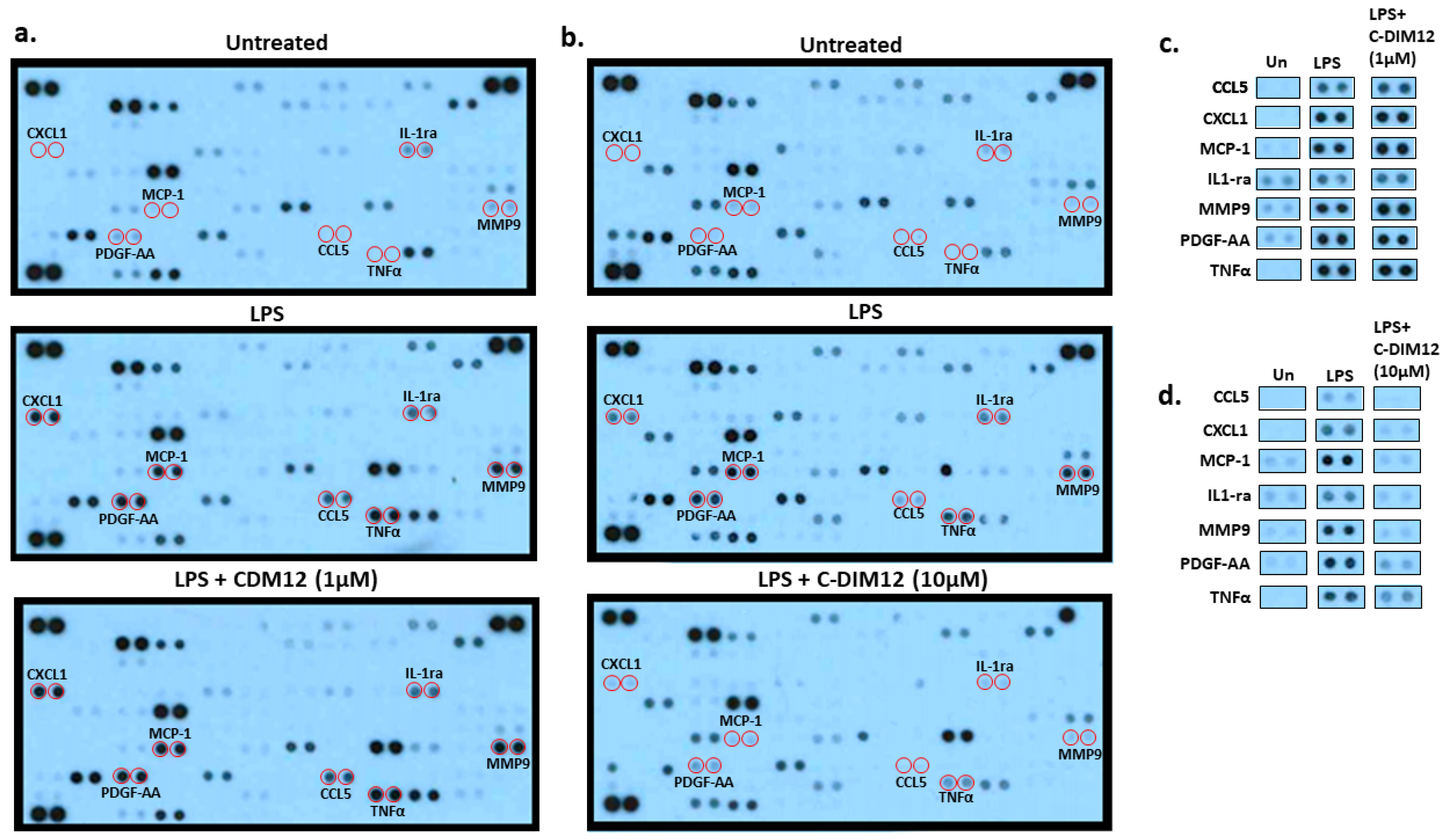


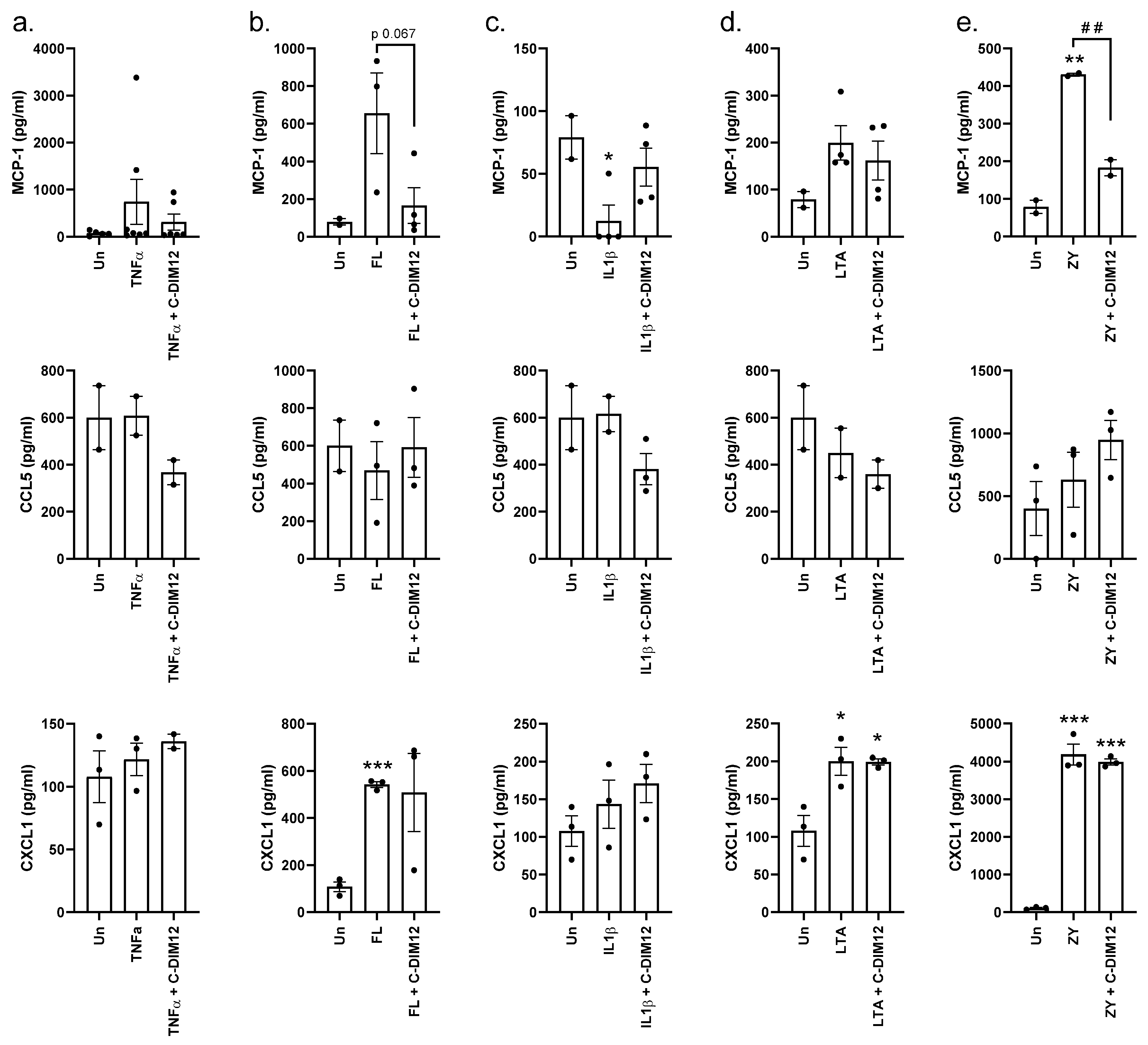



4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| α-KG | α-ketoglutarate |
| CCL5 | Chemokine ligand 5 |
| Csn-B | Cytosporone B |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 |
| C-DIM12 | 3-[(4-Chlorophenyl)-(1H-indol-3-yl)methyl]-1H-indole |
| FL | Flagellin |
| KO | Knock out |
| IL-1β | Interleukin-1 beta |
| IL-1R | Interleukin-1 receptor |
| IL-1ra | Interleukin-1 receptor alpha |
| LPS | Lipopolysaccharide |
| LTA | Lipoteichoic acid |
| MCP-1 | Monocyte chemoattract protein-1 |
| MMP9 | Matrix metalloproteinase 9 |
| MPP+ | 1-methyl-4-phenylpyridinium |
| MPTP | 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine |
| MyD88 | Myeloid differentiation primary response 88 |
| NBRE | NR4A binding motifs |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NR4A | Orphan nuclear receptor subfamily 4A |
| NRE | NF-κB binding motifs |
| PDGF-AA | Platelet-derived growth factor-AA |
| TLR | Toll-Like Receptor |
| TNFα | Tumour necrosis factor alpha |
| TNFR | Tumour necrosis factor receptor |
| ZY | Zymosan |
References
- Kurakula, K.; Koenis, D.S.; van Tiel, C.M.; de Vries, C.J. NR4A nuclear receptors are orphans but not lonesome. Biochim. Biophys. Acta 2014, 1843, 2543–2555. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.P.; Crean, D. Molecular Interactions between NR4A Orphan Nuclear Receptors and NF-κB Are Required for Appropriate Inflammatory Responses and Immune Cell Homeostasis. Biomolecules 2015, 5, 1302–1318. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Bruemmer, D. NR4A Orphan Nuclear Receptors in Cardiovascular Biology. Drug Discov. Today Dis. Mech. 2009, 6, e43–e48. [Google Scholar] [CrossRef]
- Ashouri, J.F.; Hsu, L.Y.; Yu, S.; Rychkov, D.; Chen, Y.; Cheng, D.A.; Sirota, M.; Hansen, E.; Lattanza, L.; Zikherman, J.; et al. Reporters of TCR signaling identify arthritogenic T cells in murine and human autoimmune arthritis. Proc. Natl. Acad. Sci. USA 2019, 116, 18517–18527. [Google Scholar] [CrossRef] [PubMed]
- Elcombe, S.E.; Naqvi, S.; Van Den Bosch, M.W.; MacKenzie, K.F.; Cianfanelli, F.; Brown, G.D.; Arthur, J.S. Dectin-1 regulates IL-10 production via a MSK1/2 and CREB dependent pathway and promotes the induction of regulatory macrophage markers. PLoS ONE 2013, 8, e60086. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Saini, A.; Chandra, V.; Nanduri, R.; Kalra, R.; Bhagyaraj, E.; Khatri, N.; Gupta, P. Nuclear Receptor Nr4a2 Promotes Alternative Polarization of Macrophages and Confers Protection in Sepsis. J. Biol. Chem. 2015, 290, 18304–18314. [Google Scholar] [CrossRef] [PubMed]
