Phenotyping Diabetes Mellitus on Aggregated Electronic Health Records from Disparate Health Systems
Abstract
1. Introduction
2. Results
| Atrial Fibrillation Cohort (n = 1194) | Random Hospitalized Sample (n = 1619) | ||||
|---|---|---|---|---|---|
| 2019 (n = 608) | 2020 (n = 586) | 2019 (n = 808) | 2020 (n = 811) | ||
| Sex, n (%) | Male | 305 (50.2%) | 310 (52.9%) | 380 (47.0%) | 401 (49.4%) |
| Female | 303 (49.8%) | 276 (47.1%) | 428 (53.0%) | 410 (50.6%) | |
| Race, n (%) | Chinese | 451 (74.2%) | 458 (78.2%) | 514 (63.6%) | 489 (60.3%) |
| Malay | 92 (15.1%) | 81 (13.8%) | 139 (17.2%) | 137 (16.8%) | |
| Indian | 29 (4.8%) | 25 (4.3%) | 84 (10.4%) | 99 (12.3%) | |
| Others | 36 (5.9%) | 22 (3.8%) | 71 (8.8%) | 86 (10.6%) | |
| Age | Mean | 72.2 | 72.4 | 47.5 | 45.8 |
| Standard deviation | 11.8 | 12.0 | 28.8 | 27.5 | |
| Diabetes | Yes | 228 (37.5%) | 229 (39.1%) | 198 (24.5%) | 169 (20.8%) |
| No | 380 (62.5%) | 357 (60.9%) | 610 (75.5%) | 642 (79.2%) | |
3. Methodology
3.1. Study Setting and Algorithm Development
3.2. Validation Population and Chart Review
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Diagnosis Code | Description of Code |
|---|---|
| 200687002 | Cellulitis in diabetic foot |
| 73211009 | Diabetes mellitus (DM) |
| 280137006 | Diabetic foot |
| 371087003 | Diabetic foot ulcer |
| 310505005 | Diabetic hyperosmolar non-ketotic state |
| 312912001 | Diabetic macular oedema |
| 399864000 | Diabetic macular oedema not clinically significant |
| 232020009 | Diabetic maculopathy |
| 25093002 | Diabetic oculopathy (eye disease) |
| 49455004 | Diabetic polyneuropathy |
| 268519009 | Diabetic—poor control |
| 127014009 | Diabetic peripheral vascular disease (angiopathy) |
| 127013003 | Diabetic renal disease |
| 4855003 | Diabetic retinopathy |
| 420789003 | Diabetic retinopathy associated with OR due to Type 1 DM |
| 232023006 | Diabetic traction retinal detachment |
| 312910009 | Diabetic vitreous hemorrhage |
| 402864004 | Diabetic wet gangrene of the foot |
| 441656006 | Hyperglycemic crisis due to OR in DM |
| 237633009 | Hypoglycemia due to DM |
| 421750000 | Ketoacidosis due to Type 2 DM |
| 420422005 | Ketoacidosis in DM |
| 426875007 | Latent autoimmune DM in adults (LADA) |
| 236499007 | Microalbuminuric diabetic nephropathy |
| 312903003 | Mild non-proliferative diabetic retinopathy |
| 312904009 | Moderate non-proliferative diabetic retinopathy |
| 230572002 | Neuropathy due to DM |
| 405749004 | Newly diagnosed diabetes |
| 390834004 | Non-proliferative diabetic retinopathy (NPDR)/Background diabetic retinopathy (BDR) |
| 59276001 | Proliferative diabetic retinopathy (PDR) |
| 236500003 | Proteinuric diabetic nephropathy |
