Abstract
The purpose of this literature review is to provide a fundamental synopsis of current research pertaining to artificial intelligence (AI) within the domain of clinical practice. Artificial intelligence has revolutionized the field of medicine and healthcare by providing innovative solutions to complex problems. One of the most important benefits of AI in clinical practice is its ability to investigate extensive volumes of data with efficiency and precision. This has led to the development of various applications that have improved patient outcomes and reduced the workload of healthcare professionals. AI can support doctors in making more accurate diagnoses and developing personalized treatment plans. Successful examples of AI applications are outlined for a series of medical specialties like cardiology, surgery, gastroenterology, pneumology, nephrology, urology, dermatology, orthopedics, neurology, gynecology, ophthalmology, pediatrics, hematology, and critically ill patients, as well as diagnostic methods. Special reference is made to legal and ethical considerations like accuracy, informed consent, privacy issues, data security, regulatory framework, product liability, explainability, and transparency. Finally, this review closes by critically appraising AI use in clinical practice and its future perspectives. However, it is also important to approach its development and implementation cautiously to ensure ethical considerations are met.
1. Introduction
Artificial intelligence (AI) refers to the emulation of human intelligence in machines designed to exhibit cognitive abilities and acquire knowledge akin to human beings [1,2]. The ancient Greeks attributed a distinctiveness to human beings by virtue of their possession of faculties of reasoning. The notion of the soul was introduced by various religious scholars, who posited it as an enduring and intrinsic essence bestowed upon humanity by a divine creator [3]. According to Plato, it is conceivable for an individual to possess intelligence while simultaneously lacking substantial knowledge about the external world or, more significantly, one’s self. Aristotle, the student of Plato, pioneered the formulation of a distinct set of principles that govern the logical aspect of human cognition. In 1936, Alan Turing authored a scholarly article wherein he elucidated the concept of “Entscheidungsproblem” and put forth the notion of “effective calculability” as a means to address this quandary. The authors established the groundwork for computational models known as algorithms [4]. The initial development of an artificial neural network (ANN) composed of electrical circuits occurred in 1943, with the aim of simulating the interactions between neurons in the brain [5]. The inception of AI took place in 1956 at Dartmouth College. After a span of three years, the initial computer research using an ANN was successfully conducted, utilizing models referred to as “Adaline” and “Madaline” [6]. Computer-aided diagnosis was initially implemented in the examination of pulmonary nodules identified in chest radiographs in 1963 [7]. Researchers made a significant observation regarding AI’s applicability in the bioscience field approximately fifteen years after its inception. This observation was particularly evident in the Dendral experiments [8]. Nevertheless, the utilization of AI in the field of medicine was constrained by technological limitations until 1998, when the United States Food and Drug Administration (FDA) granted approval for the first mammography computer-aided detection (CAD) system [9]. A schematic representation of some important milestones in the evolution of AI is depicted in Figure 1.
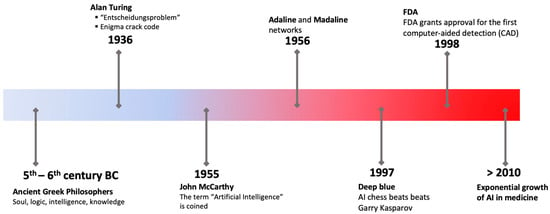
Figure 1.
The progression of concepts in artificial intelligence and significant milestones.
Today, as stated by the expert group on AI within the European Commission’s digital strategy, AI systems refer to software and potentially hardware systems. These systems are designed to operate in either physical or digital conditions, with the ability to perceive their surroundings through data acquisition. In recent years, there has been significant progress in AI, leading to its widespread adoption across various industries such as healthcare, finance, transportation, drug discovery, and quite recently in pharmacokinetics [10,11,12,13,14,15,16,17,18,19,20,21,22]. Over the past few years, notable progress has been made in the field of AI, characterized by the emergence of algorithms and computer programs that exhibit human-like cognitive abilities.
One area where AI has shown great promise is in clinical practice [23]. The incorporation of AI into clinical settings introduces a range of advantages and challenges, accompanied by notable implications for ethical and legal considerations [1]. AI holds the potential to enhance diagnostic precision, streamline administrative tasks, and personalize treatment plans. Through the analysis of extensive medical data, AI systems can discern patterns and correlations that may elude human observation, leading to more accurate and timely interventions [1,2]. Moreover, AI has the capacity to contribute to cost-effective healthcare solutions, ultimately improving overall patient outcomes. The integration of AI technology facilitates informed clinical decision-making processes, thereby promising advancements such as quicker and more accurate diagnoses, personalized treatment plans, and reduced healthcare costs. While the potential benefits of AI in clinical practice are substantial, ethical and legal complexities emerge. The utilization of AI in clinical decision-making raises concerns about transparency, accountability, and the potential bias within algorithms. Safeguarding patient privacy and ensuring data security becomes crucial, necessitating robust ethical guidelines and legal frameworks. Achieving a delicate equilibrium between fostering innovation and protecting patient rights requires thoughtful consideration of the ethical implications of AI in clinical practice, coupled with the development of adaptable legal frameworks capable of keeping pace with technological advancements in the healthcare sector. Addressing AI integration’s ethical and legal challenges in clinical practice mandates a comprehensive approach encompassing legal frameworks and regulations, transparent and explainable AI, ethical guidelines and standards, regular audits and assessments, incentives for ethical practices, and international collaboration.
This literature review aims to provide a fundamental synopsis of current research on AI within the domain of clinical practice. Apart from the widespread role of AI in diagnostic methods, the applications of AI in several medical specialties like cardiology, anesthesiology, surgery, pneumology, neurology, urology, gynecology, hematology, and pediatrics are also discussed. It should be emphasized that the purpose of this review is not solely to provide a synopsis of a specific field (e.g., specialty) but rather to attempt to offer an overview of the current applications of AI in medicine.
2. Materials and Methods
The scope of this investigation was confined to articles written in English and subjected to peer review that fulfilled at least one of the following prerequisites: (a) being published within the timeframe of the last ten years and (b) being seminal papers in the field of AI that built what we know today as artificial intelligence.
A literature search was conducted utilizing the PubMed and Scopus databases from 14 July 2023 to 31 August 2023. Additionally, textbooks on AI were consulted. Two sets of keywords were utilized to recognize terms within the title, abstract, and keywords of the articles.
- The initial set of keywords encompassed terms associated with artificial intelligence, such as “artificial intelligence”, “machine learning”, and “deep learning”. Nevertheless, it is highly probable that research using these methodologies will incorporate terms such as “artificial intelligence” or “machine learning” in their abstracts or keywords;
- The subsequent set of keywords encompassed concepts associated with the application in clinical practice and the legal status. In this case, composite searches were performed using the terms “Artificial intelligence” AND the medical specialty: “cardiology”, “surgery”, “anesthesiology”, “gastroenterology and hepatology”, “pneumonology”, “nephrology”, “urology”, “dermatology”, “orthopedics”, “neurology”, “gynecology, “ophthalmology”, “pediatrics”, “hematology”, “intensive care unit”, “diagnostic methods”, “legal status”, “liability”, “regulatory framework”.
Following the elimination of duplicate entries, a thorough assessment of the titles and abstracts of the identified articles was made in order to ascertain their suitability for inclusion:
The selection criteria for the evaluation of studies were systematically applied. After eliminating duplicate articles, the author assessed each study based on the following criteria: (i) journal, (ii) authorship, (iii) publication date, (iv) study design, (v) methods of analysis, (vi) results, and (vii) conclusions. The eligibility criteria encompassed articles written in English pertinent to the review objectives. An initial screening of abstracts was conducted, excluding studies that did not align with the eligibility criteria. To enhance data quality, all studies meeting the inclusion criteria underwent a comprehensive evaluation, focusing on aspects such as rationale, method design, results, discussion, and conclusions. Studies exhibiting any bias in methodology, results, or data interpretation that could impact the overall outcome were subsequently excluded.
The exclusion criteria encompassed the following: (a) studies that exclusively focused on the advancement and verification of clinical AI algorithms without any tangible implementation and (b) AI applications that predominantly provided automation functionalities, such as the automated delivery and monitoring of insulin, as opposed to offering decision support.
3. Results
3.1. General
Artificial intelligence has revolutionized the field of medicine and healthcare by providing innovative solutions to complex problems [1,5]. There are various types of AI, including deep learning (DL), machine learning (ML), and natural language (Figure 2). DL is a subset of artificial intelligence that focuses on training neural networks to learn and make decisions in a manner similar to the human brain (Figure 3). DL algorithms are designed to learn and improve from experience automatically, without the need for explicit programming [24,25,26]. This ability to analyze large amounts of data and extract meaningful patterns has made DL a powerful tool in fields such as image recognition and autonomous driving.
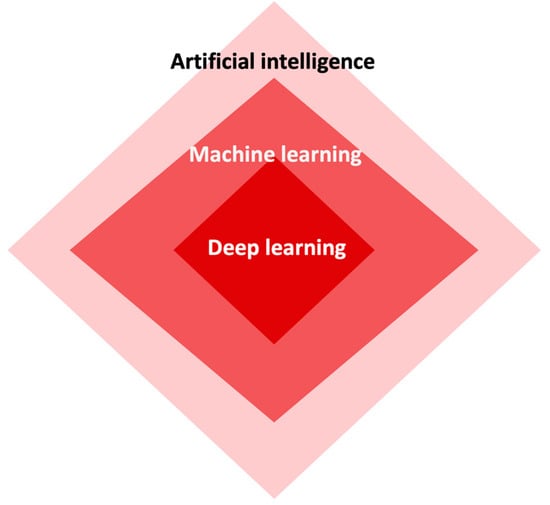
Figure 2.
The interconnectedness among artificial intelligence, machine learning, and deep learning.
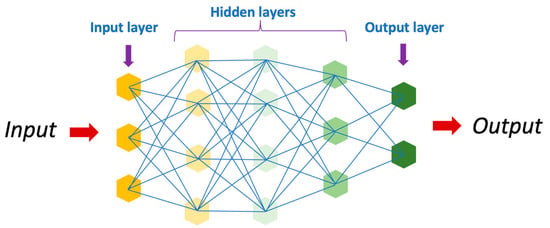
Figure 3.
Schematic representation of an artificial neural network.
Machine learning primarily focuses on advancing algorithms and models that empower computers to acquire knowledge and generate predictions or decisions autonomously without the need for explicit programming [2]. ML can be broadly classified into several categories, such as supervised, unsupervised, and reinforcement learning (refer to Table 1). In supervised learning, an algorithm learns from labeled data to make predictions or decisions [2]. This approach trains the algorithm on a dataset comprising input variables and their corresponding output variables. The goal is to enable the algorithm to understand the relationship between the input and output variables, thereby facilitating precise predictions for novel and unobserved data instances. Various supervised learning algorithms are commonly used, including linear regression, logistic regression, support vector machines, and decision trees.

Table 1.
A common classification of machine learning algorithms.
Unsupervised learning represents a distinct subfield within machine learning, where the algorithm functions without the presence of labeled data [2,24,27,28]. Instead, its purpose is to autonomously identify patterns, structures, or relationships within the data. This learning type proves highly advantageous when a definitive target variable is absent or when the goal is to extract valuable insights from the data without predetermined predictions. Unsupervised learning algorithms include various methods, such as clustering algorithms like k-means and hierarchical clustering, as well as dimensionality reduction techniques like PCA and factor analysis.
The primary objective of reinforcement learning is to train autonomous agents to effectively make a series of decisions within a given environment, aiming to optimize the total cumulative reward obtained [29]. Unlike supervised learning, where the agent is provided with labeled data, or unsupervised learning, where the agent learns patterns and structures from unlabeled data, reinforcement learning operates on the principle of trial and error. Examples of reinforcement learning approaches include the value-based methods (e.g., Q-learning and SARSA), the policy-based methods (e.g., policy gradient and reinforce), and model-based methods (e.g., Monte Carlo tree search).
Natural language processing (NLP) is a field of study that centers on examining and understanding the interplay between computer systems and human language [2]. The field of study pertains to the advancement of algorithms and methodologies that facilitate machines in comprehending, interpreting, and producing human language in a manner that possesses significance and utility. NLP has become increasingly important in our digital age, as it allows computers to process and analyze vast amounts of text data, such as emails, social media posts, and news articles, to extract valuable insights and information.
One of the primary advantages of AI in clinical practice is its ability to rapidly and accurately analyze extensive volumes of data. This capability has given rise to a variety of applications that have not only improved patient outcomes but also lessened the workload on healthcare professionals [30]. In this section, we will explore some of the most promising applications of AI in clinical practice. The evolution of AI has undergone significant changes over the last few decades. The advent of machine learning (ML) and deep learning (DL) has expanded applications in the field of artificial intelligence in medicine, paving the way for personalized medicine rather than relying solely on algorithmic approaches. The use of predictive models holds promise for applications in disease diagnosis, forecasting therapeutic response, and potentially advancing the field of preventive medicine in the coming years. AI has the potential to enhance diagnostic precision, optimize the workflow of healthcare providers and clinical operations, facilitate more effective monitoring of diseases and therapies, improve the precision of medical procedures, and ultimately enhance patient outcomes.
3.2. Cardiology
The application of sophisticated computational algorithms and machine learning techniques in the field of cardiology is commonly referred to as AI. This approach aims to analyze and interpret cardiac data in a more advanced and efficient manner. It involves the development of intelligent systems that can learn from data, make predictions, and offer valuable insights to assist in diagnosing, treating, and managing cardiovascular diseases.
At present, two distinct positions for AI exist in the domain of cardiovascular imaging [31]. Automation refers to the process of replacing human involvement in various tasks, including but not limited to image segmentation and the assessment of structural and functional parameters. Another significant aspect is the identification of insights that hold clinical significance. The majority of documented applications were primarily centered around the implementation of task automation. Furthermore, there have been reports on developing algorithms capable of acquiring cardiac measurements.
AI has significantly impacted various facets of cardiovascular imaging, covering the entire spectrum from initial data acquisition to the final reporting phase [32,33]. Examples of this impact include the use of AI in advancing computed tomography and magnetic resonance imaging techniques for measuring lumen diameter, recognizing coronary calcium score, and identifying obstructive coronary disease. Furthermore, AI has been instrumental in automating processes such as acquisition, segmentation, and report generation [34,35]. In contrast to the methodologies mentioned earlier, a notable concern arises regarding the substantial observer variability observed in the interpretation of echocardiograms. AI holds the potential to address this issue by mitigating inter-observer variability and enhancing diagnostic precision within the field of echocardiography.
In recent years, numerous studies have been conducted to investigate cardiomyopathy screening, with a particular focus on the utilization of AI in conjunction with electrocardiography (ECG) for enhanced diagnostic capabilities [36,37]. The feasibility of the joint use of AI/ECG screening for amyloidosis, cardiomyopathy, and dilated cardiomyopathy remains intact, even in cases of mild left ventricular dysfunction [38,39]. The application of AI/ECG in regular clinical practice has increased the identification of left ventricular systolic dysfunction. In imaging, AI is utilized to automatically evaluate the thickness and properties of the myocardium to distinguish between different types of cardiomyopathies [39,40]. However, there is currently a lack of research investigating the prognostic potential of this AI technology. AI is also being utilized in cardiomyopathy genomics, particularly for predicting the pathogenicity of genetic variants and determining their clinical relevance [41,42,43].
