The Microbiota Is Not an Organ: Introducing the Muco-Microbiotic Layer as a Novel Morphofunctional Structure
Abstract
1. Brief Introduction to the History and Terminology Related to the Term “Microbiota” and about Its Definition in Scientific Literature
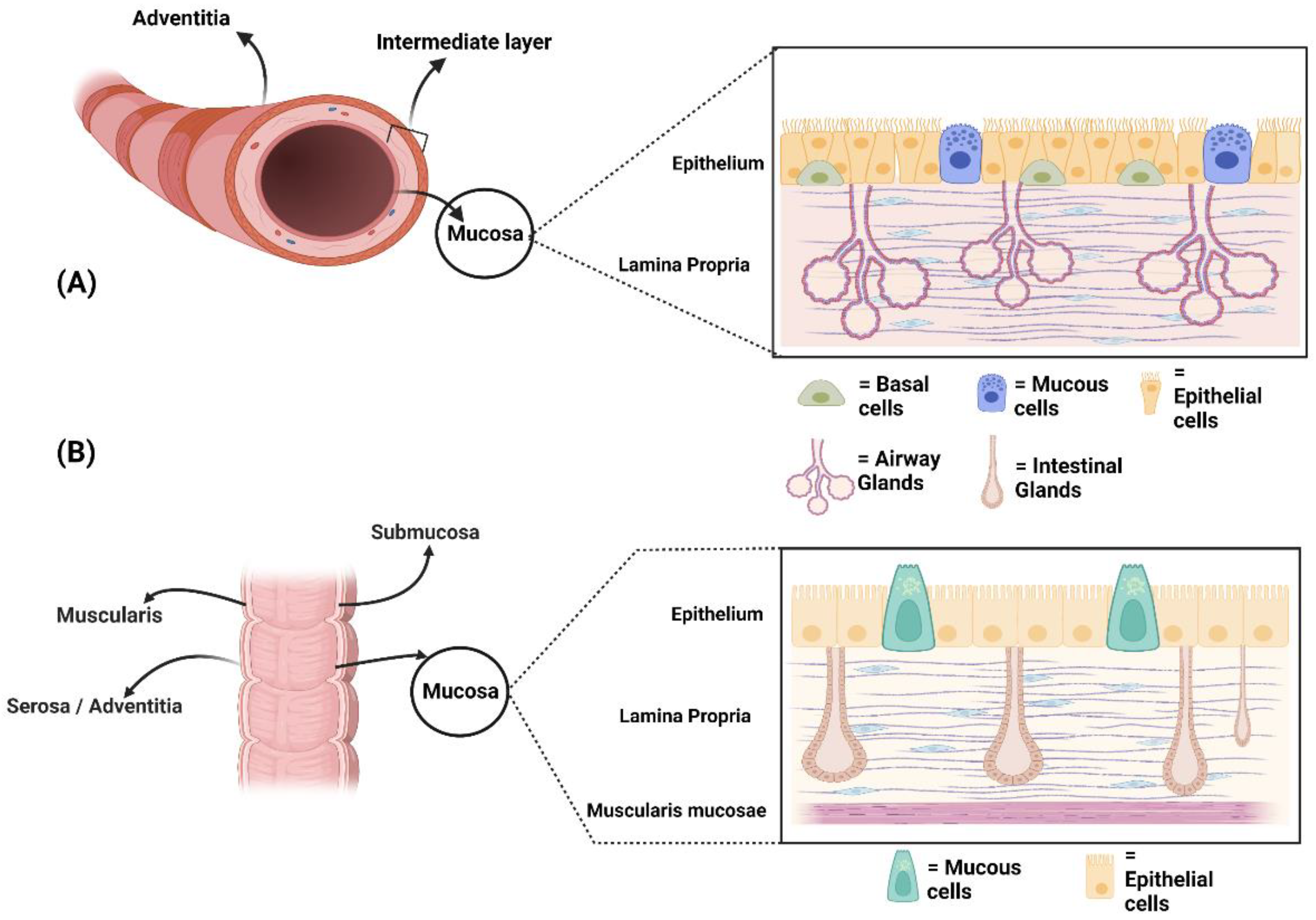
2. Microscopic Anatomy of Human Airways and Bowel: Similarities and Differences
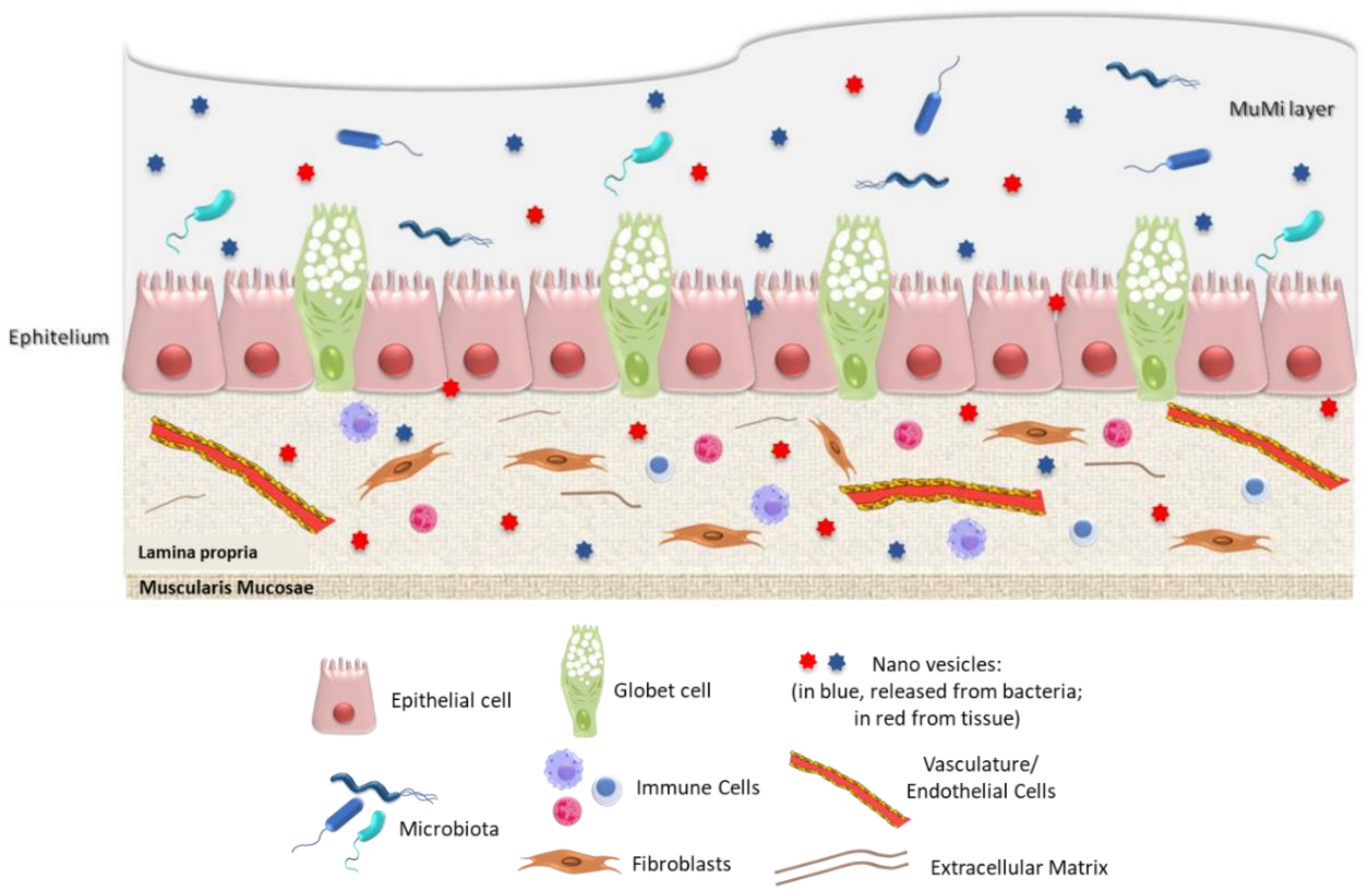
3. The “Ghost” Layer
4. Extracellular Vesicles: Physiology and Pathophysiology
5. Airway Composition
6. Bowel Mucus Composition
7. Bowel Microbiota Composition
| Anatomy | Density | Kingdoms | Phylum | Genus | (%) | References | |
|---|---|---|---|---|---|---|---|
| URT (upper respiratory tract) | Nasal cavity | 103 | Bacteria | Actinomycetota | Corynebacterium spp. | 20–25% | [16,73,74,76,77,78,107,132,133,134] |
| Bacillota | Staphylococcus spp. | 7–12% | |||||
| Streptococcus spp. | 8.5–13% | ||||||
| Pseudomonadota | Haemophilus spp. | 2–7% | |||||
| Moraxella spp. | 13–18% | ||||||
| Nasopharynx | 103 | Actinomycetota | Corynebacterium spp. | 6–11% | |||
| Bacillota | Staphylococcus spp. | 12–17% | |||||
| Streptococcus spp. | 10.5–15% | ||||||
| Pseudomonadota | Moraxella spp. | 14–19% | |||||
| Oropharynx | 106 | Actinomycetota | Rothia spp. | 5.5–10% | |||
| Bacteroidota | Prevotella spp. | 12–17% | |||||
| Bacillota | Streptococcus spp. | 23–28,5% | |||||
| Lactobacillus spp. | 1–5% | ||||||
| Veillonella spp. | 4–9% | ||||||
| Fusobacteriota | Leptotrichia spp. | 4–9% | |||||
| LRT (lower respiratory tract | Lungs | 102 | Bacteria | Bacteroidota | Prevotella spp. | 75–80% | |
| Bacillota | Lactobacillus spp. | 78–83% | |||||
| Streptococcus spp. | |||||||
| Veillonella spp. | |||||||
| Pseudomonadota | Halomonas spp. | 41–46% | |||||
| Domain | Kingdoms | Phylum | Genus | (%) | References |
|---|---|---|---|---|---|
| Procaryota | Bacteria | Bacteroidota | Bacteroides | 73.13 ± 22.16% | [136,137,138,139] |
| Prevotella | |||||
| Bacillota | Clostridium | 22.2 ± 18.66% | |||
| Lactobacillus | |||||
| Actinomycetota | Bifidobacterium | 1.67 ± 2.94 | |||
| Fusobacteriota | Fusobacterium | 19 ± 21.4% | |||
| Pseudomonadota | Escherichia | 2.15 ± 10.39% | |||
| Shigella | |||||
| Verrucomicrobiota | Akkermansia | 1 ± 2% | |||
| Spirochaetota | Brachyspira | 5 ± 7.8% | |||
| Archea | Euryarchaeota | Halobacterium | 0.1 ± 21.3% | [140,141] | |
| Domain | Kingdoms | Phylum | Family | (%) | References |
| Eukaryota | Fungi | Ascomycota | Aspergillaceae | 47.8 ± 99.5% | |
| Debaryomycetaceae | [142,143,144] | ||||
| Dipodascaceae | |||||
| Saccharomycetaceae | |||||
| Basidiomycota | Tremellaceae | 47.8 ± 99.5% | |||
| Malasseziaceae | |||||
| Virii | Bacteriophage | Uroviricota | Demerecviridae | 80 ± 84% | [139,145,146,147] |
| Ackermannviridae | |||||
| Autographiviridae |
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prescott, S.L. History of medicine: Origin of the term microbiome and why it matters. Hum. Microbiome J. 2017, 4, 24–25. [Google Scholar] [CrossRef]
- Aya, V.; Flórez, A.; Perez, L.; Ramírez, J.D. Association between physical activity and changes in intestinal microbiota composition: A systematic review. PLoS ONE 2021, 16, e0247039. [Google Scholar] [CrossRef] [PubMed]
- Riccio, P.; Rossano, R. The human gut microbiota is neither an organ nor a commensal. FEBS Lett. 2020, 594, 3262–3271. [Google Scholar] [CrossRef] [PubMed]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, E.; Zilber-Rosenberg, I. The hologenome concept of evolution after 10 years. Microbiome 2018, 6, 78. [Google Scholar] [CrossRef]
- Triviño, V.; Suárez, J. Holobionts: Ecological communities, hybrids, or biological individuals? A metaphysical perspective on multispecies systems. Stud. Hist. Philos. Biol. Biomed. Sci. 2020, 84, 101323. [Google Scholar] [CrossRef]
- Richter, A.; Puddicombe, S.M.; Lordan, J.L.; Bucchieri, F.; Wilson, S.J.; Djukanovic, R.; Dent, G.; Holgate, S.T.; Davies, D.E. The contribution of interleukin (IL)-4 and IL-13 to the epithelial-mesenchymal trophic unit in asthma. Am. J. Respir. Cell Mol. Biol. 2001, 25, 385–391. [Google Scholar] [CrossRef]
- Bucchieri, F.; Pitruzzella, A.; Fucarino, A.; Gammazza, A.M.; Bavisotto, C.C.; Marcianò, V.; Cajozzo, M.; Lo Iacono, G.; Marchese, R.; Zummo, G.; et al. Functional characterization of a novel 3D model of the epithelial-mesenchymal trophic unit. Exp. Lung Res. 2017, 43, 82–92. [Google Scholar] [CrossRef]