| 312905005 | Severe non-proliferative diabetic retinopathy |
| 46635009 | Type 1 DM Insulin-Dependent Diabetes Mellitus (IDDM) |
| 44054006 | Type 2 DM Non-Insulin-Dependent Diabetes Mellitus (NIDDM) |
| 443694000 | Type 2 DM uncontrolled |
| 190331003 | Type 2 DM with hyperosmolar coma |
| Diagnosis Code | Description of Code |
|---|---|
| 25000 | DM without mention of complication, T2 or unspecified type, not stated as uncontrolled |
| Laboratory Test | Components of Blood | Threshold Values $ | |
|---|---|---|---|
| mmol/L | mg/dL | ||
| Fasting glucose | Plasma/Serum/Venous | ≥7.0 | ≥126 |
| Glucose Tolerance Test (GTT)—Fasting | - | ≥7.0 | ≥126 |
| Random glucose | Plasma/Serum/Venous | ≥11.1 | ≥200 |
| Oral Glucose Tolerance Test (OGTT)—1 h | - | ≥10.0 | ≥180 |
| Glucose 1 h post-prandial | - | ≥10.0 | ≥180 |
| Glucose (60 min) | Plasma/Serum | ≥10.0 | ≥180 |
| Oral Glucose Tolerance Test (OGTT)—2 h | - | ≥11.1 | ≥200 |
| Glucose 2 h post-prandial | - | ≥11.1 | ≥200 |
| Glucose (120 min) | Plasma/Serum | ≥11.1 | ≥200 |
| Laboratory Test | Threshold Values | |
|---|---|---|
| % | mmol/mol | |
| HbA1c | ≥6.5 | ≥48 |
| Drug Class | Active Ingredient | Brand Name | |
|---|---|---|---|
| Biguanide | Metformin | Adimet | |
| Diabetmin | |||
| Diabetmin XR | |||
| Diamet | |||
| Formet | |||
| Glucient | |||
| Meijumet | |||
| Thiazolidinedione | Pioglitazone | Actos | |
| Sulfonylureas | Glipizide | Beapizide | |
| Diacon | |||
| Diactin | |||
| Dibizide | |||
| Glynase | |||
| Melizide | |||
| Minidiab | |||
| Sunglucon | |||
| Gliclazide | Diamicron | ||
| Diamicron MR | |||
| Dianorm | |||
| Diapro | |||
| Gliavis | |||
| Gliclada | |||
| Glimicron | |||
| Glizide | |||
| Glynade | |||
| Medoclazide | |||
| Melicron | |||
| Mexan | |||
| Sun-gliclazide | |||
| Sun-glizide | |||
| Glimepiride | Amaryl | ||
| Dialosa | |||
| Diapride | |||
| Glibenclamide | Benil | ||
| Clamide | |||
| Daonil | |||
| Glyboral | |||
| Tolbutamide | Tobumide | ||
| Tolmide | |||
| Meglitinide | Repaglinide | Novonorm | |
| Dipeptidyl peptidase-4 (DPP-4) inhibitors | Linagliptin | Trajenta | |
| Saxagliptin | Onglyza | ||
| Sitagliptin | Januvia | ||
| Vildagliptin | Galvus | ||
| GLP-1 Agonists (Incretin mimetics) | Dulaglutide | Trulicity | |
| Liraglutide | Saxenda | ||
| Victoza | |||
| Semaglutide | Ozempic | ||
| Rybelsus | |||
| α-Glucosidase inhibitors | Acarbose | Garbose | |
| Glucobay | |||
| Sodium-glucose co-transporter-2 (SGLT-2) inhibitor | Canagliflozin | Invokana | |
| Ertugliflozin | Steglatro | ||
| Short-acting insulins (Bolus insulins) | Insulin aspart | Fiasp | |
| Novorapid | |||
| Insulin glulisine | Apidra Solostar | ||
| Insulin lispro | Humalog | ||