- Crean, D.; Cummins, E.P.; Bahar, B.; Mohan, H.; McMorrow, J.P.; Murphy, E.P. Adenosine Modulates NR4A Orphan Nuclear Receptors to Attenuate Hyperinflammatory Responses in Monocytic Cells. J. Immunol. 2015, 195, 1436–1448. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, C.; de Gaetano, M.; Giffney, H.E.; Bahar, B.; Cummins, E.P.; Brennan, E.P.; Barry, M.; Belton, O.; Godson, C.G.; Murphy, E.P.; et al. NR4A Receptors Differentially Regulate NF-κB Signaling in Myeloid Cells. Front. Immunol. 2017, 8, 7. [Google Scholar] [CrossRef]
- Pei, L.; Castrillo, A.; Chen, M.; Hoffmann, A.; Tontonoz, P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J. Biol. Chem. 2005, 280, 29256–29262. [Google Scholar] [CrossRef]
- Afzali, M.F.; Popichak, K.A.; Burton, L.H.; Klochak, A.L.; Wilson, W.J.; Safe, S.; Tjalkens, R.B.; Legare, M.E. A novel diindolylmethane analog, 1,1-bis(3′-indolyl)-1-(p-chlorophenyl) methane, inhibits the tumor necrosis factor-induced inflammatory response in primary murine synovial fibroblasts through a Nurr1-dependent mechanism. Mol. Immunol. 2018, 101, 46–54. [Google Scholar] [CrossRef]
- McEvoy, A.N.; Murphy, E.A.; Ponnio, T.; Conneely, O.M.; Bresnihan, B.; FitzGerald, O.; Murphy, E.P. Activation of nuclear orphan receptor NURR1 transcription by NF-kappa B and cyclic adenosine 5’-monophosphate response element-binding protein in rheumatoid arthritis synovial tissue. J. Immunol. 2002, 168, 2979–2987. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Ran, J.; Xu, L.; Tong, Z.; Adel Abdo, M.S.; Ma, C.; Xu, K.; He, Y.; Wu, Z.; Chen, Z.; et al. Reactivation of NR4A1 Restrains Chondrocyte Inflammation and Ameliorates Osteoarthritis in Rats. Front. Cell Dev. Biol. 2020, 8, 158. [Google Scholar] [CrossRef] [PubMed]
- Lappas, M. Effect of spontaneous term labour on the expression of the NR4A receptors nuclear receptor related 1 protein (Nurr1), neuron-derived clone 77 (Nur77) and neuron-derived orphan receptor 1 (NOR1) in human fetal membranes and myometrium. Reprod. Fertil. Dev. 2016, 28, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Regulation of NF-κB by TNF family cytokines. Semin. Immunol. 2014, 26, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Haddad, M. The Impact of CB1 Receptor on Nuclear Receptors in Skeletal Muscle Cells. Pathophysiol. Off. J. Int. Soc. Pathophysiol. 2021, 28, 457–470. [Google Scholar] [CrossRef]
- Giffney, H.E.; Cummins, E.P.; Murphy, E.P.; Brayden, D.J.; Crean, D. Protein kinase D, ubiquitin and proteasome pathways are involved in adenosine receptor-stimulated NR4A expression in myeloid cells. Biochem. Biophys. Res. Commun. 2021, 555, 19–25. [Google Scholar] [CrossRef]