- Evans, M.J.; Winkle, L.S.V.; Fanucchi, M.V.; Plopper, C.G. The Attenuated Fibroblast Sheath of the Respiratory Tract Epithelial–Mesenchymal Trophic Unit. Am. J. Respir. Cell Mol. Biol. 1999, 21, 655–657. [Google Scholar] [CrossRef]
- Sangiorgi, C.; Vallese, D.; Gnemmi, I.; Bucchieri, F.; Balbi, B.; Brun, P.; Leone, A.; Giordano, A.; Conway de Macario, E.; Macario, A.J.; et al. HSP60 activity on human bronchial epithelial cells. Int. J. Immunopathol. Pharm. 2017, 30, 333–340. [Google Scholar] [CrossRef]
- Sunadome, H.; Matsumoto, H.; Petrova, G.; Kanemitsu, Y.; Tohda, Y.; Horiguchi, T.; Kita, H.; Kuwabara, K.; Tomii, K.; Otsuka, K.; et al. IL4Rα and ADAM33 as genetic markers in asthma exacerbations and type-2 inflammatory endotype. Clin. Exp. Allergy 2017, 47, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Minoo, P.; King, R.J. Epithelial-Mesenchymal Interactions in Lung Development. Annu. Rev. Physiol. 1994, 56, 13–45. [Google Scholar] [CrossRef] [PubMed]
- Bartis, D.; Mise, N.; Mahida, R.Y.; Eickelberg, O.; Thickett, D.R. Epithelial–mesenchymal transition in lung development and disease: Does it exist and is it important? Thorax 2014, 69, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Ward, C.; Eapen, M.S.; Myers, S.; Hallgren, O.; Levine, H.; Sohal, S.S. Epithelial–mesenchymal transition, a spectrum of states: Role in lung development, homeostasis, and disease. Dev. Dyn. 2018, 247, 346–358. [Google Scholar] [CrossRef]
- Hansel, N.N.; Paré, P.D.; Rafaels, N.; Sin, D.D.; Sandford, A.; Daley, D.; Vergara, C.; Huang, L.; Elliott, W.M.; Pascoe, C.D.; et al. Genome-Wide Association Study Identification of Novel Loci Associated with Airway Responsiveness in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Cell Mol. Biol. 2015, 53, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Al-Muhsen, S.; Johnson, J.R.; Hamid, Q. Remodeling in asthma. J. Allergy Clin. Immunol. 2011, 128, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Pulvirenti, G.; Parisi, G.F.; Giallongo, A.; Papale, M.; Manti, S.; Savasta, S.; Licari, A.; Marseglia, G.L.; Leonardi, S. Lower Airway Microbiota. Front. Pediatrics 2019, 7, 393. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Rajilić-Stojanović, M.; de Vos, W.M. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014, 38, 996–1047. [Google Scholar] [CrossRef]
- D’Argenio, V.; Salvatore, F. The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta 2015, 451, 97–102. [Google Scholar] [CrossRef]
- Wagner Mackenzie, B.; Chang, K.; Zoing, M.; Jain, R.; Hoggard, M.; Biswas, K.; Douglas, R.G.; Taylor, M.W. Longitudinal study of the bacterial and fungal microbiota in the human sinuses reveals seasonal and annual changes in diversity. Sci. Rep. 2019, 9, 17416. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Mac Aogáin, M.; Fam, K.D.; Chia, K.L.; Binte Mohamed Ali, N.A.; Yap, M.M.C.; Yap, E.P.H.; Chotirmall, S.H.; Lim, C.L. Airway microbiome composition correlates with lung function and arterial stiffness in an age-dependent manner. PLoS ONE 2019, 14, e0225636. [Google Scholar] [CrossRef] [PubMed]
- Yamanishi, S.; Pawankar, R. Current advances on the microbiome and role of probiotics in upper airways disease. Curr. Opin. Allergy Clin. Immunol. 2020, 20, 30–35. [Google Scholar] [CrossRef]
- Man, W.H.; Scheltema, N.M.; Clerc, M.; van Houten, M.A.; Nibbelke, E.E.; Achten, N.B.; Arp, K.; Sanders, E.A.M.; Bont, L.J.; Bogaert, D. Infant respiratory syncytial virus prophylaxis and nasopharyngeal microbiota until 6 years of life: A subanalysis of the MAKI randomised controlled trial. Lancet Respir. Med. 2020, 8, 1022–1031. [Google Scholar] [CrossRef]
- Simrén, M.; Barbara, G.; Flint, H.J.; Spiegel, B.M.; Spiller, R.C.; Vanner, S.; Verdu, E.F.; Whorwell, P.J.; Zoetendal, E.G. Intestinal microbiota in functional bowel disorders: A Rome foundation report. Gut 2013, 62, 159–176. [Google Scholar] [CrossRef] [PubMed]
- Hillman, E.T.; Lu, H.; Yao, T.; Nakatsu, C.H. Microbial Ecology along the Gastrointestinal Tract. Microbes Environ. 2017, 32, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Eun, C.S.; Kwak, M.J.; Han, D.S.; Lee, A.R.; Park, D.I.; Yang, S.K.; Kim, Y.S.; Kim, J.F. Does the intestinal microbial community of Korean Crohn’s disease patients differ from that of western patients? BMC Gastroenterol. 2016, 16, 28. [Google Scholar] [CrossRef]