3.3. Surgery
The application of AI and ML models holds significant potential in the field of surgery. These models demonstrate promising applications in both the preoperative phase, accurately diagnosing pancreatic conditions, and the postoperative phase, evaluating prognosis and predicting complications [44,45,46]. AI has also proven beneficial in assisting bariatric surgeries. The increasing integration of AI technologies in various healthcare subspecialties has led to promising developments in their application within bariatric surgery [47,48]. The management of patients who are candidates for bariatric surgery is a complex subject. The evaluation process requires the involvement of a multidisciplinary team comprising professionals from various fields, including internists, psychiatrists, general surgeons, and anesthesiologists. Physicians across various medical specialties engage in the comprehensive assessment of patients before, during, and after surgical procedures, a task that presents considerable difficulties due to the intricate nature of individuals afflicted with obesity [49].
Numerous potential applications of AI exist during the intraoperative period. It has the potential to be utilized in the management of pharmacotherapy, hemodynamic optimization, neuromuscular block monitoring, and anesthesia depth assessment [50]. One of the most notable reports pertains to predicting the early distribution kinetics of propofol. Indeed, the volume of drug distribution in individuals with obesity is subject to modification. Specifically, there is an increase in blood volume and cardiac output, alongside alterations in plasma transport proteins. A study utilized AI to handle the induction phase’s kinetics effectively [51]. This was achieved through the utilization of a comprehensive pharmacokinetic dataset with high resolution. A comparative analysis was undertaken to evaluate the performance of a traditional four-compartment model, a recirculatory model, and a gated recurrent unit neural network. The study concluded that both a recirculatory model and a gated recurrent unit ANN demonstrated similar performance, surpassing a compartmental model in accurately representing high-resolution pharmacokinetic data of propofol [51].
In the same context, plastic surgeons frequently encounter clinical scenarios that lack definitive solutions. Achieving an ideal treatment approach necessitates utilizing a comprehensive decision model that effectively incorporates various influential factors, including clinical and demographic data. Before the advent of AI, the decision tree analysis technique was commonly used for constructing such models. The localization of significant anatomical landmarks in medical imaging plays a crucial role in preoperative planning and postoperative outcome evaluation [52]. Nevertheless, the current identification process is carried out either manually or by running the inserted auxiliaries, resulting in a time-consuming and imprecise procedure. In order to enhance the precision of landmark localization on the distal femur surface, scientists devised an algorithm that initially transformed three-dimensional images into three distinct sets of two-dimensional images [52]. Subsequently, the algorithm acquired the ability to recognize landmarks within these images and subsequently integrated these outcomes to accurately determine the spatial coordinates of the identified landmarks in three dimensions.
3.4. Anesthesiology
The application of AI has yielded remarkable outcomes in anesthesia and operating room management [53,54]. Throughout each phase of the perioperative process—specifically the preoperative [55,56,57], intraoperative [16,17,18,19,20], and postoperative phases [42,44]—distinct tasks can be executed using diverse techniques. The effectiveness of a neural network designed to identify esophageal intubation becomes unnecessary in the presence of continuous capnography [58,59]. In this case, a reliable clinical examination has revealed a previously concealed and highly detrimental complication. The use of video laryngoscopy requires the adjustment of an ML model designed to predict challenging intubation based on patient appearance. The expansion of airway management technology has resulted in an increased spectrum of acceptable outcomes in terms of laryngeal visualization.
Since the 1950s, the concept of an algorithm autonomously regulating the depth of anesthesia using EEG recordings has been a subject of ongoing research. Anesthesiologists have explored this possibility for a considerable period, but it continues to be an active area of investigation.
3.5. Gastroenterology and Hepatology
The field of gastroenterology and hepatology is witnessing significant growth in the potential implementation of AI and ML techniques. In recent years, there has been a burgeoning body of research focusing on examining AI applications in various medical contexts, particularly involving the utilization of computer-aided diagnosis (CAD). These applications encompass the use of CAD in diagnosing premalignant and malignant gastrointestinal lesions, predicting treatment response in patients with inflammatory bowel disease, conducting histopathological analysis of biopsy specimens, assessing the severity of liver fibrosis in individuals with chronic liver disease, developing models for liver transplant allocation, and exploring other related areas [60].
The domain of esophageal cancer prevention and early detection shows significant potential for advancements through the utilization of AI. Substantial research advancements have been made in this field, with a notable portion of esophageal cancer research in the United States dedicated to investigating technologies, including those involving AI, aimed at enhancing the early detection and treatment of Barrett’s esophagus and esophageal adenocarcinoma [61,62].
AI possesses the capacity to assume a significant role in the decision-making process for the treatment of inflammatory bowel disease by accurately predicting treatment response at an earlier stage and providing guidance for personalized therapy selection. Within the field of inflammatory bowel disease, researchers have made advancements in the development of AI/ML computer vision tools. These tools have been specifically designed to assess the severity of diseases through endoscopic examination. The main goals of their study involve the differentiation of colitis from neoplasia and the distinction between sporadic adenomas and non-neoplastic lesions. AI algorithms have undergone training to forecast the response to treatment and assess the likelihood of disease recurrence [63,64]. There are numerous potential applications for AI and ML in the domain of hepatology. The objectives above encompass the assessment of hepatic fibrosis progression, the identification of non-alcoholic fatty liver disease, the recognition of individuals at risk for hepatocellular carcinoma development, and the enhancement of protocols for organ transplantation [65,66].
The prevention and control of colorectal cancer represent significant public health endeavors undertaken by gastroenterologists. The progress made in the field of ML has resulted in the utilization of computer vision techniques to assist in the detection of polyps during colonoscopy procedures. Empirical evidence has demonstrated the efficacy of CAD systems in enhancing the adenoma detection rate [67,68,69,70].
3.6. Pneumonology
AI, specifically the utilization of DL and ML algorithms for pattern recognition, holds significant promise for various applications within the field of pulmonary medicine. These applications encompass image analysis, decision-making processes, and the prediction of prognoses [5,6,7]. Lung cancer is a prevalent malignant neoplasm characterized by significant clinical morbidity and mortality rates [71]. Lung nodules are the prevailing imaging manifestations observed during the initial phase of lung cancer, posing challenges to manual film interpretation. AI recognition technology can conduct multi-parameter cluster analysis and streamline image processing, thereby assisting medical professionals in the early detection of lung cancer [72]. In recent years, reports have indicated that AI systems have demonstrated the capability to identify malignant pulmonary nodules by analyzing chest computed tomography (CT) images [73]. The model has been developed using DL technology, and AI is utilized for the analysis of CT films in order to support medical professionals in enhancing the accuracy of lung cancer screening. Another study constructed a predictive model by applying logistic regression analysis, integrating specific tumor markers into the model [74]. The study’s results demonstrated that the developed predictive model showed significantly better performance when compared to the basic combined detection strategy involving tumor markers.
Research has demonstrated that AI can potentially enhance surgical risk prediction, thereby facilitating the selection of the most optimal surgical approach [75,76]. An example of a cognitive computing system, IBM Watson for Oncology, utilizes AI techniques for data analysis and image conversion. Its primary objective is to assist medical professionals in efficiently identifying crucial information within patients’ medical records, presenting pertinent evidence, and facilitating the exploration of potential treatment options [77]. The application of deep neural networks in the identification of respiratory illnesses, specifically in chest radiographs and CT scans, has resulted in a noteworthy enhancement in diagnostic precision compared to subjective characteristics like tumor speculation, as well as objective characteristics such as shape and texture acquired through image analysis software [78].
3.7. Nephrology
The concept of progressive immunoglobulin refers to the gradual development and maturation of immunoglobulins and IgA nephropathy (IgAN) is an acknowledged etiology of renal failure. However, the ability of the nephrologist to anticipate the occurrence of kidney failure among patients at the time of diagnosis is challenging. Nevertheless, the capacity to discern these individuals would prove advantageous in terms of prognostication and treatment purposes. It has been postulated the existence of a function that establishes a relationship between clinical and biological parameters, such as age, sex, blood pressure, proteinuria, serum creatinine level, and anti-hypertensive treatments, at the time of IgAN diagnosis and the likelihood of developing progressive IgAN [79]. The researchers devised and executed the development of an ANN with the purpose of approximating the aforementioned function. The findings indicated that the ANN demonstrated superior accuracy in predicting the onset of progressive IgAN compared to experienced nephrologists [79]. Specifically, the ANN achieved correct predictions in 87% of cases, whereas the nephrologists achieved a lower accuracy rate of 69.4%. Furthermore, the ANN exhibited a higher sensitivity of 86.4% compared to the nephrologists’ sensitivity of 72%, indicating its ability to correctly identify true positive cases. Similarly, the ANN displayed a higher specificity of 87.5% compared to the nephrologists’ specificity of 66%, indicating its capacity to accurately identify true negative cases. These approaches can potentially be used in a wide range of progressive diseases, thereby aiding clinicians in the process of patient staging and management.
AI models have been applied for various purposes, including predicting the rate of decline in glomerular filtration rate in individuals with autosomal dominant polycystic kidney disease, enhancing anemia management in hemodialysis patients, estimating an appropriate duration for dialysis to achieve the desired level of urea removal, determining the optimal dry weight in patients undergoing hemodialysis, and identifying specific pathogens responsible for bacterial infections in patients with Parkinson’s disease [80,81,82,83].
3.8. Urology
AI is predominantly used in the field of urology, particularly in the domain of genitourinary malignancies. In a study, AI was utilized to predict the outcomes of prostate biopsies, with a specific focus on prostate cancer. ML algorithms were applied to analyze recurrence-free probability and diagnostic evaluation for bladder cancer. There have been anecdotal reports concerning the staging and prediction of disease recurrence in cases of kidney and testis cancer. Recently, AI has found application in non-oncological diseases, specifically in areas such as stones and functional urology.
In recent decades, numerous scholarly investigations have examined the utilization of AI in the management of prostate cancer. These studies align with the contemporary paradigm of precision medicine and surgery [84]. Prostate cancer diagnosis encompasses a broad range of applications, which have experienced numerous advancements in recent years [85]. A seminal study was conducted in 1994 to determine the potential utility of ANN in predicting biopsy outcomes in males displaying abnormal prostate-specific antigen levels. Additionally, the study aimed to assess the effectiveness of ANN in predicting treatment outcomes following radical prostatectomy [85,86]. A study demonstrated the predictive accuracy of two distinct AI systems [87]. These systems were specifically designed using Vienna-based multicenter European referral database data. These AI systems aim to facilitate the early detection of prostate cancer in males. Another study found that a DL survival model exhibited the ability to predict the timeframe for urinary continence recovery after Robot-Assisted Radical Prostatectomy [88]. This prediction was achieved by incorporating Anatomical Pathology Markers (APMs) and patient-related factors. Furthermore, this particular model has successfully identified APMs of top surgeons that can effectively classify surgeons, surpassing the predictive ability of surgeon experience alone. The APMs were able to differentiate surgeons based on the quality of urinary continence recovery observed in their patients, distinguishing between those with superior and inferior outcomes.
In a seminal study twenty years ago, the authors conducted a comparative analysis of AI and Cox regression models to predict disease recurrence following surgery [89]. The results of the study demonstrated that Cox regression models exhibited superior performance in this regard. In conjunction with the increasing range of surgical indications for metastatic kidney cancer, a study was conducted to assess the predictive capacity of AI in determining the prognosis of patients with metastatic renal cell carcinoma who initiate systemic therapy [90]. The researchers provided their AI system with a dataset consisting of information from 175 patients who had undergone nephrectomy of the primary tumor prior to receiving systemic therapy. The objective of this study was to forecast the overall survival rate three years after initiating the initial treatment, utilizing parameters that are accessible at the commencement of first-line therapy. AI has demonstrated the potential to achieve a prediction accuracy of 95% in forecasting overall survival rates. This performance surpasses regression models, indicating the potential future application of AI as a risk stratification tool.
A urinary tract infection is a common bacterial infection that affects the urinary system, including the bladder and urethra. A notable study focused on urinary tract infections where an AI system was developed to assist in the diagnosis of such infections [91]. The study involved individuals diagnosed with either cystitis or nonspecific urethritis. Subjects underwent various procedures, including a medical history assessment, physical examination, analysis of urine samples, and the use of ultrasonography. The findings demonstrated the efficacy of AI in diagnosing urinary tract infections based solely on erythrocyte values in conjunction with symptoms such as suprapubic pain, pollakiuria, and urinalysis results. The AI model exhibited a remarkably high accuracy rate of 98.3%, suggesting it could serve as a cost-effective alternative to expensive laboratory and ultrasound tests.
Functional urology refers to the branch of urology that focuses on studying and managing the urinary tract. The exploration of AI potential applications has also extended to the domain of functional urology. A study compared an AI model and multiple linear regression in terms of their effectiveness in replacing preoperative urodynamic evaluation in women diagnosed with pelvic organ prolapse [92]. A total of 804 women diagnosed with pelvic organ prolapse were subjected to examination, revealing that both multivariate logistic regression and AI were determined to be less effective than urodynamic studies in evaluating urinary dysfunction. A kidney transplant is a surgical procedure in which a healthy kidney from a donor is transplanted into a recipient. Over the past few years, there has been a growing interest in utilizing AI predictive tools in kidney transplantation. Similarly, the potential application of AI in identifying risk factors and co-variates that contribute to the failure of renal transplantation has been explored [93]. The AI approach was compared with the traditional logistic regression model. The AI method demonstrated superior accuracy compared to logistic regression, as evidenced by data analysis from 378 patients.
3.9. Dermatology
Identifying skin diseases primarily relies on the apparent attributes exhibited by the lesions. However, dermatology encompasses a vast collection of over 2000 distinct types of dermatological diseases. Certain skin lesions associated with various diseases may exhibit similarities, posing challenges in accurate diagnosis and treatment [94,95]. Notably, there is a significant shortage of dermatologists, particularly in developing countries and remote regions, where increased medical resources, professional consultations, and clinical support are urgently needed [96,97].
The convergence of rapid iteration in big data, advancements in image recognition technology, and the global proliferation of smartphones present transformative potential for diagnosing and treating skin diseases [98,99]. AI, in particular, has the capacity to offer prompt diagnoses, facilitating a wider range of treatment options and enhancing accessibility, especially for marginalized regions and individuals with limited resources [100]. The integration of AI technology and algorithms has the potential to establish itself rapidly as a standard approach in the field of diagnosis and evaluation. The examination of the structure and form of a skin abnormality is a fundamental aspect of dermatological diagnosis. Advancements in AI have led to significant improvements in facial recognition and aesthetic analysis, rendering them more dependable [101].
The inception of AI in the field of dermatopathology can be traced back to 1987 when a text-based system known as TEGUMENT was utilized on a personal computer. The system was specifically developed with the purpose of classifying 986 histopathological diagnoses based on light microscopic images. It demonstrated a diagnostic accuracy of 91.8% compared to the assessments made by a qualified dermatopathologist [102]. During that particular time frame, the absence of necessary equipment and technologies for whole slide imaging led to the belief that the notion of human-independent image analysis was not viable. In recent years, the accurate classification of routine diagnoses by machine-based systems has become a tangible reality [103].