| Regular (soluble/neutral) insulin | Actrapid | ||
| Humulin R | |||
| Long-acting insulins (Basal insulins) | Insulin degludec | Ryzodeg | |
| Tresiba | |||
| Insulin detemir | Levemir | ||
| Insulin glargine | Basalog one | ||
| Lantus Solostar | |||
| Semglee | |||
| Toujeo Solostar | |||
| Neutral Protamine Hagedorn (NPH)/isophane insulin | Humulin N | ||
| Insulatard | |||
| Mixed insulins | Insulin aspart and insulin aspart protamine crystals | Novomix | |
| Insulin lispro and lispro protamine | Humalog mix | ||
| Regular insulin and insulin isophane | Humulin 30/70 | ||
| Regular insulin and isophane insulin | Mixtard | ||
| Combination Medications | Vildagliptin | Metformin | Galvus Met |
| Empagliflozin | Linagliptin | Glyxambi | |
| Glibenclamide | Metformin HCL | Glucovance | |
| Metformin/Metformin XR | Sitagliptin | Janumet/Janumet XR | |
| Metformin XR | Saxagliptin | Kombiglyze | |
| Linagliptin | Metformin HCL | Trajenta Duo | |
| Insulin glargine | Lixisenatide | Soliqua | |
| Sitagliptin | Ertugliflozin | Steglujan | |
| Dapagliflozin | Metformin/Metformin XR | Xigduo/Xigduo XR | |
| Annotator ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 0.99 | 1 | 0.99 | 0.94 | 1 | 0.93 | 0.99 | 0.99 | 0.95 | 0.95 | 1 | 0.99 | |
| 2 | 1 | 1 | 0.99 | 1 | 0.99 | 0.94 | 1 | 0.93 | 0.99 | 0.99 | 0.95 | 0.95 | 1 | 0.99 | |
| 3 | 1 | 1 | 0.99 | 1 | 0.99 | 0.94 | 1 | 0.93 | 0.99 | 0.99 | 0.95 | 0.95 | 1 | 0.99 | |
| 4 | 0.99 | 0.99 | 0.99 | 0.99 | 0.98 | 0.95 | 0.99 | 0.92 | 0.98 | 0.98 | 0.94 | 0.96 | 0.99 | 0.98 | |
| 5 | 1 | 1 | 1 | 0.99 | 0.99 | 0.94 | 1 | 0.93 | 0.99 | 0.99 | 0.95 | 0.95 | 1 | 0.99 | |
| 6 | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 | 0.93 | 0.99 | 0.92 | 0.98 | 0.98 | 0.96 | 0.94 | 0.99 | 0.98 | |
| 7 | 0.94 | 0.94 | 0.94 | 0.95 | 0.94 | 0.93 | 0.94 | 0.92 | 0.93 | 0.93 | 0.89 | 0.96 | 0.94 | 0.93 | |
| 8 | 1 | 1 | 1 | 0.99 | 1 | 0.99 | 0.94 | 0.93 | 0.99 | 0.99 | 0.95 | 0.95 | 1 | 0.99 | |
| 9 | 0.93 | 0.93 | 0.93 | 0.92 | 0.93 | 0.92 | 0.92 | 0.93 | 0.94 | 0.92 | 0.88 | 0.95 | 0.93 | 0.92 | |
| 10 | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 | 0.98 | 0.93 | 0.99 | 0.94 | 0.98 | 0.94 | 0.96 | 0.99 | 0.98 | |
| 11 | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 | 0.98 | 0.93 | 0.99 | 0.92 | 0.98 | 0.94 | 0.94 | 0.99 | 0.98 | |
| 12 | 0.95 | 0.95 | 0.95 | 0.94 | 0.95 | 0.96 | 0.89 | 0.95 | 0.88 | 0.94 | 0.94 | 0.9 | 0.95 | 0.94 | |
| 13 | 0.95 | 0.95 | 0.95 | 0.96 | 0.95 | 0.94 | 0.96 | 0.95 | 0.95 | 0.96 | 0.94 | 0.9 | 0.95 | 0.94 | |
| 14 | 1 | 1 | 1 | 0.99 | 1 | 0.99 | 0.94 | 1 | 0.93 | 0.99 | 0.99 | 0.95 | 0.95 | 0.99 | |
| 15 | 0.99 | 0.99 | 0.99 | 0.98 | 0.99 | 0.98 | 0.93 | 0.99 | 0.92 | 0.98 | 0.98 | 0.94 | 0.94 | 0.99 |