- Fulde, M.; Sommer, F.; Chassaing, B.; van Vorst, K.; Dupont, A.; Hensel, M.; Basic, M.; Klopfleisch, R.; Rosenstiel, P.; Bleich, A.; et al. Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature 2018, 560, 489–493. [Google Scholar] [CrossRef]
- Maynard, C.; Weinkove, D. The Gut Microbiota and Ageing. Sub-Cell. Biochem. 2018, 90, 351–371. [Google Scholar] [CrossRef]
- Cebula, A.; Seweryn, M.; Rempala, G.A.; Pabla, S.S.; McIndoe, R.A.; Denning, T.L.; Bry, L.; Kraj, P.; Kisielow, P.; Ignatowicz, L. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013, 497, 258–262. [Google Scholar] [CrossRef]
- Martens, K.; Pugin, B.; De Boeck, I.; Spacova, I.; Steelant, B.; Seys, S.F.; Lebeer, S.; Hellings, P.W. Probiotics for the airways: Potential to improve epithelial and immune homeostasis. Allergy 2018, 73, 1954–1963. [Google Scholar] [CrossRef] [PubMed]
- Caverly, L.J.; Huang, Y.J.; Sze, M.A. Past, Present, and Future Research on the Lung Microbiome in Inflammatory Airway Disease. Chest 2019, 156, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.; Lo, B.C.; Núñez, G. Host-microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 2020, 20, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Fais, S.; O’Driscoll, L.; Borras, F.E.; Buzas, E.; Camussi, G.; Cappello, F.; Carvalho, J.; Cordeiro da Silva, A.; Del Portillo, H.; El Andaloussi, S.; et al. Evidence-Based Clinical Use of Nanoscale Extracellular Vesicles in Nanomedicine. ACS Nano 2016, 10, 3886–3899. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Rondelli, V.; Di Cola, E.; Koutsioubas, A.; Alongi, J.; Ferruti, P.; Ranucci, E.; Brocca, P. Mucin Thin Layers: A Model for Mucus-Covered Tissues. Int. J. Mol. Sci. 2019, 20, 3712. [Google Scholar] [CrossRef]
- Johansson, M.E.V.; Larsson, J.M.H.; Hansson, G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host—microbial interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 4659–4665. [Google Scholar] [CrossRef]
- Cappello, F.; Rappa, F.; Canepa, F.; Carini, F.; Mazzola, M.; Tomasello, G.; Bonaventura, G.; Giuliana, G.; Leone, A.; Saguto, D.; et al. Probiotics Can Cure Oral Aphthous-Like Ulcers in Inflammatory Bowel Disease Patients: A Review of the Literature and a Working Hypothesis. Int. J. Mol. Sci. 2019, 20, 5026. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Sabrina David, F.R.; Antonella Marino Gammazza, Alberto Giuseppe Fucarino, Celeste Caruso Bavisotto, Alessandro Pitruzzella, Claudia Campanella. Exosomes: Can doctors still ignore their existence? EuroMediterranean Biomed. J. 2013, 8, 4. [Google Scholar] [CrossRef]
- Campanella, C.; Caruso Bavisotto, C.; Logozzi, M.; Marino Gammazza, A.; Mizzoni, D.; Cappello, F.; Fais, S. On the Choice of the Extracellular Vesicles for Therapeutic Purposes. Int. J. Mol. Sci. 2019, 20, 236. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Caruso Bavisotto, C.; Marino Gammazza, A.; Campanella, C.; Bucchieri, F.; Cappello, F. Extracellular heat shock proteins in cancer: From early diagnosis to new therapeutic approach. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Kooijmans, S.A.A.; Schiffelers, R.M.; Zarovni, N.; Vago, R. Modulation of tissue tropism and biological activity of exosomes and other extracellular vesicles: New nanotools for cancer treatment. Pharmacol. Res. 2016, 111, 487–500. [Google Scholar] [CrossRef]
- Tickner, J.A.; Urquhart, A.J.; Stephenson, S.A.; Richard, D.J.; O’Byrne, K.J. Functions and therapeutic roles of exosomes in cancer. Front. Oncol. 2014, 4, 127. [Google Scholar] [CrossRef]
- Graziano, F.; Iacopino, D.G.; Cammarata, G.; Scalia, G.; Campanella, C.; Giannone, A.G.; Porcasi, R.; Florena, A.M.; Conway de Macario, E.; Macario, A.J.L.; et al. The Triad Hsp60-miRNAs-Extracellular Vesicles in Brain Tumors: Assessing Its Components for Understanding Tumorigenesis and Monitoring Patients. Appl. Sci. 2021, 11, 2867. [Google Scholar] [CrossRef]