- Haddad, M. The Impact of Adenosine A2B Receptors Modulation on Nuclear Receptors (NR4A) Gene Expression. Biomed. Pharmacol. J. 2016, 9, 177–185. [Google Scholar] [CrossRef]
- Chintharlapalli, S.; Burghardt, R.; Papineni, S.; Ramaiah, S.; Yoon, K.; Safe, S. Activation of Nur77 by selected 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes induces apoptosis through nuclear pathways. J. Biol. Chem. 2005, 280, 24903–24914. [Google Scholar] [CrossRef]
- Dubois, C.; Hengerer, B.; Mattes, H. Identification of a potent agonist of the orphan nuclear receptor Nurr1. ChemMedChem 2006, 1, 955–958. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Tello, P.; Lin, H.; Khan, P.; de Vera, I.M.S.; Kamenecka, T.M.; Kojetin, D.J. Assessment of NR4A Ligands That Directly Bind and Modulate the Orphan Nuclear Receptor Nurr1. J. Med. Chem. 2020, 63, 15639–15654. [Google Scholar] [CrossRef] [PubMed]
- Safe, S.; Shrestha, R.; Mohankumar, K. Orphan nuclear receptor 4A1 (NR4A1) and novel ligands. Essays Biochem. 2021, 65, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Du, X.; Chen, H.; Liu, J.; Zhao, B.; Huang, D.; Li, G.; Xu, Q.; Zhang, M.; Weimer, B.C.; et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat. Chem. Biol. 2008, 4, 548–556. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, B.R.; Miller, J.A.; Hansen, R.J.; Lunghofer, P.J.; Safe, S.; Gustafson, D.L.; Colagiovanni, D.; Tjalkens, R.B. Neuroprotective efficacy and pharmacokinetic behavior of novel anti-inflammatory para-phenyl substituted diindolylmethanes in a mouse model of Parkinson’s disease. J. Pharmacol. Exp. Ther. 2013, 345, 125–138. [Google Scholar] [CrossRef] [PubMed]
- De Miranda, B.R.; Popichak, K.A.; Hammond, S.L.; Jorgensen, B.A.; Phillips, A.T.; Safe, S.; Tjalkens, R.B. The Nurr1 Activator 1,1-Bis(3′-Indolyl)-1-(p-Chlorophenyl)Methane Blocks Inflammatory Gene Expression in BV-2 Microglial Cells by Inhibiting Nuclear Factor κB. Mol. Pharmacol. 2015, 87, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Le, S.; Wang, K.; Su, Y.; Chen, S.; Wu, C.; Chen, J.; Chen, S.; Zhang, A.; Xia, J. Cytosporone B (Csn-B), an NR4A1 agonist, attenuates acute cardiac allograft rejection by inducing differential apoptosis of CD4+T cells. Int. Immunopharmacol. 2022, 104, 108521. [Google Scholar] [CrossRef]
- Inamoto, T.; Papineni, S.; Chintharlapalli, S.; Cho, S.D.; Safe, S.; Kamat, A.M. 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol. Cancer Ther. 2008, 7, 3825–3833. [Google Scholar] [CrossRef]