| Atrial Fibrillation Cohort (n = 1194) | |||||
|---|---|---|---|---|---|
| Actual DM Group (n = 457) | Diagnosis Codes and/or Laboratory Tests and/or Medications (n = 597) | Diagnosis Codes and/or Laboratory Tests (n = 574) | Diagnosis Codes and/or Medications (n = 456) | ||
| Sex, n (%) | Male | 247 (54.0%) | 314 (52.6%) | 303 (52.8%) | 247 (54.2%) |
| Female | 210 (46.0%) | 283 (47.4%) | 271 (47.2%) | 209 (45.8%) | |
| Race, n (%) | Chinese | 328 (71.8%) | 436 (73.0%) | 424 (73.9%) | 333 (73.0%) |
| Malay | 83 (18.2%) | 105 (17.6%) | 96 (16.7%) | 79 (17.3%) | |
| Indian | 26 (5.7%) | 34 (5.7%) | 33 (5.7%) | 25 (5.5%) | |
| Others | 20 (4.4%) | 22 (3.7%) | 21 (3.7%) | 19 (4.2%) | |
| Age | Mean | 72.3 | 73.6 | 73.7 | 72.3 |
| Standard deviation | 11.2 | 11.1 | 11.2 | 11.2 | |
| Median | 73.0 | 74.0 | 75.0 | 73.0 | |
| Interquartile range | 16.0 | 16.0 | 15.0 | 16.0 | |
| Random hospitalized sample (n = 1619) | |||||
| Actual DM group (n = 367) | Diagnosis codes and/or laboratory tests and/or medications (n = 584) | Diagnosis codes and/or laboratory tests (n = 573) | Diagnosis codes and/or medications (n = 382) | ||
| Sex, n (%) | Male | 197 (53.7%) | 319 (54.6%) | 315 (55.0%) | 198 (51.8%) |
| Female | 170 (46.3%) | 265 (45.4%) | 258 (45.0%) | 184 (48.2%) | |
| Race, n (%) | Chinese | 237 (64.6%) | 404 (69.2%) | 398 (69.5%) | 251 (65.7%) |
| Malay | 55 (15.0%) | 71 (12.2%) | 71 (12.4%) | 56 (14.7%) | |
| Indian | 50 (13.6%) | 73 (12.5%) | 68 (11.9%) | 54 (14.1%) | |
| Others | 25 (6.8%) | 36 (6.2%) | 36 (6.3%) | 21 (5.5%) | |
| Age | Mean | 67.7 | 66.2 | 66.6 | 67.1 |
| Standard deviation | 13.8 | 17.1 | 16.9 | 15.0 | |
| Median | 69.0 | 68.0 | 68.0 | 68.5 | |
| Interquartile range | 17.0 | 20.0 | 20.0 | 17.0 | |
References
- Upadhyaya, S.G.; Murphree, D.H.; Ngufor, C.G.; Knight, A.M.; Cronk, D.J.; Cima, R.R.; Curry, T.B.; Pathak, J.; Carter, R.E.; Kor, D.J. Automated Diabetes Case Identification Using Electronic Health Record Data at a Tertiary Care Facility. Mayo Clin. Proc. Innov. Qual. Outcomes 2017, 1, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Kagawa, R.; Kawazoe, Y.; Ida, Y.; Shinohara, E.; Tanaka, K.; Imai, T.; Ohe, K. Development of Type 2 Diabetes Mellitus Phenotyping Framework Using Expert Knowledge and Machine Learning Approach. J. Diabetes Sci. Technol. 2017, 11, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Weerahandi, H.M.; Horwitz, L.I.; Blecker, S.B. Diabetes Phenotyping Using the Electronic Health Record. J. Gen. Intern. Med. 2020, 35, 3716–3718. [Google Scholar] [CrossRef] [PubMed]