- Vitale, A.M.; Santonocito, R.; Vergilio, G.; Marino Gammazza, A.; Campanella, C.; Conway de Macario, E.; Bucchieri, F.; Macario, A.J.L.; Caruso Bavisotto, C. Brain Tumor-Derived Extracellular Vesicles as Carriers of Disease Markers: Molecular Chaperones and MicroRNAs. Appl. Sci. 2020, 10, 6961. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Cipolla, C.; Graceffa, G.; Barone, R.; Bucchieri, F.; Bulone, D.; Cabibi, D.; Campanella, C.; Marino Gammazza, A.; Pitruzzella, A.; et al. Immunomorphological Pattern of Molecular Chaperones in Normal and Pathological Thyroid Tissues and Circulating Exosomes: Potential Use in Clinics. Int. J. Mol. Sci. 2019, 20, 4496. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Cappello, F.; Macario, A.J.L.; Conway de Macario, E.; Logozzi, M.; Fais, S.; Campanella, C. Exosomal HSP60: A potentially useful biomarker for diagnosis, assessing prognosis, and monitoring response to treatment. Expert Rev. Mol. Diagn. 2017, 17, 815–822. [Google Scholar] [CrossRef]
- Cappello, F.; Logozzi, M.; Campanella, C.; Bavisotto, C.C.; Marcilla, A.; Properzi, F.; Fais, S. Exosome levels in human body fluids: A tumor marker by themselves? Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2017, 96, 93–98. [Google Scholar] [CrossRef]
- Campanella, C.; Rappa, F.; Sciumè, C.; Marino Gammazza, A.; Barone, R.; Bucchieri, F.; David, S.; Curcurù, G.; Caruso Bavisotto, C.; Pitruzzella, A.; et al. Heat shock protein 60 levels in tissue and circulating exosomes in human large bowel cancer before and after ablative surgery. Cancer 2015, 121, 3230–3239. [Google Scholar] [CrossRef] [PubMed]
- Campanella, C.; D’Anneo, A.; Marino Gammazza, A.; Caruso Bavisotto, C.; Barone, R.; Emanuele, S.; Lo Cascio, F.; Mocciaro, E.; Fais, S.; Conway De Macario, E.; et al. The histone deacetylase inhibitor SAHA induces HSP60 nitration and its extracellular release by exosomal vesicles in human lung-derived carcinoma cells. Oncotarget 2016, 7, 28849–28867. [Google Scholar] [CrossRef] [PubMed]
- Lässer, C.; Alikhani, V.S.; Ekström, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef]
- Sánchez-Vidaurre, S.; Eldh, M.; Larssen, P.; Daham, K.; Martinez-Bravo, M.J.; Dahlén, S.E.; Dahlén, B.; van Hage, M.; Gabrielsson, S. RNA-containing exosomes in induced sputum of asthmatic patients. J. Allergy Clin. Immunol. 2017, 140, 1459.e1452–1461.e1452. [Google Scholar] [CrossRef]
- Admyre, C.; Grunewald, J.; Thyberg, J.; Gripenbäck, S.; Tornling, G.; Eklund, A.; Scheynius, A.; Gabrielsson, S. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur. Respir. J. 2003, 22, 578–583. [Google Scholar] [CrossRef]
- Pastor, L.; Vera, E.; Marin, J.M.; Sanz-Rubio, D. Extracellular Vesicles from Airway Secretions: New Insights in Lung Diseases. Int. J. Mol. Sci. 2021, 22, 583. [Google Scholar] [CrossRef]
- Leoni, G.; Neumann, P.A.; Kamaly, N.; Quiros, M.; Nishio, H.; Jones, H.R.; Sumagin, R.; Hilgarth, R.S.; Alam, A.; Fredman, G.; et al. Annexin A1-containing extracellular vesicles and polymeric nanoparticles promote epithelial wound repair. J. Clin. Investig. 2015, 125, 1215–1227. [Google Scholar] [CrossRef]
- Zhang, X.; Deeke, S.A.; Ning, Z.; Starr, A.E.; Butcher, J.; Li, J.; Mayne, J.; Cheng, K.; Liao, B.; Li, L.; et al. Metaproteomics reveals associations between microbiome and intestinal extracellular vesicle proteins in pediatric inflammatory bowel disease. Nat. Commun. 2018, 9, 2873. [Google Scholar] [CrossRef]
- Mitsuhashi, S.; Feldbrügge, L.; Csizmadia, E.; Mitsuhashi, M.; Robson, S.C.; Moss, A.C. Luminal Extracellular Vesicles (EVs) in Inflammatory Bowel Disease (IBD) Exhibit Proinflammatory Effects on Epithelial Cells and Macrophages. Inflamm. Bowel Dis. 2016, 22, 1587–1595. [Google Scholar] [CrossRef]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Avila-Calderón, E.D.; Araiza-Villanueva, M.G.; Cancino-Diaz, J.C.; López-Villegas, E.O.; Sriranganathan, N.; Boyle, S.M.; Contreras-Rodríguez, A. Roles of bacterial membrane vesicles. Arch. Microbiol. 2015, 197, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Olaya-Abril, A.; Prados-Rosales, R.; McConnell, M.J.; Martín-Peña, R.; González-Reyes, J.A.; Jiménez-Munguía, I.; Gómez-Gascón, L.; Fernández, J.; Luque-García, J.L.; García-Lidón, C.; et al. Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J. Proteom. 2014, 106, 46–60. [Google Scholar] [CrossRef] [PubMed]
- Roier, S.; Zingl, F.G.; Cakar, F.; Durakovic, S.; Kohl, P.; Eichmann, T.O.; Klug, L.; Gadermaier, B.; Weinzerl, K.; Prassl, R.; et al. A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 2016, 7, 10515. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.R.; Hong, S.W.; Choi, E.B.; Lee, W.H.; Kim, Y.S.; Jeon, S.G.; Jang, M.H.; Gho, Y.S.; Kim, Y.K. Staphylococcus aureus-derived extracellular vesicles induce neutrophilic pulmonary inflammation via both Th1 and Th17 cell responses. Allergy 2012, 67, 1271–1281. [Google Scholar] [CrossRef]