In a research study, 11 DL algorithms were developed to identify and classify whole slide images of dermal nevus, seborrheic keratoses, and nodular basal cell carcinoma [104]. The visual representations underwent a process of pixelation, resulting in the disintegration of the images, which were subsequently subjected to data analysis. A DL algorithm was developed for pathology that incorporates whole slide imaging. The algorithm effectively categorizes these images into four distinct diagnostic groups: basaloid, squamoid, melanocytic, and other. The implemented system utilizes a series of three consecutive convolutional neural networks to determine the diagnosis with the highest probability.
Distinguishing between malignant and benign lesions holds the highest importance for dermatopathologists due to the consequential divergence in therapeutic decisions. In this context, a study used a sample of 695 melanocytic neoplasms to distinguish between melanoma and nevus by means of classification [105]. The study included a comprehensive representation of all stages of melanoma, as well as various types of nevi. In the present investigation, it was observed that the convolutional neural network exhibited a statistically significant superiority over the pathologists in terms of accurately diagnosing nevi and melanoma through histopathological analysis. The observed discordance rate of 25–26% among dermatopathologists was found to be comparable to the aforementioned similarity.
In another research study, the objective was to assess the precision of a DL algorithm in diagnosing three dermatopathological conditions through the utilization of whole-slide imaging [106]. The study’s findings indicated that the AI system demonstrated high accuracy, correctly classifying several types of carcinomas. In contrast to the straightforward binary classification involved in diagnosing melanoma and distinguishing it from pigmented nevi, the diagnosis of non-melanoma skin cancers presents a more challenging task. This challenge stems from the intricate categorization of these conditions and the inclusion of various benign and malignant diseases, along with inflammatory dermatoses, within the differential diagnoses. A study was conducted to assess the effectiveness of convolutional neural networks in precisely detecting and diagnosing non-pigmented lesions [107]. The findings were compared with the diagnoses rendered by a cohort of 95 clinicians, which included 62 dermatologists with appropriate qualifications. Convolutional neural networks did not exhibit superior accuracy in diagnosing medical conditions compared to human experts. However, they demonstrated greater accuracy in diagnosing prevalent skin cancers. Conversely, convolutional neural networks exhibited lower accuracy than clinicians in diagnosing uncommon non-pigmented malignancies, specifically amelanotic melanoma.
3.10. Orthopedics
Supervised ML can be applied to classify individuals into pain phenotypes based on brain MRI, considering the high prevalence of long-standing pain in the UK, estimated to be between 30% and 50% [108]. The absence of tissue pathology that corresponds to pain, as well as the dependence on self-reported measures for subgroup classification, pose a significant challenge in identifying the neural correlates of pain and provide a comprehensive overview of ML applications used in the context of chronic pain, which encompasses pain conditions beyond musculoskeletal disorders [109]. The authors specifically highlight using ML techniques to classify individuals into distinct pain phenotypes based on predictive models.
Another study established a correlation between frontal plane knee biomechanics and the ability to predict the risk of knee injuries [110]. In this study, inertial sensor data were used to categorize the performance of single-leg squats based on the extent of knee valgus [111]. The study sample consisted of 14 participants, and a total of 140 images were analyzed. Additionally, the researchers sought the opinions of three expert raters regarding the potential risk associated with the observed performances. Supervised learning was applied to perform classification among three distinct classes, namely “poor”, “moderate”, and “good”. The study’s findings indicate that the accuracy levels were observed to be significantly high when performing a 2-class classification task. However, when the complexity of the classification task was increased to a 3-class classification, the accuracy levels experienced a notable reduction of approximately 30%. There is a scarcity of instances where unsupervised learning techniques have been utilized within the domain of musculoskeletal research. According to a study, the chronic pain challenge assesses the likelihood of chronic pain based on assigned weights for various health behaviors [112]. The study included both supervised and unsupervised methods to demonstrate the precise prediction of pain levels, as measured by the visual analog scale and the Oswestry Disability Index. These predictions are made based on the corresponding scores for depression, nutrition, and physical activity. Nevertheless, although this emphasizes the potential of ML to categorize the risk of chronicity using patient-reported data, the effectiveness of unsupervised learning by itself has not been confirmed.
3.11. Neurology
Neuroimaging plays a pivotal role in clinical practice and scientific inquiry, facilitating the examination of the brain in various states of well-being and pathology. Similar to several other domains, neuroimaging is enhanced by the utilization of sophisticated analysis methodologies in order to harness imaging data effectively to investigate the brain and its functionality. In recent times, ML has made significant contributions. Additionally, it has played a crucial role in the prompt identification of acute conditions like stroke and in monitoring imaging changes over time. As our capacity to visualize and examine the brain progresses, so does our comprehension of its complex interconnections and their significance in making therapeutic decisions.
Despite being in the early stages of development, AI’s utilization in neuro-oncology exhibits considerable potential. It is highly probable that AI algorithms will enhance our comprehension of brain tumors and play a pivotal role in fostering advancements in the field of neuro-oncology. The field of neuro-oncology has experienced a growing emphasis on the integration of molecular markers for the purpose of guiding therapeutic interventions [113]. AI algorithms have demonstrated notable efficacy in the noninvasive identification of significant molecular markers from diagnostic imaging, exhibiting remarkable accuracy. In various institutional datasets, AI algorithms have successfully determined the mutational status of several markers [114,115]. Moreover, it has been demonstrated that algorithms based on AI can effectively identify investigational molecular markers, even in smaller cohorts of patients [116].
The utilization of AI for the analysis of diagnostic imaging has proven to be beneficial in the clinical management of brain tumors. The utilization of AI to automate labor-intensive tasks holds great potential in the field of neuro-oncology. Multiple studies have demonstrated the efficacy of DL techniques in identifying brain metastases measuring in the millimeter range through MRI imaging. Furthermore, it has been observed that comparable DL models have demonstrated significant efficacy in the automated segmentation of tumors, thereby enhancing the efficiency of radiation therapy treatment planning [117,118,119]. AI has demonstrated potential in accurately differentiating various central nervous system malignancies without the need for invasive procedures, achieving comparable results to those of expert neuroradiologists [120,121]. The extensive application of these AI algorithms could prove to be highly beneficial in resource-constrained environments that lack access to specialized neuroradiologists.
3.12. Gynecology
Despite encountering various obstacles, the integration of AI in obstetrics and gynecology has exhibited remarkable progress. The utilization of AI in various domains has proven to be highly effective in addressing persistent issues related to diagnosis and treatment. According to a study, AI has the potential to enhance knowledge and provide assistance to medical professionals in the fields of gynecology and obstetrics [122]. The latest applications of AI models in gynecology involve identifying endometrial carcinoma, in vitro fertilization, uterine sarcoma, cervical intraepithelial neoplasia, and advancing anticancer medication [123,124].
The integration of AI technology into ultrasonography has the capacity to enhance the adoption of medical ultrasound in various clinical environments, facilitating its broader application by healthcare professionals. Therefore, the utilization of AI in the field of ultrasonography for prenatal care has the potential to assist medical professionals in efficiently prioritizing and accurately diagnosing the anatomical structures of pregnant individuals. In certain medical applications such as obstetric pelvic and echocardiography ultrasonography, where visual analysis and measurement play a crucial role, the utilization of video clips can provide a comprehensive set of structured data. This enables spatiotemporal analysis and enhances the advantages of ANNs [125]. A study investigated the efficacy of AI algorithms in ultrasonic diagnosis for pregnant patients with brain tumors. They specifically focused on evaluating the accuracy rate of this diagnostic approach [126]. The diagnostic accuracy achieved through the utilization of AI was recorded at 94.50%. Another research study was conducted, which involved a prospective and descriptive approach. The study focused on a sample of approximately 244 pregnant women in their first pregnancy trimester. The registered female participants were specifically queried regarding their utilization of iron, folic acid, potassium iodide, and multivitamin supplements throughout their pregnancies. The utilization of an ANN model that incorporates variables related to pregnancy checks, intake of iodized salt, iodized supplements, and iodine-rich foods can be used to predict iodine deficiency during the early stages of pregnancy. This predictive model can assist experts in making a more feasible diagnosis [127]. In their study, Sakai et al. utilized a newly developed DL-based explainable graph chart diagram representation to aid in fetal cardiac ultrasound screening. This screening process is known to have a relatively low rate of detecting congenital heart disease during the second-trimester stages, primarily due to the challenges associated with mastering the technique [128]. Consequently, the utilization of AI in the second and third trimesters of pregnancy for diagnostic purposes, specifically using diagram representation, enhances screening performance. The accuracy rate for experts increases from 96% to 97.50%, while non-experts improve from 82% to 89% [129].
3.13. Ophthalmology
The utilization of AI in diagnosing and managing ocular disease has become increasingly popular due to research findings emphasizing its potential to enhance personalized medicine and improve healthcare outcomes [130]. Numerous AI algorithms are currently under development for managing patients diagnosed with diabetes mellitus [131].
Due to advancements in the management of diabetes mellitus, there has been an enhancement in the monitoring of patients, resulting in a higher incidence of diabetic retinopathy and diabetic macular edema. The primary cause of significant visual impairment and blindness among individuals of working age is the presence of diabetic macular edema that has not been diagnosed or treated [132]. Hence, it is imperative to conduct extensive screening for diabetic retinopathy on a large scale in order to identify potentially detrimental alterations at an early phase, thereby facilitating effective management and treatment strategies.
Considering the prevailing patterns of population growth and the significant incidence of diabetic retinopathy and diabetic macular edema within the community, it is inevitable that automated screening and diagnosis will become increasingly prevalent in ophthalmic healthcare settings. Efforts have been made to explore automated retinal screening techniques for the diagnosis of diabetic retinopathy to enhance patient management and mitigate the societal impact. Various AI, ML, and DL methodologies have been used for the automated diagnosis and grading of diabetic retinopathy. The most efficacious automated systems have been developed based on comprehensive investigations conducted within the last three years. Recent research on diabetic retinopathy has shown that AI techniques have exhibited significant accuracy, sensitivity, and specificity in identifying and diagnosing diabetic retinopathy [133].
Automated application systems have the potential to enhance doctors’ comprehension of diabetic retinopathy predictions and enhance the practicality of intelligent diagnostic models in real-world clinical settings [133]. Based on the aforementioned studies, it was observed that the automated analysis of retinal images exhibited a high level of accuracy, validity, sensitivity, and specificity in detecting diabetic retinopathy. Furthermore, the diagnostic performance of AI techniques was deemed clinically acceptable and demonstrated high reproducibility when applied to the validation data set.
Age-related macular degeneration is a chronic ocular condition that is recognized as a prominent contributor to visual impairment [134]. Prognostications AI algorithms exist to generate personalized predictions in age-related macular degeneration. These algorithms can make predictions regarding the presence of drusen beneath the retina in individuals with age-related macular degeneration. The AI algorithms offer automated detection capabilities for identifying drusen, fluid, and geographic atrophy in relation to age-related macular degeneration lesions. These algorithms leverage fundus images and spectral-domain optical coherence tomograph to enhance the diagnosis and treatment [135]. The utilization of AI in the automated detection of drusen holds promise for enhancing the diagnostic capabilities of ophthalmologists in the early and efficient assessment of fundus images [136]. The application of AI techniques in diagnosing and grading age-related macular degeneration has been extensively explored. Recent studies have shown that these automated approaches exhibit notable efficacy, demonstrating high accuracy, sensitivity, and specificity levels in detecting age-related macular degeneration [137].
Glaucoma, which ranks as the second most prevalent factor leading to visual impairment on a global scale, is distinguished by the gradual degeneration of retinal ganglion cells and the permanent depletion of axons within the optic nerve. The timely identification and management of glaucoma is of paramount significance in the prevention of preventable visual impairment. AI techniques have demonstrated exceptional efficacy in efficiently classifying glaucomatous and healthy eyes. Ophthalmologists have the ability to utilize these automated results as a reference point, enabling them to enhance their decision-making process within clinical practice. The utilization of automated AI applications has demonstrated significant efficacy and holds promise in addressing the imminent challenge of diabetic retinopathy, age-related macular degeneration, and glaucoma screenings in both developed and developing nations [138].
3.14. Pediatrics
Imaging techniques play a paramount role in the management of pediatric neurologic, neurosurgical, and neuro-oncological conditions [139]. Multi-parametric MRI techniques are gaining popularity, particularly when combined with radiogenomic analyses that establish connections between imaging features and molecular biomarkers associated with diseases. Nevertheless, incorporating this approach into regular clinical practice continues to be challenging. AI techniques can model extensive datasets related to childhood neurologic disease, including radiologic, biological, and clinical data. This capability enables the integration of such information into prognostic modeling systems at an early stage. Consequently, AI techniques offer a viable solution to address this issue [139].
In certain applications within the field of pediatric neuroradiology, ANNs have demonstrated notable efficacy in a focused manner. This concept is most effectively demonstrated through the utilization of ventricular size determination to categorize children into either a normal or hydrocephalic group. In a recent study, an analysis was performed on hydrocephalus and controls [140]. They achieved an accuracy score of 94.6% for hydrocephalus and 85.6% for controls using a training set of T2-weighted MRI images from around 399 children. Previous studies have reported comparable achievements in the field of pediatric hydrocephalus through the implementation of evolutionary modifications in ANN methodologies [141].
The application of a support vector machine for the categorization of children into normal or hypoxic-ischemic brain injury groups, based on the measurement of corpus callosum widths, yielded a classification accuracy of 95% [142]. Another study utilized a comparable methodology to examine a group of adolescents who had experienced traumatic brain injury. Specifically, they utilized edge-density imaging and support vector machines to classify the participants into two categories: normal and mild traumatic brain injury [143]. The aforementioned method, which achieved a precision rate of 94%, demonstrated superior performance compared to neurocognitive testing in this aspect [25]. Support vector machines have demonstrated successful classification of various magnetic resonance imaging abnormalities in the fetal brain. These abnormalities encompass functional connectivity, brain maturity, and severe fetal abnormalities. The classification accuracies achieved by support vector machines in these studies range from 79% to 84% [144].
Support vector machines (SVMs) have been utilized in magnetic resonance imaging texture analysis to examine brain tumors. This machine learning application aims to quantitatively analyze imaging data to generate an image texture that is generally imperceptible to human visual perception [145]. Texture analysis in clinical practice is advantageous for clinicians because it can incorporate comprehensive imaging data of the entire tumor. This approach takes into consideration the presence of intra-tumor heterogeneity, which may not be adequately represented by a single biopsy site or even multiple biopsy sites [145].
A study expanded on the application of texture analysis by integrating both linear discriminant analysis and a probabilistic neural network [146]. Their objective was to categorize posterior fossa tumors, specifically medulloblastoma, pilocytic astrocytoma, and ependymoma. The combined techniques achieved an accuracy ranging from 86% to 93% through a validation process. The utilization of AI in diagnosis offers a potential enhancement to the effectiveness of diagnoses.