- Pulakazhi Venu, V.K.; Alston, L.; Iftinca, M.; Tsai, Y.C.; Stephens, M.; Warriyar, K.V.V.; Rehal, S.; Hudson, G.; Szczepanski, H.; von der Weid, P.Y.; et al. Nr4A1 modulates inflammation-associated intestinal fibrosis and dampens fibrogenic signaling in myofibroblasts. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 321, G280–G297. [Google Scholar] [CrossRef]
- De Miranda, B.R.; Popichak, K.A.; Hammond, S.L.; Miller, J.A.; Safe, S.; Tjalkens, R.B. Novel para-phenyl substituted diindolylmethanes protect against MPTP neurotoxicity and suppress glial activation in a mouse model of Parkinson’s disease. Toxicol. Sci. Off. J. Soc. Toxicol. 2015, 143, 360–373. [Google Scholar] [CrossRef]
- Ismaiel, M.; Murphy, B.; Aldhafiri, S.; Giffney, H.E.; Thornton, K.; Mukhopadhya, A.; Keogh, C.E.; Fattah, S.; Mohan, H.M.; Cummins, E.P.; et al. The NR4A agonist, Cytosporone B, attenuates pro-inflammatory mediators in human colorectal cancer tissue ex vivo. Biochem. Biophys. Res. Commun. 2021, 554, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Li, X.M.; Wang, J.R.; Gan, W.J.; Jiang, F.Q.; Liu, Y.; Zhang, X.D.; He, X.S.; Zhao, Y.Y.; Lu, X.X.; et al. NUR77 exerts a protective effect against inflammatory bowel disease by negatively regulating the TRAF6/TLR-IL-1R signalling axis. J. Pathol. 2016, 238, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Chanput, W.; Mes, J.J.; Wichers, H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014, 23, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Phelan, D.E.; Shigemura, M.; Aldhafiri, S.; Mota, C.; Hall, T.J.; Sznajder, J.I.; Murphy, E.P.; Crean, D.; Cummins, E.P. Transcriptional Profiling of Monocytes Deficient in Nuclear Orphan Receptors NR4A2 and NR4A3 Reveals Distinct Signalling Roles Related to Antigen Presentation and Viral Response. Front. Immunol. 2021, 12, 676644. [Google Scholar] [CrossRef] [PubMed]
- Promega. Available online: https://worldwide.promega.com/products/cell-health-assays/cell-viability-and-cytotoxicity-assays/realtime_glo-mt-cell-viability-assay/?catNum=G9711 (accessed on 5 December 2023).
- Hammond, S.L.; Safe, S.; Tjalkens, R.B. A novel synthetic activator of Nurr1 induces dopaminergic gene expression and protects against 6-hydroxydopamine neurotoxicity in vitro. Neurosci. Lett. 2015, 607, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, X.; Tu, Y.; Ding, D.; Yi, Q.; Sun, X.; Wang, Y.; Wang, K.; Zhu, M.; Mao, J. Dysfunctional Nurr1 promotes high glucose-induced Müller cell activation by up-regulating the NF-κB/NLRP3 inflammasome axis. Neuropeptides 2020, 82, 102057. [Google Scholar] [CrossRef]
- Bonta, P.I.; Matlung, H.L.; Vos, M.; Peters, S.L.; Pannekoek, H.; Bakker, E.N.; de Vries, C.J. Nuclear receptor Nur77 inhibits vascular outward remodelling and reduces macrophage accumulation and matrix metalloproteinase levels. Cardiovasc. Res. 2010, 87, 561–568. [Google Scholar] [CrossRef]