- Park, K.S.; Lee, J.; Jang, S.C.; Kim, S.R.; Jang, M.H.; Lötvall, J.; Kim, Y.K.; Gho, Y.S. Pulmonary inflammation induced by bacteria-free outer membrane vesicles from Pseudomonas aeruginosa. Am. J. Respir. Cell Mol. Biol. 2013, 49, 637–645. [Google Scholar] [CrossRef]
- Stentz, R.; Carvalho, A.L.; Jones, E.J.; Carding, S.R. Fantastic voyage: The journey of intestinal microbiota-derived microvesicles through the body. Biochem. Soc. Trans. 2018, 46, 1021–1027. [Google Scholar] [CrossRef]
- Durant, L.; Stentz, R.; Noble, A.; Brooks, J.; Gicheva, N.; Reddi, D.; O’Connor, M.J.; Hoyles, L.; McCartney, A.L.; Man, R.; et al. Bacteroides thetaiotaomicron-derived outer membrane vesicles promote regulatory dendritic cell responses in health but not in inflammatory bowel disease. Microbiome 2020, 8, 88. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Invernizzi, R.; Lloyd, C.M.; Molyneaux, P.L. Respiratory microbiome and epithelial interactions shape immunity in the lungs. Immunology 2020, 160, 171–182. [Google Scholar] [CrossRef]
- Knowles, M.R.; Boucher, R.C. Mucus clearance as a primary innate defense mechanism for mammalian airways. J. Clin. Investig. 2002, 109, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, e00037. [Google Scholar] [CrossRef] [PubMed]
- Rao, R.; Dsouza, J.M.; Mathew, J.L. Comparison of microbiota in the upper versus lower respiratory tract in children during health and respiratory disease: Protocol for a systematic review. Syst. Rev. 2021, 10, 253. [Google Scholar] [CrossRef] [PubMed]
- Marsland, B.J.; Trompette, A.; Gollwitzer, E.S. The Gut-Lung Axis in Respiratory Disease. Ann. Am. Thorac. Soc. 2015, 12 (Suppl. 2), S150–S156. [Google Scholar] [CrossRef]
- Lynch, S.V. The Lung Microbiome and Airway Disease. Ann. Am. Thorac. Soc. 2016, 13 (Suppl. 2), S462–S465. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Falkowski, N.R.; Huffnagle, G.B.; Curtis, J.L. Bacterial Topography of the Healthy Human Lower Respiratory Tract. mBio 2017, 8, e02287-16. [Google Scholar] [CrossRef]
- Sahin-Yilmaz, A.; Naclerio, R.M. Anatomy and physiology of the upper airway. Proc. Am. Thorac. Soc. 2011, 8, 31–39. [Google Scholar] [CrossRef]
- Man, W.H.; de Steenhuijsen Piters, W.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Goto, T. Airway Microbiota as a Modulator of Lung Cancer. Int. J. Mol. Sci. 2020, 21, 3044. [Google Scholar] [CrossRef]
- Elgamal, Z.; Singh, P.; Geraghty, P. The Upper Airway Microbiota, Environmental Exposures, Inflammation, and Disease. Medicina 2021, 57, 823. [Google Scholar] [CrossRef]
- Larsen, G.L.; Holt, P.G. The concept of airway inflammation. Am. J. Respir. Crit. Care Med. 2000, 162, S2–S6. [Google Scholar] [CrossRef]
- Zhou-Suckow, Z.; Duerr, J.; Hagner, M.; Agrawal, R.; Mall, M.A. Airway mucus, inflammation and remodeling: Emerging links in the pathogenesis of chronic lung diseases. Cell Tissue Res. 2017, 367, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.E.; Lynch, S.V. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe 2015, 17, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Vinolo, M.A.; Rodrigues, H.G.; Nachbar, R.T.; Curi, R. Regulation of inflammation by short chain fatty acids. Nutrients 2011, 3, 858–876. [Google Scholar] [CrossRef]
- Chiu, C.J.; Huang, M.T. Asthma in the Precision Medicine Era: Biologics and Probiotics. Int. J. Mol. Sci. 2021, 22, 4528. [Google Scholar] [CrossRef]
- Yang, K.; Dong, W. Perspectives on Probiotics and Bronchopulmonary Dysplasia. Front. Pediatrics 2020, 8, 570247. [Google Scholar] [CrossRef]
- Sestito, S.; D’Auria, E.; Baldassarre, M.E.; Salvatore, S.; Tallarico, V.; Stefanelli, E.; Tarsitano, F.; Concolino, D.; Pensabene, L. The Role of Prebiotics and Probiotics in Prevention of Allergic Diseases in Infants. Front. Pediatrics 2020, 8, 583946. [Google Scholar] [CrossRef]
- Herath, M.; Hosie, S.; Bornstein, J.C.; Franks, A.E.; Hill-Yardin, E.L. The Role of the Gastrointestinal Mucus System in Intestinal Homeostasis: Implications for Neurological Disorders. Front. Cell. Infect. Microbiol. 2020, 10, 248. [Google Scholar] [CrossRef]