Decision trees have also been used in another significant capacity within the field of ML in the context of pediatric neuroimaging. Specifically, they have been utilized for the purpose of data analysis in order to provide insights and information regarding neuroimaging in clinical trials. An instance can be observed in a study where a decision tree classifier was used within a randomized controlled trial conducted on children diagnosed with autism who were undergoing treatment with simvastatin [147]. The study utilized a random forest classifier to effectively categorize children from the control group who had undergone simvastatin treatment [147]. The classifier achieved a classification accuracy of 79%. This observation suggests the potential benefits that such applications may offer in the future.
3.15. Hematology
AI has been used to examine various types of medical data, including hematopathological, radiographic, laboratory, genomic, pharmacological, and chemical data. The purpose of using AI in these analyses is to enhance the accuracy and effectiveness of diagnosis, outcome prediction, and treatment planning and to expand our understanding of benign and malignant hematology.
Recent advancements in CNN-based models have shown the ability to effectively differentiate between various types of leukocytes on peripheral smears, indicating their potential for automating routine pathology practices [148]. Ongoing research is being conducted in the field of automated interpretation of bone marrow specimens [149]. CNNs have also exhibited their usefulness in characterizing qualitative and quantitative variations within specific cell lineages, such as the morphology of erythrocytes and the textural alterations observed in sickle cell disease [150]. The aforementioned achievements encompass the differential diagnosis of various diseases, as evidenced by the capacity of models to accurately diagnose acute myeloid leukemia, distinguish between different causes of bone marrow failure, and function as a screening tool for lymphoma in settings with limited resources [151].
AI has been applied in various domains to enhance diagnostic processes’ dependability, convenience, and efficacy. Previous studies have shown that CNN methods have proven effective in diagnosing multiple myeloma solely using mass spectrometry data obtained from peripheral blood [152,153]. Personalized models have been shown to possess a high diagnostic capability when distinguishing between challenging conditions, such as different causes of bone marrow failure, is difficult. This is achieved by integrating patient demographics, laboratory data, and fundamental genetic information. Previous studies have also utilized similar methods to differentiate between peripheral leukemia and lymphoma [154].
The task of prognosis is widely recognized as challenging, and even commonly used clinical prognostication tools exhibit notable variability within different risk categories [155]. AI, possessing advanced capabilities in processing nonlinear and intricate data, promises to deliver more sophisticated and individualized prognostications. The aforementioned methodologies have been used within the field of benign hematology to enhance the accuracy of risk assessment for central catheter thrombosis. These methodologies have successfully identified individuals with a low risk of developing thrombosis [155]. AI has been utilized to categorize patients undergoing hematopoietic stem cell transplants into low and high-risk groups for acute graft-versus-host disease. This classification has important implications for making informed decisions regarding the administration of immunosuppressive treatments to these individuals [156]. Previous studies have also been conducted in the field of autologous transplants for multiple myeloma. AI has been utilized in the field of malignant hematology to enhance the initial assessment of risk stratification for acute myeloid leukemia and myelodysplastic syndromes [157,158]. In post-treatment scenarios, where minimal residual disease is considered a negative prognostic factor, AI has exhibited the capability to attain performance comparable to that of humans. This achievement has the potential to simplify and establish a consistent approach to handling and analyzing this kind of data [159].
3.16. Intensive Care Unit
ML models have been applied within the intensive care unit (ICU) setting to anticipate pathologies such as acute kidney injury, identify symptoms such as delirium, and suggest appropriate therapeutic interventions such as vasopressors and fluid administration in cases of sepsis. The timely identification and management of sepsis is of paramount importance due to its potential to significantly decrease mortality rates. Although the management of early sepsis involves source control and the administration of broad-spectrum antibiotics, the detection of sepsis during this phase of the illness poses considerable challenges [159]. Identifying sepsis becomes increasingly feasible as the condition advances, while the treatment poses considerable challenges. Due to the diverse nature of sepsis, the existing diagnostic and prognostic methods pose a significant challenge in early sepsis detection and accurate prognosis estimation. This difficulty further complicates determining an appropriate treatment strategy for individual patients [160]. AI prediction models have the potential to provide significant value for patients diagnosed with sepsis. AI models possess the capacity to enhance the timely identification of individuals requiring antibiotic treatment. Certain AI prediction models appear to outperform existing diagnostic methods; however, these models exhibit notable limitations, such as including predictor variables like blood pressure in the present sepsis definition. The assessment of AI models’ performance is exaggerated in this context. These models exhibit limited generalizability. The existence of unresolved concerns has resulted in a significant disparity between the advancement of algorithms and their practical implementation in clinical settings.
The growing utilization of Electronic Health Records within the ICU is driving the dissemination of data science and ML techniques in the critical care setting. Hemodynamic data derived from monitors, infusion data obtained from infusion pumps, and respiratory data collected from ventilators generate substantial data volumes. These datasets can be compared to other sources of big data, such as omics data encompassing genomics or proteomics. A study devised a computational model utilizing reinforcement learning techniques to propose optimal treatment strategies dynamically for adult ICU patients [161]. A study was conducted wherein pervasive monitoring and ML techniques were applied to continuously evaluate delirium and agitation in a cohort of 22 patients admitted to an ICU [162]. The patients were categorized based on the Confusion Assessment Method for the ICU scale. The researchers utilized cameras and accelerometers to capture and document facial expressions and movements. Three accelerometers were strategically positioned on the patient’s wrist, ankle, and arm to discern and classify their posture. The researchers applied a pre-existing neural network model to perform facial recognition and detect facial expressions using individual facial features.
3.17. Diagnostic Methods
AI has the capability to fundamentally transform the methodologies applied in disease diagnosis and treatment. Through the examination of patient data, including medical history, symptoms, and test results, AI algorithms can provide clinicians with more precise and tailored diagnoses for individual patients. This has the potential to facilitate the identification of diseases at an earlier stage and enhance the efficacy of treatment strategies. Additionally, AI can assist in identifying potential drug interactions and adverse reactions, ensuring that patients receive the safest and most effective treatments.
Ultrasound (US) has gained widespread global adoption as a primary imaging modality in various clinical domains, owing to the continuous advancements in ultrasonic technologies and the established digital health infrastructure. Breast cancer is widely recognized as a prevalent form of cancer among women globally and continues to be the second most significant contributor to cancer-related mortality [163]. The predominant utilization of DL in breast US, as observed in the surveyed literature, pertains to diagnosing and categorizing breast masses [164]. The utilization of DL techniques for the analysis of abdominopelvic imaging has various applications within the United States. A significant portion of these applications have been specifically directed towards the examination of the liver [165]. Their research revealed that this approach exhibited superior accuracy compared to two-dimensional shear wave elastography and certain biomarkers in the evaluation of advanced fibrosis and cirrhosis in patients infected with the hepatitis B virus. In a study, the authors devised a CNN approach to predict the METAVIR score, a semi-quantitative measure of liver fibrosis [166]. The training dataset consisted of several thousands of US images obtained from two tertiary academic referral centers. This approach demonstrated a notable level of precision in forecasting the METAVIR score, surpassing radiologists’ diagnostic capabilities in identifying liver fibrosis. In their study, Ta et al. developed a computer-aided diagnosis system for the classification of malignant and benign focal liver lesions using contrast-enhanced ultrasound cine recordings [167]. The researchers found that the accuracy of this method was comparable to that of an expert reader.
Deep learning algorithms have been utilized in various applications, including identifying muscle diseases [168], determining cone positioning, and segmentation of muscle imaging [169]. The diagnostic accuracy for neuro-muscular diseases was improved by using a CNN-based method, which enhanced the assessment and classification of inflammatory muscle diseases [168]. The progress in the field of AI/ML tools for the interpretation of imaging is experiencing rapid acceleration. This can be attributed to several factors, including the availability of extensive digitized image datasets, the accessibility of open-source algorithms, advancements in computing power, the emergence of cloud services, and the continuous development of DL techniques [170].
The utilization of automation in tasks frequently performed by radiologists, such as identifying rib fractures and lung nodules using CT scans, reassessing pleural effusion size via sequential chest radiographs, or conducting mammographic screening, shows potential as a favorable strategy. This has the potential to enable radiologists to dedicate additional time to more advanced interpretive tasks that may not be amenable to automation, as well as participate in endeavors such as multidisciplinary team meetings. The utilization of AI in the triage procedure demonstrates a notable ability to efficiently assign priority to critical cases that require immediate reporting. These cases may include CT scans that unveil the presence of pulmonary embolism, chest radiographs that indicate pneumothorax, or head CTs that reveal hemorrhage. The aforementioned methodology possesses the capacity to reduce patient morbidity and expedite the duration of hospitalization in emergency departments. The utilization of DL systems in synthetic MRI enables the post-processing and reconstruction of MR image data, reducing image acquisition time without significant deterioration in image quality. This advancement can potentially enhance efficiency, decrease expenses, and enhance accessibility [171].
CNNs are a fundamental element within networks utilized in orthopedic and musculoskeletal radiology [172]. In recent times, the concept of generative adversarial networks (GANs) has been introduced. GANs are models designed to generate novel data closely resembling the original dataset. These models comprise two distinct networks that engage in a competitive game-like training process. A total of 12 enhanced GAN models incorporating CNNs have been successfully utilized in the domain of radiographic images [173].
Extensive research has been conducted in the field of oral and maxillofacial radiology to investigate the potential of AI in the diagnosis of various conditions. The aforementioned conditions encompass dental caries, periodontal disease, osteosclerosis, odontogenic cysts, and tumors, as well as ailments that impact the maxillary sinus or temporomandibular joints.
AI has been used in the field of dentistry for image analysis, encompassing a range of tasks, including tooth segmentation and localization, assessment of bone quality for osteoporosis, determination of bone age through hand-wrist radiographs, and localization of cephalometric landmarks [172]. DL systems utilizing CNN architectures have been successfully applied in the domain of dentistry. Notably, a novel system has been developed that incorporates both three-dimensional cone beam computed tomography images and two-dimensional images [172].
The application of DL techniques has been used to identify and categorize teeth within both cone beam computed tomography images and panoramic images. The utilization of classification systems for teeth enables dentists to make clinical decisions and streamline their charting process by using automated computer-aided design outputs. These outputs facilitate the automatic completion of digital patient records [174].
4. Discussion
4.1. General
The aim of this literature review is to provide a comprehensive overview of existing research on AI in clinical practice. In addition to its extensive use in diagnostic techniques, AI integration has been explored in various medical disciplines, including cardiology, anesthesiology, surgery, pneumology, neurology, urology, gynecology, hematology, and pediatrics. Given the continuous influx of new articles and the exponential increase in published papers, this review focuses on indicative articles to illustrate AI’s robust penetration and wide-ranging applications in clinical practice.
One of the major advantages of AI in clinical practice is its ability to improve diagnostic accuracy and treatment outcomes [10,11,12,13,14,15,16,17,18,19,20,21,22]. With AI-powered algorithms, healthcare providers can analyze large amounts of patient data and identify patterns that may not be immediately apparent to human clinicians. This can help to identify diseases earlier, resulting in expedited medical intervention and improved prognoses for individuals. Additionally, AI can predict which treatments are likely to be most effective for a given patient, allowing for personalized medicine that considers individual patient characteristics. AI can help reduce healthcare costs by streamlining administrative processes and reducing unnecessary tests [23].
In a study, ML techniques were applied in the analysis of high-throughput genome sequencing data, aiming to enhance comprehension of disease processes and the development of therapeutic modalities [175]. In this study, cutting-edge ML algorithms, including random forest, support vector machine radial kernel, adaptive boost, averaged neural network, and gradient boosting machine, were applied. The goal was to stratify patients with head and neck squamous cell carcinoma into early and late clinical stages and to predict the risk based on the expression profiles of miRNAs [175]. Also, quite recently, variational autoencoders (i.e., a type of neural network) were introduced as a method to effectively augment the sample size of clinical studies, thereby mitigating costs, time constraints, dropouts, and ethical considerations [176]. In a study, the efficacy of variational autoencoders in the context of data augmentation was demonstrated through the utilization of simulations encompassing multiple scenarios [176]. Also, in the field of bioequivalence studies, several ML methods were utilized to solve the old problem of defining the appropriate absorption rate metric [20,21,22]. Through the joint utilization of ML algorithms, non-linear mixed effect modeling, and Monte Carlo simulations, a new metric termed “average slope” was defined and introduced. It was proven that the currently used Cmax (i.e., the maximum observed plasma concentration) is unsuitable for expressing the absorption rate. On the contrary, the newly defined measure (average slope) comprises the desired properties of absorption rate, has the appropriate units of measurement (i.e., concentration units per time), exhibits ease of estimation directly from the concentration-time data of the drug, and all ML algorithms showed its supremacy over all other metrics used or proposed in bioequivalence [20,21,22].
Although the advantages of AI in clinical practice are evident, a number of obstacles exist that necessitate attention and resolution [177]. One of the foremost considerations revolves around the possibility for AI to sustain and propagate bias within the healthcare sector. If the algorithms used in AI systems are trained on biased data, they may produce biased results, leading to disparities in healthcare outcomes for certain patient populations. Also, there is a lack of transparency, interpretability, and explainability of the AI algorithms since the latter can be considered as a black box. This issue will be discussed later. Additionally, there is a risk that AI could dehumanize healthcare, with patients feeling disconnected from their care providers and reduced to a set of data points. There are also fears around the ethical implications of using AI in healthcare, particularly. The impact of integrating AI technologies into the field of medicine will be most pronounced among present and prospective medical trainees. Consequently, it is imperative for medical schools and graduate medical education programs to modify their curriculum in order to instruct current and future generations of physicians on the conscientious utilization of these potent and transformative technologies [178]. Integrating AI in medicine introduces several legal and ethical considerations, including medical liability issues such as training and competence, transparency/traceability, explainability, personal health data, regulatory compliance, product liability, and malpractice insurance. The fundamental aspects of these issues are delineated below.
4.2. Training of Healthcare Professionals
Healthcare professionals using AI systems need appropriate training to ensure competence in their use. Failure to properly understand and operate AI systems may lead to medical errors and subsequent liability. Furthermore, prospective medical students will be required to develop new skills, encompassing the concept of “knowledge capture, not knowledge retention”. This implies a shift from a curriculum focused on rote memorization to one that prioritizes critical thinking. The field of AI places significant emphasis on two primary domains: collaboration and management of AI applications, as well as a deeper comprehension of probabilities and their consequential application in clinical decision-making involving patients and families. The aforementioned domains aim to acquire knowledge pertaining to the efficient and ethical utilization of AI, while also promoting the practical application of AI technologies in the healthcare field [179].