- Bonta, P.I.; van Tiel, C.M.; Vos, M.; Pols, T.W.; van Thienen, J.V.; Ferreira, V.; Arkenbout, E.K.; Seppen, J.; Spek, C.A.; van der Poll, T.; et al. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 2288–2294. [Google Scholar] [CrossRef]
- De Paoli, F.; Eeckhoute, J.; Copin, C.; Vanhoutte, J.; Duhem, C.; Derudas, B.; Dubois-Chevalier, J.; Colin, S.; Zawadzki, C.; Jude, B.; et al. The neuron-derived orphan receptor 1 (NOR1) is induced upon human alternative macrophage polarization and stimulates the expression of markers of the M2 phenotype. Atherosclerosis 2015, 241, 18–26. [Google Scholar] [CrossRef]
- Hamers, A.A.; Argmann, C.; Moerland, P.D.; Koenis, D.S.; Marinković, G.; Sokolović, M.; de Vos, A.F.; de Vries, C.J.; van Tiel, C.M. Nur77-deficiency in bone marrow-derived macrophages modulates inflammatory responses, extracellular matrix homeostasis, phagocytosis and tolerance. BMC Genom. 2016, 17, 162. [Google Scholar] [CrossRef] [PubMed]
- Hamers, A.A.; van Dam, L.; Teixeira Duarte, J.M.; Vos, M.; Marinković, G.; van Tiel, C.M.; Meijer, S.L.; van Stalborch, A.M.; Huveneers, S.; Te Velde, A.A.; et al. Deficiency of Nuclear Receptor Nur77 Aggravates Mouse Experimental Colitis by Increased NFκB Activity in Macrophages. PLoS ONE 2015, 10, e0133598. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.N.; Shaked, I.; Hubbeling, H.G.; Punt, J.A.; Wu, R.; Herrley, E.; Zaugg, C.; Pei, H.; Geissmann, F.; Ley, K.; et al. NR4A1 (Nur77) deletion polarizes macrophages toward an inflammatory phenotype and increases atherosclerosis. Circ. Res. 2012, 110, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Kurakula, K.; Vos, M.; Logiantara, A.; Roelofs, J.J.; Nieuwenhuis, M.A.; Koppelman, G.H.; Postma, D.S.; van Rijt, L.S.; de Vries, C.J. Nuclear Receptor Nur77 Attenuates Airway Inflammation in Mice by Suppressing NF-κB Activity in Lung Epithelial Cells. J. Immunol. 2015, 195, 1388–1398. [Google Scholar] [CrossRef] [PubMed]
- Lappas, M. The NR4A receptors Nurr1 and Nur77 are increased in human placenta from women with gestational diabetes. Placenta 2014, 35, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Mix, K.S.; Attur, M.G.; Al-Mussawir, H.; Abramson, S.B.; Brinckerhoff, C.E.; Murphy, E.P. Transcriptional repression of matrix metalloproteinase gene expression by the orphan nuclear receptor NURR1 in cartilage. J. Biol. Chem. 2007, 282, 9492–9504. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.T.; Lakshmi, S.P.; Banno, A.; Jadhav, S.K.; Pulikkal Kadamberi, I.; Kim, S.C.; Reddy, R.C. Cigarette smoke downregulates Nur77 to exacerbate inflammation in chronic obstructive pulmonary disease (COPD). PLoS ONE 2020, 15, e0229256. [Google Scholar] [CrossRef]