- Sicard, J.F.; Le Bihan, G.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell. Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Rodríguez-Piñeiro, A.M.; Schütte, A.; Ermund, A.; Boysen, P.; Bemark, M.; Sommer, F.; Bäckhed, F.; Hansson, G.C.; Johansson, M.E. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015, 16, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Allaire, J.M.; Morampudi, V.; Crowley, S.M.; Stahl, M.; Yu, H.; Bhullar, K.; Knodler, L.A.; Bressler, B.; Jacobson, K.; Vallance, B.A. Frontline defenders: Goblet cell mediators dictate host-microbe interactions in the intestinal tract during health and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 314, G360–G377. [Google Scholar] [CrossRef] [PubMed]
- Pelaseyed, T.; Bergström, J.H.; Gustafsson, J.K.; Ermund, A.; Birchenough, G.M.; Schütte, A.; van der Post, S.; Svensson, F.; Rodríguez-Piñeiro, A.M.; Nyström, E.E.; et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 2014, 260, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Hansson, G.C. Immunological aspects of intestinal mucus and mucins. Nat. Rev. Immunol. 2016, 16, 639–649. [Google Scholar] [CrossRef]
- Etzold, S.; Juge, N. Structural insights into bacterial recognition of intestinal mucins. Curr. Opin. Struct. Biol. 2014, 28, 23–31. [Google Scholar] [CrossRef]
- Arike, L.; Hansson, G.C. The Densely O-Glycosylated MUC2 Mucin Protects the Intestine and Provides Food for the Commensal Bacteria. J. Mol. Biol. 2016, 428, 3221–3229. [Google Scholar] [CrossRef]
- Juge, N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol. 2012, 20, 30–39. [Google Scholar] [CrossRef]
- Douillard, F.o.P.; Ribbera, A.; J�rvinen, H.M.; Kant, R.; Pietil�, T.E.; Randazzo, C.; Paulin, L.; Laine, P.K.; Caggia, C.; Ossowski, I.v.; et al. Comparative Genomic and Functional Analysis of Lactobacillus casei and Lactobacillus rhamnosus Strains Marketed as Probiotics. Appl. Environ. Microbiol. 2013, 79, 1923–1933. [Google Scholar] [CrossRef]
- Kankainen, M.; Paulin, L.; Tynkkynen, S.; Ossowski, I.v.; Reunanen, J.; Partanen, P.; Satokari, R.; Vesterlund, S.; Hendrickx, A.P.A.; Lebeer, S.; et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human—Mucus binding protein. Proc. Natl. Acad. Sci. USA 2009, 106, 17193–17198. [Google Scholar] [CrossRef]
- Slížová, M.; Nemcová, R.; Mad’ar, M.; Hadryová, J.; Gancarčíková, S.; Popper, M.; Pistl, J. Analysis of biofilm formation by intestinal lactobacilli. Can. J. Microbiol. 2015, 61, 437–446. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.; Tremaroli, V.; Nielsen, J.; Bäckhed, F. Assessing the human gut microbiota in metabolic diseases. Diabetes 2013, 62, 3341–3349. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Palacio, S.D.; Montes, S.A.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef] [PubMed]
- Kinross, J.M.; Darzi, A.W.; Nicholson, J.K. Gut microbiome-host interactions in health and disease. Genome Med. 2011, 3, 14. [Google Scholar] [CrossRef]
- Wang, H.; Dai, W.; Feng, X.; Zhou, Q.; Wang, H.; Yang, Y.; Li, S.; Zheng, Y. Microbiota Composition in Upper Respiratory Tracts of Healthy Children in Shenzhen, China, Differed with Respiratory Sites and Ages. BioMed Res. Int. 2018, 2018, 6515670. [Google Scholar] [CrossRef]
- Wang, G.; Huang, S.; Wang, Y.; Cai, S.; Yu, H.; Liu, H.; Zeng, X.; Zhang, G.; Qiao, S. Bridging intestinal immunity and gut microbiota by metabolites. Cell. Mol. Life Sci. 2019, 76, 3917–3937. [Google Scholar] [CrossRef]
- Sánchez, B.; Delgado, S.; Blanco-Míguez, A.; Lourenço, A.; Gueimonde, M.; Margolles, A. Probiotics, gut microbiota, and their influence on host health and disease. Mol. Nutr. Food Res. 2017, 61, 1600240. [Google Scholar] [CrossRef]
- Hill, M.J. Intestinal flora and endogenous vitamin synthesis. Eur. J. Cancer Prev. Off. J. Eur. Cancer Prev. Organ. 1997, 6 (Suppl. 1), S43–S45. [Google Scholar] [CrossRef]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- Scheppach, W. Effects of short chain fatty acids on gut morphology and function. Gut 1994, 35, S35–S38. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wu, L.; Huntington, N.D.; Zhang, X. Crosstalk Between Gut Microbiota and Innate Immunity and Its Implication in Autoimmune Diseases. Front. Immunol. 2020, 11, 282. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Chirlaque, C.; Aranda, C.J.; Ocón, B.; Capitán-Cañadas, F.; Ortega-González, M.; Carrero, J.J.; Suárez, M.D.; Zarzuelo, A.; Sánchez de Medina, F.; Martínez-Augustin, O. Germ-free and Antibiotic-treated Mice are Highly Susceptible to Epithelial Injury in DSS Colitis. J. Crohns Colitis 2016, 10, 1324–1335. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, Y.; Kashyap, P.C. Germ-Free Mice Model for Studying Host–Microbial Interactions. In Mouse Models for Drug Discovery: Methods and Protocols; Proetzel, G., Wiles, M.V., Eds.; Springer: New York, NY, USA, 2016; pp. 123–135. [Google Scholar]