- Spratt, S.E.; Pereira, K.; Granger, B.B.; Batch, B.C.; Phelan, M.; Pencina, M.; Miranda, M.L.; Boulware, E.; Lucas, J.E.; Nelson, C.L.; et al. Assessing electronic health record phenotypes against gold-standard diagnostic criteria for diabetes mellitus. J. Am. Med. Inform. Assoc. 2017, 24, e121–e128. [Google Scholar] [CrossRef] [PubMed]
- Richesson, R.L.; Rusincovitch, S.A.; Wixted, D.; Batch, B.C.; Feinglos, M.N.; Miranda, M.L.; Hammond, W.E.; Califf, R.M.; Spratt, S.E. A comparison of phenotype definitions for diabetes mellitus. J. Am. Med. Inform. Assoc 2013, 20, e319–e326. [Google Scholar] [CrossRef] [PubMed]
- Psaty, B.M.; Breckenridge, A.M. Mini-Sentinel and regulatory science--big data rendered fit and functional. N. Engl. J. Med. 2014, 370, 2165–2167. [Google Scholar] [CrossRef] [PubMed]
- Voss, E.A.; Makadia, R.; Matcho, A.; Martijn, S.; Knoll, C.; Schuemie, M.; DeFalco, F.J.; Londhe, A.; Zhu, V.; Ryan, P.B. Feasibility and utility of applications of the common data model to multiple, disparate observational health databases. J. Am. Med. Inform. Assoc. 2015, 22, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Klann, J.G.; Abend, A.; Raghavan, V.A.; Mandl, K.D.; Murphy, S.N. Data interchange using i2b2. J. Am. Med. Inform. Assoc. 2016, 23, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Fleurence, R.L.; Curtis, L.H.; Califf, R.M.; Platt, R.; Selby, J.V.; Brown, J.S. Launching PCORnet, a national patient-centered clinical research network. J. Am. Med. Inform. Assoc. 2014, 21, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Schneeweiss, S. Learning from big health care data. N. Engl. J. Med. 2014, 370, 2161–2163. [Google Scholar] [CrossRef] [PubMed]
- Bourke, A.; Bate, A.; Sauer, B.C.; Brown, J.S.; Hall, G.C. Evidence generation from healthcare databases: Recommendations for managing change. Pharmacoepidemiol. Drug. Saf. 2016, 25, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.C.; Lam, C.S.P.; Matchar, D.B.; Zee, Y.K.; Wong, J.E.L. Singapore’s health-care system: Key features, challenges, and shifts. Lancet 2021, 398, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, C.B.; Gerds, T.A.; Olesen, J.B.; Kristensen, S.L.; Lamberts, M.; Lip, G.Y.; Gislason, G.H.; Køber, L.; Torp-Pedersen, C. Atrial fibrillation and risk of stroke: A nationwide cohort study. Europace 2016, 18, 1689–1697. [Google Scholar] [CrossRef] [PubMed]
- Chao, T.F.; Lip, G.Y.; Liu, C.J.; Tuan, T.C.; Chen, S.J.; Wang, K.L.; Lin, Y.J.; Chang, S.L.; Lo, L.W.; Hu, Y.F.; et al. Validation of a Modified CHA2DS2-VASc Score for Stroke Risk Stratification in Asian Patients with Atrial Fibrillation: A Nationwide Cohort Study. Stroke 2016, 47, 2462–2469. [Google Scholar] [CrossRef] [PubMed]
- TRUST. Improving Health Outcomes through Trusted Data Exchange. Available online: https://trustplatform.sg/ (accessed on 2 February 2023).