- Rodríguez-Calvo, R.; Ferrán, B.; Alonso, J.; Martí-Pàmies, I.; Aguiló, S.; Calvayrac, O.; Rodríguez, C.; Martínez-González, J. NR4A receptors up-regulate the antiproteinase alpha-2 macroglobulin (A2M) and modulate MMP-2 and MMP-9 in vascular smooth muscle cells. Thromb. Haemost. 2015, 113, 1323–1334. [Google Scholar] [CrossRef]
- Saijo, K.; Winner, B.; Carson, C.T.; Collier, J.G.; Boyer, L.; Rosenfeld, M.G.; Gage, F.H.; Glass, C.K. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 2009, 137, 47–59. [Google Scholar] [CrossRef]
- Sekiya, T.; Kashiwagi, I.; Inoue, N.; Morita, R.; Hori, S.; Waldmann, H.; Rudensky, A.Y.; Ichinose, H.; Metzger, D.; Chambon, P.; et al. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat. Commun. 2011, 2, 269. [Google Scholar] [CrossRef]
- Sekiya, T.; Kashiwagi, I.; Yoshida, R.; Fukaya, T.; Morita, R.; Kimura, A.; Ichinose, H.; Metzger, D.; Chambon, P.; Yoshimura, A. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat. Immunol. 2013, 14, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.R.; Gan, W.J.; Li, X.M.; Zhao, Y.Y.; Li, Y.; Lu, X.X.; Li, J.M.; Wu, H. Orphan nuclear receptor Nur77 promotes colorectal cancer invasion and metastasis by regulating MMP-9 and E-cadherin. Carcinogenesis 2014, 35, 2474–2484. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.L.; Popichak, K.A.; Li, X.; Hunt, L.G.; Richman, E.H.; Damale, P.U.; Chong, E.K.P.; Backos, D.S.; Safe, S.; Tjalkens, R.B. The Nurr1 Ligand,1,1-bis(3′-Indolyl)-1-(p-Chlorophenyl)Methane, Modulates Glial Reactivity and Is Neuroprotective in MPTP-Induced Parkinsonism. J. Pharmacol. Exp. Ther. 2018, 365, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Donadelli, R.; Abbate, M.; Zanchi, C.; Corna, D.; Tomasoni, S.; Benigni, A.; Remuzzi, G.; Zoja, C. Protein traffic activates NF-kB gene signaling and promotes MCP-1-dependent interstitial inflammation. Am. J. Kidney Dis. Off. J. Natl. Kidney Found. 2000, 36, 1226–1241. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rangan, G.K.; Goodwin, B.; Tay, Y.C.; Harris, D.C. Lipopolysaccharide-induced MCP-1 gene expression in rat tubular epithelial cells is nuclear factor-kappaB dependent. Kidney Int. 2000, 57, 2011–2022. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zeng, Y.; Huang, X.; Qin, Y.; Luo, W.; Xiang, S.; Sooranna, S.R.; Pinhu, L. Nur77 attenuates endothelin-1 expression via downregulation of NF-κB and p38 MAPK in A549 cells and in an ARDS rat model. Am. J. Physiology Lung Cell. Mol. Physiol. 2016, 311, L1023–L1035. [Google Scholar] [CrossRef]