4.3. Transparency, Traceability, and Explainability
Drawing inspiration from various disciplines, the domains of transparency and traceability in the context of healthcare and individual patients necessitate adherence to more rigorous criteria [180]. From a legal standpoint, it is necessary for data to adhere to all applicable laws, regulations, and additional legal obligations throughout its entire lifecycle, including acquisition, storage, transfer, processing, and analysis. Furthermore, it is imperative for the law, its interpretation, and its implementation to continually adjust in response to the ever-changing advancements in technology [181]. Numerous AI algorithms, particularly those based on deep learning models, function as enigmatic “black boxes”, rendering it difficult to clarify the rationale behind their decisions. In situations where the validity of a medical decision generated by an AI system is scrutinized, the absence of transparency can give rise to legal complications. Even when meeting all of these apparent prerequisites, the question persists as to whether the utilization of AI-driven solutions and tools necessitates the need for explainability. In essence, doctors and patients must possess knowledge regarding the outcomes presented and an understanding of the qualities and attributes upon which these outcomes are founded, as well as the corresponding underlying assumptions. Furthermore, the inclusion of additional stakeholders may necessitate a comprehensive comprehension and explication of algorithms and models. From a Western legal standpoint, three fundamental areas have been identified for the purpose of elucidating the concept of explainability [182]. These areas include (a) informed consent, (b) certification and approval in accordance with the regulations set forth by the FDA and the Medical Device Regulation (MDR), and (c) Liability.
4.4. Liability and Regulatory Framework
The certification and approval bodies responsible for medical devices have been relatively sluggish in implementing regulations pertaining to explainable AI and its impact on developing and marketing such products. The FDA significantly promotes the continuous development and enhancement of AI-based medical products through its comprehensive total product lifecycle approach. The concept of explainability is not explicitly referenced; however, there is a requirement for an appropriate level of transparency and clarity in the output and algorithm intended for users [183,184]. The primary focus of this inquiry pertains to the functionalities of the software and the alterations it has undergone throughout its evolution. The MDR does not explicitly address the requirement for explainability in relation to medical devices utilizing AI and ML, specifically. In this context, it is important to emphasize the significance of accountability and transparency [185]. Specifically, these requirements pertain to the provision of information that enables the tracing, transparency, and explication of the development process of ML and DL models that contribute to medical treatment. There is a strong probability that in forthcoming times, a more refined delineation of these prerequisites will emerge, necessitating manufacturers of AI-driven medical devices/software to furnish exhaustive details pertaining to the training and evaluation of the models, the data utilized, and the overarching methodologies used in their creation.
The integration of AI in healthcare frequently entails the utilization of highly sensitive patient data. Suppose a data security or privacy breach results in unauthorized access or use of patient information; healthcare providers and AI developers may face legal repercussions. This underscores the critical importance of robust data protection measures and adherence to privacy regulations in developing and deploying AI applications within the healthcare sector [185]. The processing of personal health data is permissible under the law only when the individual has provided explicit consent for its utilization. The current standard for utilizing patient data in AI applications is informed consent, as there is a lack of overarching legislation governing the use of personal data and information [185]. Healthcare organizations and AI developers must comply with various data protection regulations, such as the Health Insurance Portability and Accountability Act (HIPAA) in the United States or the General Data Protection Regulation (GDPR) in Europe [186,187]. Failure to comply with these regulations can lead to legal consequences and fines. Clear policies should be established regarding data ownership and patient consent. Patients should be informed about how their data will be used, who will have access to them, and for what purposes. Obtaining informed consent is essential for ethical and legal reasons. Implementing robust encryption methods for both data in transit and data at rest helps protect patient information from unauthorized access. Encryption adds an extra layer of security to prevent sensitive data from being intercepted or accessed by unauthorized parties. Access to healthcare AI systems and the data they process should be restricted and monitored. Role-based access controls should be implemented to ensure that only authorized personnel can access sensitive information. Also, healthcare AI systems should use secure methods for storing and transmitting data, while periodic security audits and assessments help identify vulnerabilities in the system.
The legal framework concerning AI in healthcare is still in a state of evolution. New laws and regulations may be introduced to address issues related to liability specifically, and healthcare professionals must stay informed about these changes. As the field of AI continues to advance, legal frameworks are likely to adapt to ensure that ethical, transparent, and responsible practices are followed in developing and deploying AI technologies in healthcare. Staying abreast of these evolving regulations is crucial for healthcare professionals to navigate the complex legal terrain and uphold accountability and patient care standards. Nevertheless, even to this day, disparities persist in international guidance between Europe and the United States regarding the legal challenges that may arise from the use of AI in healthcare. These regions adopt distinct approaches to addressing these challenges. The European Union (EU) has emerged as a leading force in the field of medical AI innovation and has acknowledged the specific difficulties that AI poses to current liability frameworks. In order to establish coherence in liability principles and ensure legal clarity, the European Commission has introduced the Artificial Intelligence Act, which represents one of the initial legal frameworks dedicated specifically to AI [186]. The European Commission endeavors to advance the secure utilization of AI in sectors with significant consequences, such as healthcare, while concurrently enhancing technological innovation. The United States lacks a comprehensive legal framework that specifically regulates AI, resulting in limited legal precedent concerning liability and medical AI. The regulatory aspect of AI in healthcare has been acknowledged by the FDA, which aims to facilitate the secure implementation of AI by developing a strategic plan to ensure ongoing supervision of AI as a medical device [187]. In order to foster a patient-centric approach, the FDA endeavors to enhance transparency by requesting manufacturers to provide detailed descriptions of the operational mechanisms of their AI devices. This initiative is aimed at facilitating a comprehensive comprehension of the advantages and potential hazards associated with such devices. The FDA also endeavors to address potential bias that may arise from training AI algorithms on specific populations or historical datasets [187]. The FDA has recently published a discussion paper entitled “Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning-Based Software as a Medical Device” with the aim of guaranteeing the safety of medical software that utilizes AI and ML technologies.
In addition, as the use of AI in healthcare becomes more prevalent, malpractice insurance considerations evolve to address potential risks associated with these technologies [188]. Given the unique risks associated with AI in healthcare, malpractice insurance policies may need to be tailored to address liabilities arising from the use of AI technologies specifically. This could include coverage for errors or malfunctions in AI algorithms that lead to adverse patient outcomes. Insurers may face challenges in underwriting policies related to AI, as the field is rapidly evolving, and assessing risks associated with emerging technologies can be complex. Insurers may need to adapt their underwriting processes to account for the specific risks posed by AI in healthcare [189]. Malpractice insurance policies related to AI must align with evolving legal and regulatory frameworks governing the use of AI in healthcare. Insurers may need to stay informed about changes in laws and regulations to ensure that their policies remain relevant and compliant. Also, insurers may require healthcare organizations to implement monitoring and reporting mechanisms for AI-related incidents. Timely reporting adverse events can facilitate a proactive response and help mitigate potential liabilities.
In the same vein, product liability in the context of AI refers to the legal responsibility of those involved in the design, development, manufacturing, distribution, and deployment of AI systems for any harm or damages caused by the AI product [190,191]. If the design of an AI system is inherently flawed and leads to harmful consequences, the designers and developers may be held liable. AI design defects could include biased algorithms, inadequate testing, or a lack of robust safety features. Manufacturing defects pertain to issues arising during an AI system’s production or deployment. These defects may lead to malfunctions, security vulnerabilities, or other problems that could result in harm. Manufacturers may be held responsible for these defects. If AI products come with inadequate warnings or instructions regarding their proper use, healthcare professionals or end-users may be unaware of potential risks. Failure to provide clear guidance on the limitations and risks of an AI system could result in liability. In cases where third-party vendors provide AI components or services, liability may extend to these vendors if their products or services contribute to harm. Determining the chain of responsibility and liability in complex AI ecosystems can be challenging. Rigorous testing and validation of AI systems are essential to identify and address potential issues before deployment. If inadequate testing contributes to harm, the parties responsible for the testing and validation process may be held liable. Maintaining detailed records of the design, development, testing, and deployment processes is important for demonstrating due diligence in the event of a product liability claim. Clear documentation can provide evidence of compliance with industry standards and best practices.
4.5. Overall
This literature review discusses the current applications of AI in various medical specialties in clinical practice. AI possesses the capacity to fundamentally transform clinical practice and enhance patient outcomes. The potential for further advancements in AI technology is vast, and the impact on patient outcomes could be significant.
Certainly, not all aspects and bibliographic references could have been included, as this literature review aims to offer a comprehensive overview of the applications of AI in as many medical specialties as possible. Consequently, it was unavoidable that some important contributions in each field could not be discussed. As an example, there has been a substantial rise in the utilization of biomarkers as early warning systems for assessing disease risk over the past decade, with extensive application evident during the recent COVID-19 pandemic [192]. In the same context, in dermatology, which is one of the fields with the widest use of AI, quite recently, a research paper has underscored the capability of machine learning to serve as a biomarker for differentiating among individuals with psoriasis, psoriatic arthritis, and those in good health [193]. Also, several investigations have been carried out, uncovering notable molecular biomarkers through miRNA expression that can differentiate between early and late stages of carcinomas [175].
It should also be emphasized that despite the concerns discussed in this review, the potential benefits of AI in healthcare cannot be ignored. AI has the ability to improve diagnosis accuracy, personalize treatment plans, and reduce healthcare costs. In the future, AI will likely become a standard tool in clinical practice, with healthcare providers working alongside AI systems to provide the best possible care for patients [185,186]. Nevertheless, to guarantee the ethical and efficient utilization of AI in the healthcare sector, we must confront these apprehensions and establish explicit protocols for advancing and integrating AI systems. The complete realization of AI’s potential to enhance healthcare outcomes can only be achieved under such circumstances. Overall, it is considered that AI has the potential to greatly improve healthcare outcomes, but it is important to address ethical concerns and establish clear guidelines for its development and implementation.
5. Conclusions
Integrating AI into clinical practice brings forth many benefits and challenges with significant implications for ethical and legal considerations. AI holds the promise of improving the precision of diagnostic accuracy, streamlining administrative tasks, and personalizing treatment plans. By analyzing vast amounts of medical data, AI systems can identify patterns and correlations that might escape human observation, leading to more precise and timely interventions. Additionally, AI can contribute to cost-effective healthcare solutions and improve overall patient outcomes. This integration of AI technology facilitates informed clinical decision-making processes. Thus, AI has the potential to enhance patient outcomes, offering faster and more accurate diagnoses, personalized treatment plans, and reduced healthcare costs. While there is still much to be explored and developed in the field of AI in clinical practice, it is crucial that we continue to invest in research and development to unlock its full potential. This approach can improve our understanding of how AI can enhance healthcare and lead to the developing of new tools and technologies that benefit patients and healthcare professionals alike. Nevertheless, it is important to approach its development and implementation cautiously and collaborate with healthcare professionals to ensure ethical considerations are met.
However, these advancements come with ethical dilemmas and legal complexities. The use of AI in clinical decision-making raises concerns about transparency, accountability, and the potential for bias in algorithms. Ensuring patient privacy and data security becomes paramount, demanding robust ethical guidelines and legal frameworks. Striking the right balance between innovation and safeguarding patient rights requires careful consideration of the ethical implications of AI in clinical practice, alongside the development of legal frameworks that can adapt to the rapid pace of technological evolution in the healthcare sector. To overcome the ethical and legal challenges associated with the integration of ai in clinical practice, a multi-faceted approach is essential that includes legal frameworks and regulations, transparent and explainable AI, ethical guidelines and standards, regular audits and assessments, incentives for ethical practices, and international collaboration.
Funding
This research received no external funding.
Conflicts of Interest
The author declares no conflict of interest.