- Lash, T.L.; Olshan, A.F. EPIDEMIOLOGY Announces the “Validation Study” Submission Category. Epidemiology 2016, 27, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.J. Validation study methods for estimating exposure proportions and odds ratios with misclassified data. J. Clin. Epidemiol. 1990, 43, 941–947. [Google Scholar] [CrossRef] [PubMed]
- Lo-Ciganic, W.; Zgibor, J.C.; Ruppert, K.; Arena, V.C.; Stone, R.A. Identifying type 1 and type 2 diabetic cases using administrative data: A tree-structured model. J. Diabetes Sci. Technol. 2011, 5, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Lipscombe, L.L.; Hwee, J.; Webster, L.; Shah, B.R.; Booth, G.L.; Tu, K. Identifying diabetes cases from administrative data: A population-based validation study. BMC Health Serv. Res. 2018, 18, 316. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.K.; Ma, J.; Ganesan, V.C.; McGill, J.B. Mistaken Identity: Missed Diagnosis of Type 1 Diabetes in an Older Adult. Med. Res. Arch. 2019, 7, 1962. [Google Scholar] [PubMed]
- Thomas, N.J.; Lynam, A.L.; Hill, A.V.; Weedon, M.N.; Shields, B.M.; Oram, R.A.; McDonald, T.J.; Hattersley, A.T.; Jones, A.G. Type 1 diabetes defined by severe insulin deficiency occurs after 30 years of age and is commonly treated as type 2 diabetes. Diabetologia 2019, 62, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
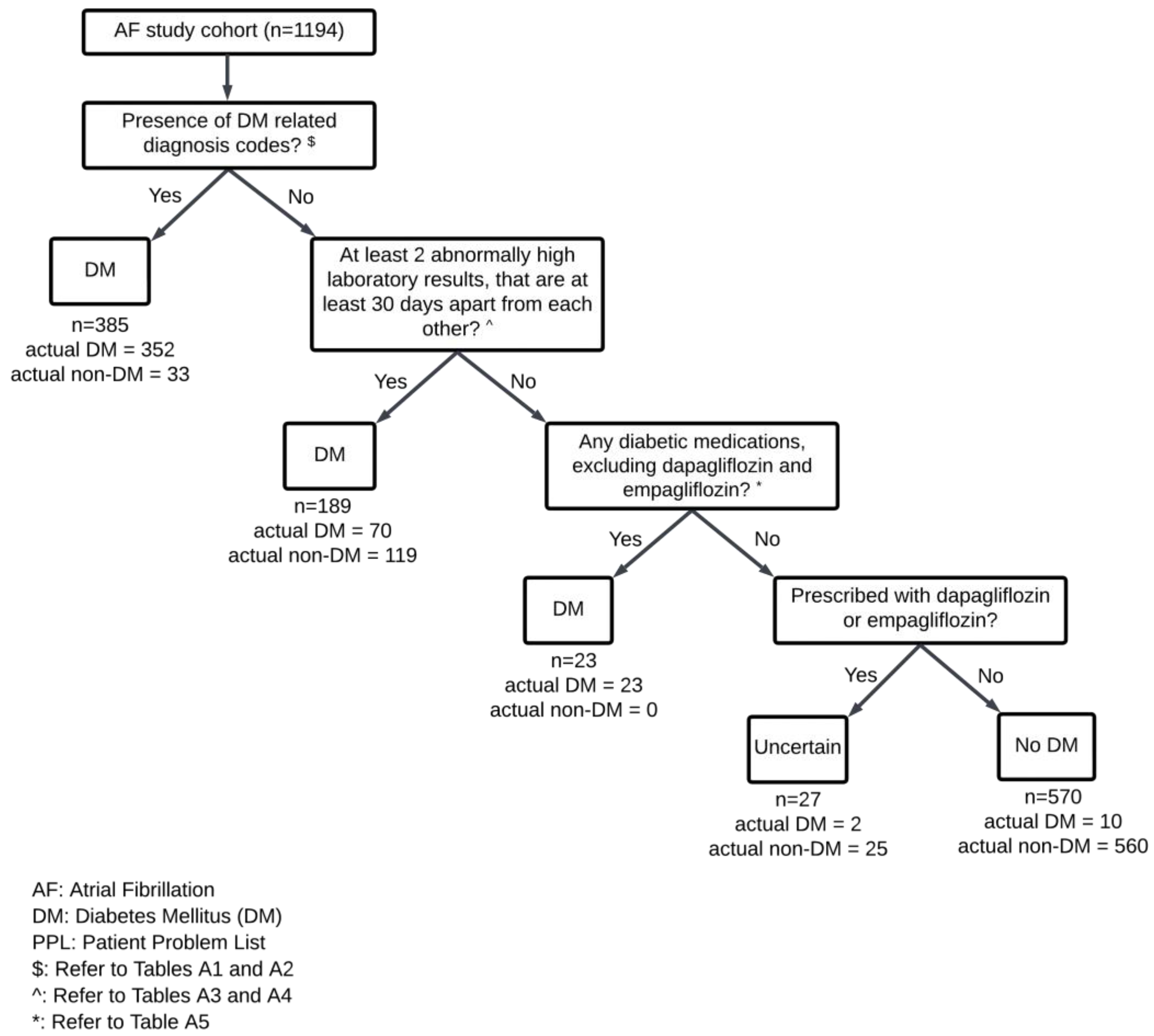
| Atrial Fibrillation Cohort (n = 1194) | Random Hospitalized Sample (n = 1619) | |||
|---|---|---|---|---|
| 2019 (n = 608) | 2020 (n = 586) | 2019 (n = 808) | 2020 (n = 811) | |
| Diabetes, yes, n | 228 | 229 | 198 | 169 |
| Sensitivity, % | 97.8 | 96.9 | 98.0 | 97.6 |
| Specificity, % | 81.1 | 77.6 | 80.3 | 83.6 |
| Positive predictive value, % | 75.6 | 73.5 | 61.8 | 61.1 |
| Negative predictive value, % | 98.4 | 97.5 | 99.2 | 99.3 |
| Data Element Checkpoint | Predicted to Have DM | Gold Standard (of Those Predicted to Have DM) | Cumulative Sensitivity (%) | ||
|---|---|---|---|---|---|
| DM | No DM (False-Positive) | ||||