- de Gaetano, M.; Tighe, C.; Gahan, K.; Zanetti, A.; Chen, J.; Newson, J.; Cacace, A.; Marai, M.; Gaffney, A.; Brennan, E.; et al. Asymmetric Synthesis and Biological Screening of Quinoxaline-Containing Synthetic Lipoxin A(4) Mimetics (QNX-sLXms). J. Med. Chem. 2021, 64, 9193–9216. [Google Scholar] [CrossRef]
- Mees, G.; Dierckx, R.; Vangestel, C.; Laukens, D.; Van Damme, N.; Van de Wiele, C. Pharmacologic activation of tumor hypoxia: A means to increase tumor 2-deoxy-2-[18F]fluoro-D-glucose uptake? Mol. Imaging 2013, 12, 49–58. [Google Scholar]
- Han, Q.W.; Shao, Q.H.; Wang, X.T.; Ma, K.L.; Chen, N.H.; Yuan, Y.H. CB2 receptor activation inhibits the phagocytic function of microglia through activating ERK/AKT-Nurr1 signal pathways. Acta Pharmacol. Sin. 2022, 43, 2253–2266. [Google Scholar] [CrossRef]
- Cesta, M.F.; Ryman-Rasmussen, J.P.; Wallace, D.G.; Masinde, T.; Hurlburt, G.; Taylor, A.J.; Bonner, J.C. Bacterial lipopolysaccharide enhances PDGF signaling and pulmonary fibrosis in rats exposed to carbon nanotubes. Am. J. Respir. Cell Mol. Biol. 2010, 43, 142–151. [Google Scholar] [CrossRef]
- Ho, H.H.; Antoniv, T.T.; Ji, J.D.; Ivashkiv, L.B. Lipopolysaccharide-induced expression of matrix metalloproteinases in human monocytes is suppressed by IFN-gamma via superinduction of ATF-3 and suppression of AP-1. J. Immunol. 2008, 181, 5089–5097. [Google Scholar] [CrossRef] [PubMed]
- Madej, M.P.; Töpfer, E.; Boraschi, D.; Italiani, P. Different Regulation of Interleukin-1 Production and Activity in Monocytes and Macrophages: Innate Memory as an Endogenous Mechanism of IL-1 Inhibition. Front. Pharmacol. 2017, 8, 335. [Google Scholar] [CrossRef] [PubMed]
- Shaked, I.; Hanna, R.N.; Shaked, H.; Chodaczek, G.; Nowyhed, H.N.; Tweet, G.; Tacke, R.; Basat, A.B.; Mikulski, Z.; Togher, S.; et al. Transcription factor Nr4a1 couples sympathetic and inflammatory cues in CNS-recruited macrophages to limit neuroinflammation. Nat. Immunol. 2015, 16, 1228–1234. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Song, J.R.; Zhao, M.J. NR4A1 regulates cerebral ischemia-induced brain injury by regulating neuroinflammation through interaction with NF-κB/p65. Biochem. Biophys. Res. Commun. 2019, 518, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Burke, S.J.; Lu, D.; Sparer, T.E.; Masi, T.; Goff, M.R.; Karlstad, M.D.; Collier, J.J. NF-κB and STAT1 control CXCL1 and CXCL2 gene transcription. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E131–E149. [Google Scholar] [CrossRef] [PubMed]