References
- Henderson, H. Artificial Intelligence: Mirrors for the Mind (Milestones in Discovery and Invention), 1st ed.; Chelsea House Publisher: New York, NY, USA, 2007; 176p. [Google Scholar]
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach, 4th ed.; Pearson: Bloomington, MN, USA, 2021; 1136p. [Google Scholar]
- Philippidis, A. Charles River Licenses ERS Genomics’ CRISPR/Cas9 Technology. Clin. OMICs 2018, 5, 7. [Google Scholar] [CrossRef]
- Turing, A.M. On Computable Numbers, with an Application to the Entscheidungsproblem. Proc. Lond. Math. Soc. 1937, s2-42, 230–265. [Google Scholar] [CrossRef]
- McCulloch, W.S.; Pitts, W. A Logical Calculus of the Ideas Immanent in Nervous Activity. Bull. Math. Biophys. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Widrow, B.; Lehr, M.A. 30 Years of Adaptive Neural Networks: Perceptron, Madaline, and Backpropagation. Proc. IEEE 1990, 78, 1415–1442. [Google Scholar] [CrossRef]
- Lodwick, G.S.; Keats, T.E.; Dorst, J.P. The Coding of Roentgen Images for Computer Analysis as Applied to Lung Cancer. Radiology 1963, 81, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Feigenbaum, E.A.; Buchanan, B.G. DENDRAL and Meta-DENDRAL: Roots of Knowledge Systems and Expert System Applications. Artif. Intell. 1993, 59, 233–240. [Google Scholar] [CrossRef]
- Goldberg, J.E.; Reig, B.; Lewin, A.A.; Gao, Y.; Heacock, L.; Heller, S.L.; Moy, L. New Horizons: Artificial Intelligence for Digital Breast Tomosynthesis. RadioGraphics 2023, 43, e220060. [Google Scholar] [CrossRef]
- Zhong, F.; Xing, J.; Li, X.; Liu, X.; Fu, Z.; Xiong, Z.; Lu, D.; Wu, X.; Zhao, J.; Tan, X.; et al. Artificial Intelligence in Drug Design. Sci. China Life Sci. 2018, 61, 1191–1204. [Google Scholar] [CrossRef]
- Bakkar, N.; Kovalik, T.; Lorenzini, I.; Spangler, S.; Lacoste, A.; Sponaugle, K.; Ferrante, P.; Argentinis, E.; Sattler, R.; Bowser, R. Artificial Intelligence in Neurodegenerative Disease Research: Use of IBM Watson to Identify Additional RNA-Binding Proteins Altered in Amyotrophic Lateral Sclerosis. Acta Neuropathol. 2017, 135, 227–247. [Google Scholar] [CrossRef]
- Segler, M.H.S.; Preuss, M.; Waller, M.P. Planning Chemical Syntheses with Deep Neural Networks and Symbolic AI. Nature 2018, 555, 604–610. [Google Scholar] [CrossRef]
- Simões, M.F.; Silva, G.; Pinto, A.C.; Fonseca, M.; Silva, N.E.; Pinto, R.M.A.; Simões, S. Artificial Neural Networks Applied to Quality-by-Design: From Formulation Development to Clinical Outcome. Eur. J. Pharm. Biopharm. 2020, 152, 282–295. [Google Scholar] [CrossRef] [PubMed]
- Gaisford, S.; Saunders, M. Essentials of Pharmaceutical Preformulation, 1st ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2013; 268p. [Google Scholar]
- Babu, N.J.; Nangia, A. Solubility Advantage of Amorphous Drugs and Pharmaceutical Cocrystals. Cryst. Growth Des. 2011, 11, 2662–2679. [Google Scholar] [CrossRef]
- Damiati, S.A.; Martini, L.G.; Smith, N.W.; Lawrence, M.J.; Barlow, D.J. Application of Machine Learning in Prediction of Hydrotrope-Enhanced Solubilisation of Indomethacin. Int. J. Pharm. 2017, 530, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.; Kabedev, A.; Parrow, A.; Bergström, C.A.S.; Larsson, P. Molecular Simulation as a Computational Pharmaceutics Tool to Predict Drug Solubility, Solubilization Processes and Partitioning. Eur. J. Pharm. Biopharm. 2019, 137, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Yang, Y.; Li, X.; Cao, D.; Ouyang, D. An Integrated Transfer Learning and Multitask Learning Approach for Pharmacokinetic Parameter Prediction. Mol. Pharm. 2018, 16, 533–541. [Google Scholar] [CrossRef]
- Ota, R.; Yamashita, F. Application of Machine Learning Techniques to the Analysis and Prediction of Drug Pharmacokinetics. J. Control. Release 2022, 352, 961–969. [Google Scholar] [CrossRef]
- Karalis, V.D. Machine Learning in Bioequivalence: Towards Identifying an Appropriate Measure of Absorption Rate. Appl. Sci. 2022, 13, 418. [Google Scholar] [CrossRef]
- Karalis, V.D. On the Interplay between Machine Learning, Population Pharmacokinetics, and Bioequivalence to Introduce Average Slope as a New Measure for Absorption Rate. Appl. Sci. 2023, 13, 2257. [Google Scholar] [CrossRef]
- Karalis, V.D. An In Silico Approach toward the Appropriate Absorption Rate Metric in Bioequivalence. Pharmaceuticals 2023, 16, 725. [Google Scholar] [CrossRef]
- Ferrara, P.; Battiato, S.; Polosa, R. Progress and Prospects for Artificial Intelligence in Clinical Practice: Learning from COVID-19. Intern. Emerg. Med. 2022, 17, 1855–1857. [Google Scholar] [CrossRef]
- Beneke, F.; Mackenrodt, M.-O. Artificial Intelligence and Collusion. IIC-Int. Rev. Intellect. Prop. Compet. Law 2018, 50, 109–134. [Google Scholar]
- Steels, L.; Brooks, R. The Artificial Life Route to Artificial Intelligence: Building Embodied, Situated Agents, 1st ed.; Routledge: London, UK, 2018; 300p. [Google Scholar]
- Bielecki, A. Models of Neurons and Perceptrons: Selected Problems and Challenges, 1st ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; 156p. [Google Scholar]
- van der Maaten, L.; Hinton, G. Visualizing Non-Metric Similarities in Multiple Maps. Mach. Learn. 2011, 87, 33–55. [Google Scholar] [CrossRef]
- Gadd, C.; Wade, S.; Shah, A.A. Pseudo-Marginal Bayesian Inference for Gaussian Process Latent Variable Models. Mach. Learn. 2021, 110, 1105–1143. [Google Scholar] [CrossRef]
- Gallego, V.; Naveiro, R.; Roca, C.; Ríos Insua, D.; Campillo, N.E. AI in Drug Development: A Multidisciplinary Perspective. Mol. Divers. 2021, 25, 1461–1479. [Google Scholar] [CrossRef] [PubMed]
- Kaul, V.; Enslin, S.; Gross, S.A. History of Artificial Intelligence in Medicine. Gastrointest. Endosc. 2020, 92, 807–812. [Google Scholar] [CrossRef] [PubMed]
- Kusunose, K. Steps to Use Artificial Intelligence in Echocardiography. J. Echocardiogr. 2020, 19, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Kusunose, K.; Haga, A.; Abe, T.; Sata, M. Utilization of Artificial Intelligence in Echocardiography. Circ. J. 2019, 83, 1623–1629. [Google Scholar] [CrossRef]
- Dey, D.; Slomka, P.J.; Leeson, P.; Comaniciu, D.; Shrestha, S.; Sengupta, P.P.; Marwick, T.H. Artificial Intelligence in Cardiovascular Imaging. J. Am. Coll. Cardiol. 2019, 73, 1317–1335. [Google Scholar] [CrossRef]
- van Hamersvelt, R.W.; Zreik, M.; Voskuil, M.; Viergever, M.A.; Išgum, I.; Leiner, T. Deep Learning Analysis of Left Ventricular Myocardium in CT Angiographic Intermediate-Degree Coronary Stenosis Improves the Diagnostic Accuracy for Identification of Functionally Significant Stenosis. Eur. Radiol. 2018, 29, 2350–2359. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, G.; Gao, Z.; Xu, C.; Zhang, Y.; Shi, R.; Keegan, J.; Xu, L.; Zhang, H.; Fan, Z.; et al. Deep Learning for Diagnosis of Chronic Myocardial Infarction on Nonenhanced Cardiac Cine MRI. Radiology 2019, 291, 606–617. [Google Scholar] [CrossRef]
- Attia, Z.I.; Kapa, S.; Lopez-Jimenez, F.; McKie, P.M.; Ladewig, D.J.; Satam, G.; Pellikka, P.A.; Enriquez-Sarano, M.; Noseworthy, P.A.; Munger, T.M.; et al. Screening for Cardiac Contractile Dysfunction Using an Artificial Intelligence–Enabled Electrocardiogram. Nat. Med. 2019, 25, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Bachtiger, P.; Petri, C.F.; Scott, F.E.; Ri Park, S.; Kelshiker, M.A.; Sahemey, H.K.; Dumea, B.; Alquero, R.; Padam, P.S.; Hatrick, I.R.; et al. Point-of-Care Screening for Heart Failure with Reduced Ejection Fraction Using Artificial Intelligence during ECG-Enabled Stethoscope Examination in London, UK: A Prospective, Observational, Multicentre Study. Lancet Digit. Health 2022, 4, e117–e125. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Rushlow, D.R.; Inselman, J.W.; McCoy, R.G.; Thacher, T.D.; Behnken, E.M.; Bernard, M.E.; Rosas, S.L.; Akfaly, A.; Misra, A.; et al. Artificial Intelligence–Enabled Electrocardiograms for Identification of Patients with Low Ejection Fraction: A Pragmatic, Randomized Clinical Trial. Nat. Med. 2021, 27, 815–819. [Google Scholar] [CrossRef] [PubMed]
- Duffy, G.; Cheng, P.P.; Yuan, N.; He, B.; Kwan, A.C.; Shun-Shin, M.J.; Alexander, K.M.; Ebinger, J.; Lungren, M.P.; Rader, F.; et al. High-Throughput Precision Phenotyping of Left Ventricular Hypertrophy With Cardiovascular Deep Learning. JAMA Cardiol. 2022, 7, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Narula, S.; Shameer, K.; Salem Omar, A.M.; Dudley, J.T.; Sengupta, P.P. Machine-Learning Algorithms to Automate Morphological and Functional Assessments in 2D Echocardiography. J. Am. Coll. Cardiol. 2016, 68, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Frazer, J.; Notin, P.; Dias, M.; Gomez, A.; Min, J.K.; Brock, K.; Gal, Y.; Marks, D.S. Disease Variant Prediction with Deep Generative Models of Evolutionary Data. Nature 2021, 599, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Li, L.; Liu, Z.; Zhao, K.; Chen, X.; Lu, M.; Yin, G.; Song, L.; Zhao, S.; Zheng, H.; et al. Deep Learning Algorithm to Improve Hypertrophic Cardiomyopathy Mutation Prediction Using Cardiac Cine Images. Eur. Radiol. 2020, 31, 3931–3940. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, J.; Heliö, K.; Saarinen, I.; Tallila, J.; Seppälä, E.H.; Tuupanen, S.; Turpeinen, H.; Kangas-Kontio, T.; Schleit, J.; Tommiska, J.; et al. Diagnostic Yield of Genetic Testing in a Heterogeneous Cohort of 1376 HCM Patients. BMC Cardiovasc. Disord. 2021, 21, 126. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Yuan, Z.; Lou, J.; Fan, Y.; Shi, A.; Huang, J.; Zhao, M.; Wu, Y. Multi-Institutional Development and External Validation of Machine Learning-Based Models to Predict Relapse Risk of Pancreatic Ductal Adenocarcinoma after Radical Resection. J. Transl. Med. 2021, 19, 281. [Google Scholar] [CrossRef]
- Kaissis, G.A.; Jungmann, F.; Ziegelmayer, S.; Lohöfer, F.K.; Harder, F.N.; Schlitter, A.M.; Muckenhuber, A.; Steiger, K.; Schirren, R.; Friess, H.; et al. Multiparametric Modelling of Survival in Pancreatic Ductal Adenocarcinoma Using Clinical, Histomorphological, Genetic and Image-Derived Parameters. J. Clin. Med. 2020, 9, 1250. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, S.; Yuan, Z.; Li, Q.; Ding, R.; Bao, X.; Zhen, T.; Fu, Z.; Fu, H.; Xing, K.; et al. Risk Factors and Socio-Economic Burden in Pancreatic Ductal Adenocarcinoma Operation: A Machine Learning Based Analysis. BMC Cancer 2020, 20, 1161. [Google Scholar] [CrossRef] [PubMed]
- Pépin, J.-L.; Letesson, C.; Le-Dong, N.N.; Dedave, A.; Denison, S.; Cuthbert, V.; Martinot, J.-B.; Gozal, D. Assessment of Mandibular Movement Monitoring With Machine Learning Analysis for the Diagnosis of Obstructive Sleep Apnea. JAMA Netw. Open 2020, 3, e1919657. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.-M.; Xue, Q.; Ye, H.-T.; Wang, Y.; Tong, J.; Ji, M.-H.; Yang, J.-J. Constructing a Prediction Model for Difficult Intubation of Obese Patients Based on Machine Learning. J. Clin. Anesth. 2021, 72, 110278. [Google Scholar] [CrossRef] [PubMed]
- Carron, M.; Safaee Fakhr, B.; Ieppariello, G.; Foletto, M. Perioperative Care of the Obese Patient. Br. J. Surg. 2020, 107, e39–e55. [Google Scholar] [CrossRef] [PubMed]
- Ermer, S.C.; Farney, R.J.; Johnson, K.B.; Orr, J.A.; Egan, T.D.; Brewer, L.M. An Automated Algorithm Incorporating Poincaré Analysis Can Quantify the Severity of Opioid-Induced Ataxic Breathing. Anesth. Analg. 2020, 130, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Ingrande, J.; Gabriel, R.A.; McAuley, J.; Krasinska, K.; Chien, A.; Lemmens, H.J.M. The Performance of an Artificial Neural Network Model in Predicting the Early Distribution Kinetics of Propofol in Morbidly Obese and Lean Subjects. Anesth. Analg. 2020, 131, 1500–1509. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, M.; Li, C.; Meng, Z.; Li, Y.; Chen, Y.; Liu, L. Image Classification for Automobile Pipe Joints Surface Defect Detection Using Wavelet Decomposition and Convolutional Neural Network. IEEE Access 2022, 10, 77191–77204. [Google Scholar] [CrossRef]
- Bellini, V.; Guzzon, M.; Bigliardi, B.; Mordonini, M.; Filippelli, S.; Bignami, E. Artificial Intelligence: A New Tool in Operating Room Management. Role of Machine Learning Models in Operating Room Optimization. J. Med. Syst. 2020, 44, 20. [Google Scholar] [CrossRef]
- Rozario, N.; Rozario, D. Can Machine Learning Optimize the Efficiency of the Operating Room in the Era of COVID-19? Can. J. Surg. 2020, 63, E527–E529. [Google Scholar] [CrossRef]
- Brennan, M.; Puri, S.; Ozrazgat-Baslanti, T.; Feng, Z.; Ruppert, M.; Hashemighouchani, H.; Momcilovic, P.; Li, X.; Wang, D.Z.; Bihorac, A. Comparing Clinical Judgment with the MySurgeryRisk Algorithm for Preoperative Risk Assessment: A Pilot Usability Study. Surgery 2019, 165, 1035–1045. [Google Scholar] [CrossRef]
- Xue, B.; Li, D.; Lu, C.; King, C.R.; Wildes, T.; Avidan, M.S.; Kannampallil, T.; Abraham, J. Use of Machine Learning to Develop and Evaluate Models Using Preoperative and Intraoperative Data to Identify Risks of Postoperative Complications. JAMA Netw. Open 2021, 4, e212240. [Google Scholar] [CrossRef] [PubMed]
- Tavolara, T.E.; Gurcan, M.N.; Segal, S.; Niazi, M.K.K. Identification of Difficult to Intubate Patients from Frontal Face Images Using an Ensemble of Deep Learning Models. Comput. Biol. Med. 2021, 136, 104737. [Google Scholar] [CrossRef] [PubMed]