| Combined atrial fibrillation cohort (n = 1194) | |||||
| With DM (457) | Diagnosis codes | 385 | 352 | 33 | 77.0 |
| Diagnosis codes and/or laboratory tests | 574 | 422 | 152 | 92.3 | |
| Diagnosis codes and/or laboratory tests and/or medications | 597 | 445 | 152 | 97.4 | |
| Combined random hospitalized sample (n = 1619) | |||||
| With DM (367) | Diagnosis codes | 329 | 306 | 23 | 83.4 |
| Diagnosis codes and/or laboratory tests | 573 | 355 | 218 | 96.7 | |
| Diagnosis codes and/or laboratory tests and/or medications | 584 | 359 | 225 | 97.8 | |
| Combined Atrial Fibrillation Cohort (n = 1194) | Combined Random Hospitalized Sample (n = 1619) | |||||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
| Sex | ||||||||
| Female | 97.1 | 78.6 | 72.1 | 98.0 | 97.6 | 85.0 | 62.6 | 99.3 |
| Male | 97.6 | 80.2 | 76.8 | 98.0 | 98.0 | 78.8 | 60.5 | 99.2 |
| Age group | ||||||||
| 64 years and below | 96.5 | 90.8 | 87.3 | 97.5 | 98.0 | 88.8 | 57.6 | 99.6 |
| 65 years and above | 97.7 | 75.8 | 71.1 | 98.2 | 97.7 | 60.6 | 64.4 | 97.3 |
| Number in Combined Atrial Fibrillation Cohort (FP = 152, FN = 12) | Number in Combined Random Hospitalized Sample (FP = 225, FN = 8) | |
|---|---|---|
| Reasons for false-positive classification | ||
| DM not mentioned in unstructured clinical notes (e.g., discharge summary), but diagnosis, laboratory tests or medications fit the DM criteria | 38 | 50 |
| Impaired fasting glucose or HbA1c in prediabetic range | 18 | 0 |
| Hyperglycemia (due to other reasons) | 0 | 3 |
| Impaired glucose tolerance | 0 | 3 |
| Gestational diabetes | 0 | 2 |
| DM on diet control | 1 | 2 |
| Total FP sampled for review | 57 | 60 |
| Reason for false-negative classification | ||
| DM mentioned in discharge summary, but no diagnosis, labs or medications fit the DM criteria | 5 | 4 |
| Total FN sampled for review | 5 | 4 |
| TP | FP | TN | FN | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|---|
| Combined atrial fibrillation cohort (n = 1194) | ||||||||