- Pocock, J.; Gómez-Guerrero, C.; Harendza, S.; Ayoub, M.; Hernández-Vargas, P.; Zahner, G.; Stahl, R.A.; Thaiss, F. Differential activation of NF-kappa B, AP-1, and C/EBP in endotoxin-tolerant rats: Mechanisms for in vivo regulation of glomerular RANTES/CCL5 expression. J. Immunol. 2003, 170, 6280–6291. [Google Scholar] [CrossRef] [PubMed]
- Thompson, W.L.; Van Eldik, L.J. Inflammatory cytokines stimulate the chemokines CCL2/MCP-1 and CCL7/MCP-3 through NFkB and MAPK dependent pathways in rat astrocytes [corrected]. Brain Res. 2009, 1287, 47–57. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Hardie, D.G. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature 2013, 493, 346–355. [Google Scholar] [CrossRef]
- Tannahill, G.M.; Curtis, A.M.; Adamik, J.; Palsson-McDermott, E.M.; McGettrick, A.F.; Goel, G.; Frezza, C.; Bernard, N.J.; Kelly, B.; Foley, N.H.; et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. [Google Scholar] [CrossRef]
- Amoasii, L.; Holland, W.; Sanchez-Ortiz, E.; Baskin, K.K.; Pearson, M.; Burgess, S.C.; Nelson, B.R.; Bassel-Duby, R.; Olson, E.N. A MED13-dependent skeletal muscle gene program controls systemic glucose homeostasis and hepatic metabolism. Genes Dev. 2016, 30, 434–446. [Google Scholar] [CrossRef]
- Amoasii, L.; Sanchez-Ortiz, E.; Fujikawa, T.; Elmquist, J.K.; Bassel-Duby, R.; Olson, E.N. NURR1 activation in skeletal muscle controls systemic energy homeostasis. Proc. Natl. Acad. Sci. USA 2019, 116, 11299–11308. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldhafiri, S.; Marai, M.; Ismaiel, M.; Murphy, B.; Giffney, H.E.; Hall, T.J.; Murphy, E.P.; Cummins, E.P.; Crean, D. The NR4A Orphan Receptor Modulator C-DIM12 Selectively Alters Inflammatory Mediators in Myeloid Cells. Receptors 2023, 2, 264-283. https://doi.org/10.3390/receptors2040018
Aldhafiri S, Marai M, Ismaiel M, Murphy B, Giffney HE, Hall TJ, Murphy EP, Cummins EP, Crean D. The NR4A Orphan Receptor Modulator C-DIM12 Selectively Alters Inflammatory Mediators in Myeloid Cells. Receptors. 2023; 2(4):264-283. https://doi.org/10.3390/receptors2040018
Chicago/Turabian StyleAldhafiri, Sarah, Mariam Marai, Mohamed Ismaiel, Brenda Murphy, Hugh E. Giffney, Thomas J. Hall, Evelyn P. Murphy, Eoin P. Cummins, and Daniel Crean. 2023. "The NR4A Orphan Receptor Modulator C-DIM12 Selectively Alters Inflammatory Mediators in Myeloid Cells" Receptors 2, no. 4: 264-283. https://doi.org/10.3390/receptors2040018
APA StyleAldhafiri, S., Marai, M., Ismaiel, M., Murphy, B., Giffney, H. E., Hall, T. J., Murphy, E. P., Cummins, E. P., & Crean, D. (2023). The NR4A Orphan Receptor Modulator C-DIM12 Selectively Alters Inflammatory Mediators in Myeloid Cells. Receptors, 2(4), 264-283. https://doi.org/10.3390/receptors2040018