- Cheney, F.W. The American Society of Anesthesiologists Closed Claims Project. Anesthesiology 1999, 91, 552–556. [Google Scholar] [CrossRef] [PubMed]
- León, M.A.; Räsänen, J. Neural Network-Based Detection of Esophageal Intubation in Anesthetized Patients. J. Clin. Monit. 1996, 12, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.C.; Connell, A.; Simonetto, D.A.; Hughes, C.; Shah, V.H. Application of Artificial Intelligence for the Diagnosis and Treatment of Liver Diseases. Hepatology 2021, 73, 2546–2563. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, R.; Requa, J.; Dao, T.; Ninh, A.; Tran, E.; Mai, D.; Lugo, M.; El-Hage Chehade, N.; Chang, K.J.; Karnes, W.E.; et al. Artificial Intelligence Using Convolutional Neural Networks for Real-Time Detection of Early Esophageal Neoplasia in Barrett’s Esophagus (with Video). Gastrointest. Endosc. 2020, 91, 1264–1271.e1. [Google Scholar] [CrossRef] [PubMed]
- Ebigbo, A.; Mendel, R.; Probst, A.; Manzeneder, J.; Prinz, F.; de Souza, L.A., Jr.; Papa, J.; Palm, C.; Messmann, H. Real-Time Use of Artificial Intelligence in the Evaluation of Cancer in Barrett’s Oesophagus. Gut 2019, 69, 615–616. [Google Scholar] [CrossRef]
- Kohli, A.; Holzwanger, E.A.; Levy, A.N. Emerging Use of Artificial Intelligence in Inflammatory Bowel Disease. World J. Gastroenterol. 2020, 26, 6923–6928. [Google Scholar] [CrossRef]
- Waljee, A.K.; Liu, B.; Sauder, K.; Zhu, J.; Govani, S.M.; Stidham, R.W.; Higgins, P.D.R. Predicting Corticosteroid-Free Biologic Remission with Vedolizumab in Crohn’s Disease. Inflamm. Bowel Dis. 2018, 24, 1185–1192. [Google Scholar] [CrossRef]
- Le Berre, C.; Sandborn, W.J.; Aridhi, S.; Devignes, M.-D.; Fournier, L.; Smaïl-Tabbone, M.; Danese, S.; Peyrin-Biroulet, L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology 2020, 158, 76–94.e2. [Google Scholar] [CrossRef]
- Spann, A.; Yasodhara, A.; Kang, J.; Watt, K.; Wang, B.; Goldenberg, A.; Bhat, M. Applying Machine Learning in Liver Disease and Transplantation: A Comprehensive Review. Hepatology 2020, 71, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Repici, A.; Badalamenti, M.; Maselli, R.; Correale, L.; Radaelli, F.; Rondonotti, E.; Ferrara, E.; Spadaccini, M.; Alkandari, A.; Fugazza, A.; et al. Efficacy of Real-Time Computer-Aided Detection of Colorectal Neoplasia in a Randomized Trial. Gastroenterology 2020, 159, 512–520.e7. [Google Scholar] [CrossRef]
- Hassan, C.; Spadaccini, M.; Iannone, A.; Maselli, R.; Jovani, M.; Chandrasekar, V.T.; Antonelli, G.; Yu, H.; Areia, M.; Dinis-Ribeiro, M.; et al. Performance of Artificial Intelligence in Colonoscopy for Adenoma and Polyp Detection: A Systematic Review and Meta-Analysis. Gastrointest. Endosc. 2021, 93, 77–85.e6. [Google Scholar]
- Lahner, E. Possible Contribution of Advanced Statistical Methods (Artificial Neural Networks and Linear Discriminant Analysis) in Recognition of Patients with Suspected Atrophic Body Gastritis. World J. Gastroenterol. 2005, 11, 5867. [Google Scholar] [CrossRef] [PubMed]
- Rotondano, G.; Cipolletta, L.; Grossi, E.; Koch, M.; Intraligi, M.; Buscema, M.; Marmo, R. Artificial Neural Networks Accurately Predict Mortality in Patients with Nonvariceal Upper GI Bleeding. Gastrointest. Endosc. 2011, 73, 2018–2026.e2. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Yang, J.; Sun, Y.; Li, C.; Wu, W.; Jin, L.; Yang, Z.; Ni, B.; Gao, P.; Wang, P.; et al. 3D Deep Learning from CT Scans Predicts Tumor Invasiveness of Subcentimeter Pulmonary Adenocarcinomas. Cancer Res. 2018, 78, 6881–6889. [Google Scholar] [CrossRef] [PubMed]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-End Lung Cancer Screening with Three-Dimensional Deep Learning on Low-Dose Chest Computed Tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Jiang, B.; Zhang, L.; Greuter, M.J.W.; de Bock, G.H.; Zhang, H.; Xie, X. Lung Nodule Detectability of Artificial Intelligence-Assisted CT Image Reading in Lung Cancer Screening. Curr. Med. Imaging 2022, 18, 327–334. [Google Scholar] [CrossRef]
- Esteva, H.; Marchevsky, A.; Núñez, T.; Luna, C.; Esteva, M. Neural Networks as a Prognostic Tool of Surgical Risk in Lung Resections. Ann. Thorac. Surg. 2002, 73, 1576–1581. [Google Scholar] [CrossRef]
- Bendixen, M.; Jørgensen, O.D.; Kronborg, C.; Andersen, C.; Licht, P.B. Postoperative Pain and Quality of Life after Lobectomy via Video-Assisted Thoracoscopic Surgery or Anterolateral Thoracotomy for Early Stage Lung Cancer: A Randomised Controlled Trial. Lancet Oncol. 2016, 17, 836–844. [Google Scholar] [CrossRef]
- Somashekhar, S.P.; Sepúlveda, M.-J.; Puglielli, S.; Norden, A.D.; Shortliffe, E.H.; Rohit Kumar, C.; Rauthan, A.; Arun Kumar, N.; Patil, P.; Rhee, K.; et al. Watson for Oncology and Breast Cancer Treatment Recommendations: Agreement with an Expert Multidisciplinary Tumor Board. Ann. Oncol. 2018, 29, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Gonem, S.; Janssens, W.; Das, N.; Topalovic, M. Applications of Artificial Intelligence and Machine Learning in Respiratory Medicine. Thorax 2020, 75, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Geddes, C.C.; Fox, J.G.; Allison, M.E.M.; Boulton-Jones, J.M.; Simpson, K. An Artificial Neural Network Can Select Patients at High Risk of Developing Progressive IgA Nephropathy More Accurately than Experienced Nephrologists. Nephrol. Dial. Transplant. 1998, 13, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Niel, O.; Boussard, C.; Bastard, P. Artificial Intelligence Can Predict GFR Decline During the Course of ADPKD. Am. J. Kidney Dis. 2018, 71, 911–912. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, C.; Molina, M.; Ponce, P.; Tothova, M.; Cattinelli, I.; Ion Titapiccolo, J.; Mari, F.; Amato, C.; Leipold, F.; Wehmeyer, W.; et al. An International Observational Study Suggests That Artificial Intelligence for Clinical Decision Support Optimizes Anemia Management in Hemodialysis Patients. Kidney Int. 2016, 90, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Friberg, I.M.; Kift-Morgan, A.; Parekh, G.; Morgan, M.P.; Liuzzi, A.R.; Lin, C.-Y.; Donovan, K.L.; Colmont, C.S.; Morgan, P.H.; et al. Machine-Learning Algorithms Define Pathogen-Specific Local Immune Fingerprints in Peritoneal Dialysis Patients with Bacterial Infections. Kidney Int. 2017, 92, 179–191. [Google Scholar] [CrossRef]
- Niel, O.; Bastard, P.; Boussard, C.; Hogan, J.; Kwon, T.; Deschênes, G. Artificial Intelligence Outperforms Experienced Nephrologists to Assess Dry Weight in Pediatric Patients on Chronic Hemodialysis. Pediatr. Nephrol. 2018, 33, 1799–1803. [Google Scholar] [CrossRef]
- Checcucci, E.; Amparore, D.; De Luca, S.; Autorino, R.; Fiori, C.; Porpiglia, F. Precision Prostate Cancer Surgery: An Overview of New Technologies and Techniques. Minerva Urol. Nefrol. 2019, 71, 487–501. [Google Scholar] [CrossRef]
- Cicione, A.; De Nunzio, C.; Manno, S.; Damiano, R.; Posti, A.; Lima, E.; Tubaro, A.; Balloni, F. An Update on Prostate Biopsy in the Era of Magnetic Resonance Imaging. Minerva Urol. Nephrol. 2018, 70, 264–274. [Google Scholar] [CrossRef]
- Snow, P.B.; Smith, D.S.; Catalona, W.J. Artificial Neural Networks in the Diagnosis and Prognosis of Prostate Cancer: A Pilot Study. J. Urol. 1994, 152 Pt 2, 1923–1926. [Google Scholar] [CrossRef]
- Djavan, B.; Remzi, M.; Zlotta, A.; Seitz, C.; Snow, P.; Marberger, M. Novel Artificial Neural Network for Early Detection of Prostate Cancer. J. Clin. Oncol. 2002, 20, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Hung, A.J.; Chen, J.; Che, Z.; Nilanon, T.; Jarc, A.; Titus, M.; Oh, P.J.; Gill, I.S.; Liu, Y. Utilizing Machine Learning and Automated Performance Metrics to Evaluate Robot-Assisted Radical Prostatectomy Performance and Predict Outcomes. J. Endourol. 2018, 32, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Kattan, M.W. Comparison of Cox Regression with Other Methods for Determining Prediction Models and Nomograms. J. Urol. 2003, 170, S6–S10. [Google Scholar] [CrossRef] [PubMed]
- Buchner, A.; Kendlbacher, M.; Nuhn, P.; Tüllmann, C.; Haseke, N.; Stief, C.G.; Staehler, M. Outcome Assessment of Patients with Metastatic Renal Cell Carcinoma Under Systemic Therapy Using Artificial Neural Networks. Clin. Genitourin. Cancer 2012, 10, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Ozkan, I.A.; Koklu, M.; Sert, I.U. Diagnosis of Urinary Tract Infection Based on Artificial Intelligence Methods. Comput. Methods Programs Biomed. 2018, 166, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Serati, M.; Salvatore, S.; Siesto, G.; Cattoni, E.; Braga, A.; Sorice, P.; Cromi, A.; Ghezzi, F.; Bolis, P. Urinary Symptoms and Urodynamic Findings in Women with Pelvic Organ Prolapse: Is There a Correlation? Results of an Artificial Neural Network Analysis. Eur. Urol. 2011, 60, 253–260. [Google Scholar] [CrossRef]
- Tapak, L.; Hamidi, O.; Amini, P.; Poorolajal, J. Prediction of Kidney Graft Rejection Using Artificial Neural Network. Healthc. Inform. Res. 2017, 23, 277. [Google Scholar] [CrossRef]
- Berk-Krauss, J.; Polsky, D.; Stein, J.A. Mole Mapping for Management of Pigmented Skin Lesions. Dermatol. Clin. 2017, 35, 439–445. [Google Scholar] [CrossRef]
- Demers, A.A.; Nugent, Z.; Mihalcioiu, C.; Wiseman, M.C.; Kliewer, E.V. Trends of Nonmelanoma Skin Cancer from 1960 through 2000 in a Canadian Population. J. Am. Acad. Dermatol. 2005, 53, 320–328. [Google Scholar] [CrossRef]
- Kaddu, S.; Soyer, H.P.; Gabler, G.; Kovarik, C. The Africa Teledermatology Project: Preliminary Experience with a Sub-Saharan Teledermatology and e-Learning Program. J. Am. Acad. Dermatol. 2009, 61, 155–157. [Google Scholar] [CrossRef]
- Gaffney, R.; Rao, B. Global Teledermatology. Glob. Dermatol. 2016, 2, 209–214. [Google Scholar] [CrossRef]
- Kaliyadan, F.; Ashique, K. Use of Mobile Applications in Dermatology. Indian J. Dermatol. 2020, 65, 371. [Google Scholar] [CrossRef]
- Freeman, K.; Dinnes, J.; Chuchu, N.; Takwoingi, Y.; Bayliss, S.E.; Matin, R.N.; Jain, A.; Walter, F.M.; Williams, H.C.; Deeks, J.J. Algorithm Based Smartphone Apps to Assess Risk of Skin Cancer in Adults: Systematic Review of Diagnostic Accuracy Studies. BMJ 2020, 368, m127. [Google Scholar] [CrossRef] [PubMed]
- Veronese, F.; Branciforti, F.; Zavattaro, E.; Tarantino, V.; Romano, V.; Meiburger, K.M.; Salvi, M.; Seoni, S.; Savoia, P. The Role in Teledermoscopy of an Inexpensive and Easy-to-Use Smartphone Device for the Classification of Three Types of Skin Lesions Using Convolutional Neural Networks. Diagnostics 2021, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Kagian, A.; Dror, G.; Leyvand, T.; Meilijson, I.; Cohen-Or, D.; Ruppin, E. A Machine Learning Predictor of Facial Attractiveness Revealing Human-like Psychophysical Biases. Vis. Res. 2008, 48, 235–243. [Google Scholar] [CrossRef]
- Potter, B.; Ronan, S.G. Computerized Dermatopathologic Diagnosis. J. Am. Acad. Dermatol. 1987, 17, 119–131. [Google Scholar] [CrossRef]
- Wells, A.; Patel, S.; Lee, J.B.; Motaparthi, K. Artificial Intelligence in Dermatopathology: Diagnosis, Education, and Research. J. Cutan. Pathol. 2021, 48, 1061–1068. [Google Scholar] [CrossRef]
- Ianni, J.D.; Soans, R.E.; Sankarapandian, S.; Chamarthi, R.V.; Ayyagari, D.; Olsen, T.G.; Bonham, M.J.; Stavish, C.C.; Motaparthi, K.; Cockerell, C.J.; et al. Tailored for Real-World: A Whole Slide Image Classification System Validated on Uncurated Multi-Site Data Emulating the Prospective Pathology Workload. Sci. Rep. 2020, 10, 3217. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Berking, C.; Klode, J.; Schadendorf, D.; Jansen, P.; Franklin, C.; Holland-Letz, T.; Krahl, D.; et al. Pathologist-Level Classification of Histopathological Melanoma Images with Deep Neural Networks. Eur. J. Cancer 2019, 115, 79–83. [Google Scholar] [CrossRef]
- Olsen, T.G.; Jackson, B.H.; Feeser, T.A.; Kent, M.N.; Moad, J.C.; Krishnamurthy, S.; Lunsford, D.D.; Soans, R.E. Diagnostic Performance of Deep Learning Algorithms Applied to Three Common Diagnoses in Dermatopathology. J. Pathol. Inform. 2018, 9, 32. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Akay, B.N.; Argenziano, G.; Blum, A.; Braun, R.P.; Cabo, H.; Gourhant, J.-Y.; Kreusch, J.; Lallas, A.; et al. Expert-Level Diagnosis of Nonpigmented Skin Cancer by Combined Convolutional Neural Networks. JAMA Dermatol. 2019, 155, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Fayaz, A.; Croft, P.; Langford, R.M.; Donaldson, L.J.; Jones, G.T. Prevalence of Chronic Pain in the UK: A Systematic Review and Meta-Analysis of Population Studies. BMJ Open 2016, 6, e010364. [Google Scholar] [CrossRef] [PubMed]
- Lötsch, J.; Ultsch, A. Machine Learning in Pain Research. Pain 2017, 159, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Culvenor, A.G.; Schache, A.G.; Vicenzino, B.; Pandy, M.G.; Collins, N.J.; Cook, J.L.; Crossley, K.M. Are Knee Biomechanics Different in Those With and Without Patellofemoral Osteoarthritis After Anterior Cruciate Ligament Reconstruction? Arthritis Care Res. 2014, 66, 1566–1570. [Google Scholar] [CrossRef] [PubMed]
- Kianifar, R.; Lee, A.; Raina, S.; Kulic, D. Automated Assessment of Dynamic Knee Valgus and Risk of Knee Injury During the Single Leg Squat. IEEE J. Transl. Eng. Health Med. 2017, 5, 2100213. [Google Scholar] [CrossRef] [PubMed]