- Grover, M.; Kashyap, P.C. Germ-free mice as a model to study effect of gut microbiota on host physiology. Neurogastroenterol. Motil. 2014, 26, 745–748. [Google Scholar] [CrossRef]
- Lazar, V.; Ditu, L.-M.; Pircalabioru, G.G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L.; Chifiriuc, M.C. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front. Immunol. 2018, 9, 1830. [Google Scholar] [CrossRef] [PubMed]
- Liévin, V.; Peiffer, I.; Hudault, S.; Rochat, F.; Brassart, D.; Neeser, J.-R.; Servin, A.L. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut 2000, 47, 646–652. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics Regulate Gut Microbiota: An Effective Method to Improve Immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef]
- Klann, E.M.; Dissanayake, U.; Gurrala, A.; Farrer, M.; Shukla, A.W.; Ramirez-Zamora, A.; Mai, V.; Vedam-Mai, V. The Gut-Brain Axis and Its Relation to Parkinson’s Disease: A Review. Front Aging Neurosci. 2022, 13, 782082. [Google Scholar] [CrossRef]
- Gustafsson, G.; Lööv, C.; Persson, E.; Lázaro, D.F.; Takeda, S.; Bergström, J.; Erlandsson, A.; Sehlin, D.; Balaj, L.; György, B.; et al. Secretion and Uptake of α-Synuclein Via Extracellular Vesicles in Cultured Cells. Cell Mol. Neurobiol. 2018, 38, 1539–1550. [Google Scholar] [CrossRef]
- Bui, T.M.; Mascarenhas, L.A.; Sumagin, R. Extracellular vesicles regulate immune responses and cellular function in intestinal inflammation and repair. Tissue Barriers 2018, 6, e1431038. [Google Scholar] [CrossRef]
- Lo Cicero, A.; Stahl, P.D.; Raposo, G. Extracellular vesicles shuffling intercellular messages: For good or for bad. Curr. Opin. Cell Biol. 2015, 35, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi Badi, S.; Moshiri, A.; Fateh, A.; Rahimi Jamnani, F.; Sarshar, M.; Vaziri, F.; Siadat, S.D. Microbiota-Derived Extracellular Vesicles as New Systemic Regulators. Front. Microbiol. 2017, 8, 1610. [Google Scholar] [CrossRef] [PubMed]
- Lagos, L.; Tandberg, J.; Kashulin-Bekkelund, A.; Colquhoun, D.J.; Sørum, H.; Winther-Larsen, H.C. Isolation and Characterization of Serum Extracellular Vesicles (EVs) from Atlantic Salmon Infected with Piscirickettsia Salmonis. Proteomes 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Moshiri, A.; Dashtbani-Roozbehani, A.; Najar Peerayeh, S.; Siadat, S.D. Outer membrane vesicle: A macromolecule with multifunctional activity. Hum. Vaccines Immunother. 2012, 8, 953–955. [Google Scholar] [CrossRef] [PubMed]
- Campbell, H.K.; Maiers, J.L.; DeMali, K.A. Interplay between tight junctions & adherens junctions. Exp. Cell Res. 2017, 358, 39–44. [Google Scholar] [CrossRef]
- Hu, G.; Gong, A.-Y.; Roth, A.L.; Huang, B.Q.; Ward, H.D.; Zhu, G.; LaRusso, N.F.; Hanson, N.D.; Chen, X.-M. Release of Luminal Exosomes Contributes to TLR4-Mediated Epithelial Antimicrobial Defense. PLOS Pathog. 2013, 9, e1003261. [Google Scholar] [CrossRef]
- Muraca, M.; Putignani, L.; Fierabracci, A.; Teti, A.; Perilongo, G. Gut microbiota-derived outer membrane vesicles: Under-recognized major players in health and disease? Discov. Med. 2015, 19, 343–348. [Google Scholar]
- Shen, Y.; Giardino Torchia, M.L.; Lawson, G.W.; Karp, C.L.; Ashwell, J.D.; Mazmanian, S.K. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 2012, 12, 509–520. [Google Scholar] [CrossRef]
- Haurat, M.F.; Elhenawy, W.; Feldman, M.F. Prokaryotic membrane vesicles: New insights on biogenesis and biological roles. Biol. Chem. 2015, 396, 95–109. [Google Scholar] [CrossRef]
- Li, K.J.; Chen, Z.L.; Huang, Y.; Zhang, R.; Luan, X.Q.; Lei, T.T.; Chen, L. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir. Res. 2019, 20, 272. [Google Scholar] [CrossRef]
- Hernández-Terán, A.; Vega-Sánchez, A.E.; Mejía-Nepomuceno, F.; Serna-Muñoz, R.; Rodríguez-Llamazares, S.; Salido-Guadarrama, I.; Romero-Espinoza, J.A.; Guadarrama-Pérez, C.; Sandoval, J.; Campos, F.; et al. Microbiota composition in the lower respiratory tract is associated with severity in patients with acute respiratory distress by influenza. medRxiv 2021. Available online: https://www.medrxiv.org/content/10.1101/2021.12.07.21267419v1 (accessed on 8 October 2022).