| Diagnosis codes and/or laboratory tests and/or medications | 445 | 152 | 585 | 12 | 97.4 | 79.4 | 74.5 | 98.0 |
| Diagnosis codes and/or laboratory tests | 422 | 152 | 585 | 35 | 92.3 | 79.4 | 73.5 | 94.4 |
| Diagnosis codes and/or medications | 420 | 36 | 701 | 37 | 91.9 | 95.1 | 92.1 | 95.0 |
| Combined random hospitalized sample (n = 1619) | ||||||||
| Diagnosis codes and/or laboratory tests and/or medications | 359 | 225 | 1027 | 8 | 97.8 | 82.0 | 61.5 | 99.2 |
| Diagnosis codes and/or laboratory tests | 355 | 218 | 1034 | 12 | 96.7 | 82.6 | 62.0 | 98.9 |
| Diagnosis codes and/or medications | 345 | 37 | 1215 | 22 | 94.0 | 97.0 | 90.3 | 98.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, H.X.; Lim, R.L.T.; Ang, P.S.; Foo, B.P.Q.; Koon, Y.L.; Neo, J.W.; Ng, A.J.J.; Tan, S.H.; Teo, D.C.H.; Tham, M.Y.; et al. Phenotyping Diabetes Mellitus on Aggregated Electronic Health Records from Disparate Health Systems. Pharmacoepidemiology 2023, 2, 223-235. https://doi.org/10.3390/pharma2030019
Tan HX, Lim RLT, Ang PS, Foo BPQ, Koon YL, Neo JW, Ng AJJ, Tan SH, Teo DCH, Tham MY, et al. Phenotyping Diabetes Mellitus on Aggregated Electronic Health Records from Disparate Health Systems. Pharmacoepidemiology. 2023; 2(3):223-235. https://doi.org/10.3390/pharma2030019
Chicago/Turabian StyleTan, Hui Xing, Rachel Li Ting Lim, Pei San Ang, Belinda Pei Qin Foo, Yen Ling Koon, Jing Wei Neo, Amelia Jing Jing Ng, Siew Har Tan, Desmond Chun Hwee Teo, Mun Yee Tham, and et al. 2023. "Phenotyping Diabetes Mellitus on Aggregated Electronic Health Records from Disparate Health Systems" Pharmacoepidemiology 2, no. 3: 223-235. https://doi.org/10.3390/pharma2030019
APA StyleTan, H. X., Lim, R. L. T., Ang, P. S., Foo, B. P. Q., Koon, Y. L., Neo, J. W., Ng, A. J. J., Tan, S. H., Teo, D. C. H., Tham, M. Y., Yap, A. J. Y., Ng, N. K. M., Loke, C. W. P., Peck, L. F., Huang, H., & Dorajoo, S. R. (2023). Phenotyping Diabetes Mellitus on Aggregated Electronic Health Records from Disparate Health Systems. Pharmacoepidemiology, 2(3), 223-235. https://doi.org/10.3390/pharma2030019