- Wang, H.; Gu, X.; Weng, Y.; Xu, T.; Fu, Z.; Peng, W.; Yu, W. Quantitative analysis of pathogens in the lower respiratory tract of patients with chronic obstructive pulmonary disease. BMC Pulm. Med. 2015, 15, 94. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ley, R.E.; Sonnenburg, J.L.; Peterson, D.A.; Gordon, J.I. Host-bacterial mutualism in the human intestine. Science 2005, 307, 1915–1920. [Google Scholar] [CrossRef]
- King, C.H.; Desai, H.; Sylvetsky, A.C.; LoTempio, J.; Ayanyan, S.; Carrie, J.; Crandall, K.A.; Fochtman, B.C.; Gasparyan, L.; Gulzar, N.; et al. Baseline human gut microbiota profile in healthy people and standard reporting template. PLoS ONE 2019, 14, e0206484. [Google Scholar] [CrossRef]
- Nayfach, S.; Páez-Espino, D.; Call, L.; Low, S.J.; Sberro, H.; Ivanova, N.N.; Proal, A.D.; Fischbach, M.A.; Bhatt, A.S.; Hugenholtz, P.; et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nat. Microbiol. 2021, 6, 960–970. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. Methanogens in human and animal intestinal Tracts. Syst. Appl. Microbiol. 1986, 7, 223–229. [Google Scholar] [CrossRef]
- Kim, J.Y.; Whon, T.W.; Lim, M.Y.; Kim, Y.B.; Kim, N.; Kwon, M.S.; Kim, J.; Lee, S.H.; Choi, H.J.; Nam, I.H.; et al. The human gut archaeome: Identification of diverse haloarchaea in Korean subjects. Microbiome 2020, 8, 114. [Google Scholar] [CrossRef]
- Kühbacher, T.; Ott, S.J.; Helwig, U.; Mimura, T.; Rizzello, F.; Kleessen, B.; Gionchetti, P.; Blaut, M.; Campieri, M.; Fölsch, U.R.; et al. Bacterial and fungal microbiota in relation to probiotic therapy (VSL#3) in pouchitis. Gut 2006, 55, 833–841. [Google Scholar] [CrossRef]
- Raimondi, S.; Amaretti, A.; Gozzoli, C.; Simone, M.; Righini, L.; Candeliere, F.; Brun, P.; Ardizzoni, A.; Colombari, B.; Paulone, S.; et al. Longitudinal Survey of Fungi in the Human Gut: ITS Profiling, Phenotyping, and Colonization. Front. Microbiol. 2019, 10, 1575. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, D.; Yu, B.; He, J.; Zheng, P.; Mao, X.; Yu, J.; Luo, J.; Tian, G.; Huang, Z.; et al. Fungi in Gastrointestinal Tracts of Human and Mice: From Community to Functions. Microb. Ecol. 2018, 75, 821–829. [Google Scholar] [CrossRef]
- Zhang, T.; Breitbart, M.; Lee, W.H.; Run, J.-Q.; Wei, C.L.; Soh, S.W.L.; Hibberd, M.L.; Liu, E.T.; Rohwer, F.; Ruan, Y. RNA Viral Community in Human Feces: Prevalence of Plant Pathogenic Viruses. PLoS Biol. 2005, 4, e3. [Google Scholar] [CrossRef] [PubMed]
- Manrique, P.; Dills, M.; Young, M.J. The Human Gut Phage Community and Its Implications for Health and Disease. Viruses 2017, 9, 141. [Google Scholar] [CrossRef] [PubMed]
- Mayneris-Perxachs, J.; Castells-Nobau, A.; Arnoriaga-Rodríguez, M.; Garre-Olmo, J.; Puig, J.; Ramos, R.; Martínez-Hernández, F.; Burokas, A.; Coll, C.; Moreno-Navarrete, J.M.; et al. Caudovirales bacteriophages are associated with improved executive function and memory in flies, mice, and humans. Cell Host Microbe 2022, 30, 340–356.e348. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fucarino, A.; Burgio, S.; Paladino, L.; Caruso Bavisotto, C.; Pitruzzella, A.; Bucchieri, F.; Cappello, F. The Microbiota Is Not an Organ: Introducing the Muco-Microbiotic Layer as a Novel Morphofunctional Structure. Anatomia 2022, 1, 186-203. https://doi.org/10.3390/anatomia1020019
Fucarino A, Burgio S, Paladino L, Caruso Bavisotto C, Pitruzzella A, Bucchieri F, Cappello F. The Microbiota Is Not an Organ: Introducing the Muco-Microbiotic Layer as a Novel Morphofunctional Structure. Anatomia. 2022; 1(2):186-203. https://doi.org/10.3390/anatomia1020019
Chicago/Turabian StyleFucarino, Alberto, Stefano Burgio, Letizia Paladino, Celeste Caruso Bavisotto, Alessandro Pitruzzella, Fabio Bucchieri, and Francesco Cappello. 2022. "The Microbiota Is Not an Organ: Introducing the Muco-Microbiotic Layer as a Novel Morphofunctional Structure" Anatomia 1, no. 2: 186-203. https://doi.org/10.3390/anatomia1020019
APA StyleFucarino, A., Burgio, S., Paladino, L., Caruso Bavisotto, C., Pitruzzella, A., Bucchieri, F., & Cappello, F. (2022). The Microbiota Is Not an Organ: Introducing the Muco-Microbiotic Layer as a Novel Morphofunctional Structure. Anatomia, 1(2), 186-203. https://doi.org/10.3390/anatomia1020019