- Navani, A.; Li, G. Chronic Pain Challenge: A Statistical Machine-Learning Method for Chronic Pain Assessment. J. Recent Adv. Pain 2016, 2, 82–86. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual Convolutional Neural Network for the Determination of IDH Status in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef]
- Chang, P.; Grinband, J.; Weinberg, B.D.; Bardis, M.; Khy, M.; Cadena, G.; Su, M.-Y.; Cha, S.; Filippi, C.G.; Bota, D.; et al. Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. Am. J. Neuroradiol. 2018, 39, 1201–1207. [Google Scholar] [CrossRef]
- Akbari, H.; Bakas, S.; Pisapia, J.M.; Nasrallah, M.P.; Rozycki, M.; Martinez-Lage, M.; Morrissette, J.J.D.; Dahmane, N.; O’Rourke, D.M.; Davatzikos, C. In Vivoevaluation of EGFRvIII Mutation in Primary Glioblastoma Patients via Complex Multiparametric MRI Signature. Neuro-Oncology 2018, 20, 1068–1079. [Google Scholar] [CrossRef]
- Laukamp, K.R.; Thiele, F.; Shakirin, G.; Zopfs, D.; Faymonville, A.; Timmer, M.; Maintz, D.; Perkuhn, M.; Borggrefe, J. Fully Automated Detection and Segmentation of Meningiomas Using Deep Learning on Routine Multiparametric MRI. Eur. Radiol. 2018, 29, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Isensee, F.; Tursunova, I.; Petersen, J.; Neuberger, U.; Bonekamp, D.; Brugnara, G.; Schell, M.; Kessler, T.; Foltyn, M.; et al. Automated Quantitative Tumour Response Assessment of MRI in Neuro-Oncology with Artificial Neural Networks: A Multicentre, Retrospective Study. Lancet Oncol. 2019, 20, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Witanto, J.N.; Pratama, K.; Lee, D.; Choi, K.; Choi, S.; Kim, K.; Kim, M.; Kim, J.; Kim, Y.; et al. P13.02.B Fully Automated Segmentation and Volumetric Measurement of Intracranial Meningioma Using Deep Learning. Neuro-Oncology 2022, 24 (Suppl. S2), ii80–ii81. [Google Scholar] [CrossRef]
- Gates, E.D.H.; Lin, J.S.; Weinberg, J.S.; Hamilton, J.; Prabhu, S.S.; Hazle, J.D.; Fuller, G.N.; Baladandayuthapani, V.; Fuentes, D.; Schellingerhout, D. Guiding the First Biopsy in Glioma Patients Using Estimated Ki-67 Maps Derived from MRI: Conventional versus Advanced Imaging. Neuro-Oncology 2019, 21, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Artzi, M.; Bressler, I.; Ben Bashat, D. Differentiation between Glioblastoma, Brain Metastasis and Subtypes Using Radiomics Analysis. J. Magn. Reson. Imaging 2019, 50, 519–528. [Google Scholar] [CrossRef]
- Emin, E.I.; Emin, E.; Papalois, A.; Willmott, F.; Clarke, S.; Sideris, M. Artificial Intelligence in Obstetrics and Gynaecology: Is This the Way Forward? In Vivo 2019, 33, 1547–1551. [Google Scholar] [CrossRef]
- Dong, H.-C.; Dong, H.-K.; Yu, M.-H.; Lin, Y.-H.; Chang, C.-C. Using Deep Learning with Convolutional Neural Network Approach to Identify the Invasion Depth of Endometrial Cancer in Myometrium Using MR Images: A Pilot Study. Int. J. Environ. Res. Public Health 2020, 17, 5993. [Google Scholar] [CrossRef]
- Arsalan, M.; Haider, A.; Choi, J.; Park, K.R. Detecting Blastocyst Components by Artificial Intelligence for Human Embryological Analysis to Improve Success Rate of In Vitro Fertilization. J. Pers. Med. 2022, 12, 124. [Google Scholar] [CrossRef]
- Wu, L.; Wei, D.; Yang, N.; Lei, H.; Wang, Y. Artificial Intelligence Algorithm-Based Analysis of Ultrasonic Imaging Features for Diagnosis of Pregnancy Complicated with Brain Tumor. J. Healthc. Eng. 2021, 2021, 4022312. [Google Scholar] [CrossRef]
- Wu, Y.; Shen, Y.; Sun, H. Intelligent Algorithm-Based Analysis on Ultrasound Image Characteristics of Patients with Lower Extremity Arteriosclerosis Occlusion and Its Correlation with Diabetic Mellitus Foot. J. Healthc. Eng. 2021, 2021, 7758206. [Google Scholar] [CrossRef]
- Burgos-Artizzu, X.P.; Coronado-Gutiérrez, D.; Valenzuela-Alcaraz, B.; Vellvé, K.; Eixarch, E.; Crispi, F.; Bonet-Carne, E.; Bennasar, M.; Gratacos, E. Analysis of Maturation Features in Fetal Brain Ultrasound via Artificial Intelligence for the Estimation of Gestational Age. Am. J. Obstet. Gynecol. MFM 2021, 3, 100462. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.; Komatsu, M.; Komatsu, R.; Matsuoka, R.; Yasutomi, S.; Dozen, A.; Shozu, K.; Arakaki, T.; Machino, H.; Asada, K.; et al. Medical Professional Enhancement Using Explainable Artificial Intelligence in Fetal Cardiac Ultrasound Screening. Biomedicines 2022, 10, 551. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; He, X.; Guo, H.; He, M.; Zhang, L.; Xian, J.; Lei, T.; Xu, Q.; Zheng, J.; Feng, J.; et al. Use of Real-time Artificial Intelligence in Detection of Abnormal Image Patterns in Standard Sonographic Reference Planes in Screening for Fetal Intracranial Malformations. Ultrasound Obstet. Gynecol. 2022, 59, 304–316. [Google Scholar] [CrossRef]
- Scanzera, A.C.; Shorter, E.; Kinnaird, C.; Valikodath, N.; Al-Khaled, T.; Cole, E.; Kravets, S.; Hallak, J.A.; McMahon, T.; Chan, R.V.P. Optometrist’s Perspectives of Artificial Intelligence in Eye Care. J. Optom. 2022, 15, S91–S97. [Google Scholar] [CrossRef] [PubMed]
- Channa, R.; Wolf, R.; Abramoff, M.D. Autonomous Artificial Intelligence in Diabetic Retinopathy: From Algorithm to Clinical Application. J. Diabetes Sci. Technol. 2020, 15, 695–698. [Google Scholar] [CrossRef] [PubMed]
- Sabanayagam, C.; Banu, R.; Chee, M.L.; Lee, R.; Wang, Y.X.; Tan, G.; Jonas, J.B.; Lamoureux, E.L.; Cheng, C.-Y.; Klein, B.E.K.; et al. Incidence and Progression of Diabetic Retinopathy: A Systematic Review. Lancet Diabetes Endocrinol. 2019, 7, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Keel, S.; Liu, C.; He, Y.; Meng, W.; Scheetz, J.; Lee, P.Y.; Shaw, J.; Ting, D.; Wong, T.Y.; et al. An Automated Grading System for Detection of Vision-Threatening Referable Diabetic Retinopathy on the Basis of Color Fundus Photographs. Diabetes Care 2018, 41, 2509–2516. [Google Scholar] [CrossRef] [PubMed]
- Ferris, F.L.; Wilkinson, C.P.; Bird, A.; Chakravarthy, U.; Chew, E.; Csaky, K.; Sadda, S.R. Clinical Classification of Age-Related Macular Degeneration. Ophthalmology 2013, 120, 844–851. [Google Scholar] [CrossRef]
- Chen, Q.; Leng, T.; Zheng, L.; Kutzscher, L.; Ma, J.; de Sisternes, L.; Rubin, D.L. Automated Drusen Segmentation and Quantification in SD-OCT Images. Med. Image Anal. 2013, 17, 1058–1072. [Google Scholar] [CrossRef]
- Schlanitz, F.G.; Baumann, B.; Kundi, M.; Sacu, S.; Baratsits, M.; Scheschy, U.; Shahlaee, A.; Mittermüller, T.J.; Montuoro, A.; Roberts, P.; et al. Drusen Volume Development over Time and Its Relevance to the Course of Age-Related Macular Degeneration. Br. J. Ophthalmol. 2016, 101, 198–203. [Google Scholar] [CrossRef]
- Peng, Y.; Dharssi, S.; Chen, Q.; Keenan, T.D.; Agrón, E.; Wong, W.T.; Chew, E.Y.; Lu, Z. DeepSeeNet: A Deep Learning Model for Automated Classification of Patient-Based Age-Related Macular Degeneration Severity from Color Fundus Photographs. Ophthalmology 2019, 126, 565–575. [Google Scholar] [CrossRef]
- Asaoka, R.; Murata, H.; Hirasawa, K.; Fujino, Y.; Matsuura, M.; Miki, A.; Kanamoto, T.; Ikeda, Y.; Mori, K.; Iwase, A.; et al. Using Deep Learning and Transfer Learning to Accurately Diagnose Early-Onset Glaucoma From Macular Optical Coherence Tomography Images. Am. J. Ophthalmol. 2019, 198, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Pringle, C.; Kilday, J.-P.; Kamaly-Asl, I.; Stivaros, S.M. The Role of Artificial Intelligence in Paediatric Neuroradiology. Pediatr. Radiol. 2022, 52, 2159–2172. [Google Scholar] [CrossRef] [PubMed]
- Quon, J.L.; Han, M.; Kim, L.H.; Koran, M.E.; Chen, L.C.; Lee, E.H.; Wright, J.; Ramaswamy, V.; Lober, R.M.; Taylor, M.D.; et al. Artificial Intelligence for Automatic Cerebral Ventricle Segmentation and Volume Calculation: A Clinical Tool for the Evaluation of Pediatric Hydrocephalus. J. Neurosurg. Pediatr. 2021, 27, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Grimm, F.; Edl, F.; Kerscher, S.R.; Nieselt, K.; Gugel, I.; Schuhmann, M.U. Semantic Segmentation of Cerebrospinal Fluid and Brain Volume with a Convolutional Neural Network in Pediatric Hydrocephalus—Transfer Learning from Existing Algorithms. Acta Neurochir. 2020, 162, 2463–2474. [Google Scholar] [CrossRef]
- Stivaros, S.M.; Radon, M.R.; Mileva, R.; Connolly, D.J.A.; Cowell, P.E.; Hoggard, N.; Wright, N.B.; Tang, V.; Gledson, A.; Batty, R.; et al. Quantification of Structural Changes in the Corpus Callosumin Children with Profound Hypoxic–Ischaemic Brain Injury. Pediatr. Radiol. 2015, 46, 73–81. [Google Scholar] [CrossRef][Green Version]
- Raji, C.A.; Wang, M.B.; Nguyen, N.; Owen, J.P.; Palacios, E.M.; Yuh, E.L.; Mukherjee, P. Connectome Mapping with Edge Density Imaging Differentiates Pediatric Mild Traumatic Brain Injury from Typically Developing Controls: Proof of Concept. Pediatr. Radiol. 2020, 50, 1594–1601. [Google Scholar] [CrossRef]
- Attallah, O.; Sharkas, M.A.; Gadelkarim, H. Fetal Brain Abnormality Classification from MRI Images of Different Gestational Age. Brain Sci. 2019, 9, 231. [Google Scholar] [CrossRef]
- Bull, J.G.; Saunders, D.E.; Clark, C.A. Discrimination of Paediatric Brain Tumours Using Apparent Diffusion Coefficient Histograms. Eur. Radiol. 2011, 22, 447–457. [Google Scholar] [CrossRef]
- Orphanidou-Vlachou, E.; Vlachos, N.; Davies, N.P.; Arvanitis, T.N.; Grundy, R.G.; Peet, A.C. Texture Analysis ofT1- andT2-Weighted MR Images and Use of Probabilistic Neural Network to Discriminate Posterior Fossa Tumours in Children. NMR Biomed. 2014, 27, 632–639. [Google Scholar] [CrossRef]
- Stivaros, S.; Garg, S.; Tziraki, M.; Cai, Y.; Thomas, O.; Mellor, J.; Morris, A.A.; Jim, C.; Szumanska-Ryt, K.; Parkes, L.M.; et al. Randomised Controlled Trial of Simvastatin Treatment for Autism in Young Children with Neurofibromatosis Type 1 (SANTA). Mol. Autism 2018, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Bi, S.; Sun, M.; Wang, Y.; Wang, D.; Yang, S. Deep Learning Approach to Peripheral Leukocyte Recognition. PLoS ONE 2019, 14, e0218808. [Google Scholar] [CrossRef] [PubMed]
- Chandradevan, R.; Aljudi, A.A.; Drumheller, B.R.; Kunananthaseelan, N.; Amgad, M.; Gutman, D.A.; Cooper, L.A.D.; Jaye, D.L. Machine-Based Detection and Classification for Bone Marrow Aspirate Differential Counts: Initial Development Focusing on Nonneoplastic Cells. Lab. Investig. 2020, 100, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Papageorgiou, D.P.; Abidi, S.Z.; Dao, M.; Zhao, H.; Karniadakis, G.E. A Deep Convolutional Neural Network for Classification of Red Blood Cells in Sickle Cell Anemia. PLoS Comput. Biol. 2017, 13, e1005746. [Google Scholar] [CrossRef]
- Alsalem, M.A.; Zaidan, A.A.; Zaidan, B.B.; Hashim, M.; Madhloom, H.T.; Azeez, N.D.; Alsyisuf, S. A Review of the Automated Detection and Classification of Acute Leukaemia: Coherent Taxonomy, Datasets, Validation and Performance Measurements, Motivation, Open Challenges and Recommendations. Comput. Methods Programs Biomed. 2018, 158, 93–112. [Google Scholar] [CrossRef] [PubMed]
- Deulofeu, M.; Kolářová, L.; Salvadó, V.; María Peña-Méndez, E.; Almáši, M.; Štork, M.; Pour, L.; Boadas-Vaello, P.; Ševčíková, S.; Havel, J.; et al. Rapid Discrimination of Multiple Myeloma Patients by Artificial Neural Networks Coupled with Mass Spectrometry of Peripheral Blood Plasma. Sci. Rep. 2019, 9, 7975. [Google Scholar] [CrossRef] [PubMed]
- Moraes, L.O.; Pedreira, C.E.; Barrena, S.; Lopez, A.; Orfao, A. A Decision-Tree Approach for the Differential Diagnosis of Chronic Lymphoid Leukemias and Peripheral B-Cell Lymphomas. Comput. Methods Programs Biomed. 2019, 178, 85–90. [Google Scholar] [CrossRef]
- Patel, S.S.; Sekeres, M.A.; Nazha, A. Prognostic Models in Predicting Outcomes in Myelodysplastic Syndromes after Hypomethylating Agent Failure. Leuk. Lymphoma 2017, 58, 2532–2539. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, F.; Xie, L.; Wang, Y.; Xiang, Q.; Yue, Z.; Feng, Y.; Yang, Y.; Li, J.; Luo, L.; et al. Machine Learning Approaches for Risk Assessment of Peripherally Inserted Central Catheter-Related Vein Thrombosis in Hospitalized Patients with Cancer. Int. J. Med. Inform. 2019, 129, 175–183. [Google Scholar] [CrossRef]
- Arai, Y.; Kondo, T.; Fuse, K.; Shibasaki, Y.; Masuko, M.; Sugita, J.; Teshima, T.; Uchida, N.; Fukuda, T.; Kakihana, K.; et al. Using a Machine Learning Algorithm to Predict Acute Graft-versus-Host Disease Following Allogeneic Transplantation. Blood Adv. 2019, 3, 3626–3634. [Google Scholar] [CrossRef]
- Nazha, A.; Komrokji, R.S.; Meggendorfer, M.; Mukherjee, S.; Al Ali, N.; Walter, W.; Hutter, S.; Padron, E.; Madanat, Y.F.; Sallman, D.A.; et al. A Personalized Prediction Model to Risk Stratify Patients with Myelodysplastic Syndromes. Blood 2018, 132 (Suppl. S1), 793. [Google Scholar] [CrossRef]
- Ko, B.-S.; Wang, Y.-F.; Li, J.-L.; Li, C.-C.; Weng, P.-F.; Hsu, S.-C.; Hou, H.-A.; Huang, H.-H.; Yao, M.; Lin, C.-T.; et al. Clinically Validated Machine Learning Algorithm for Detecting Residual Diseases with Multicolor Flow Cytometry Analysis in Acute Myeloid Leukemia and Myelodysplastic Syndrome. EBioMedicine 2018, 37, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Dellinger, R.P.; Levy, M.M.; Rhodes, A.; Annane, D.; Gerlach, H.; Opal, S.M.; Sevransky, J.E.; Sprung, C.L.; Douglas, I.S.; Jaeschke, R.; et al. Surviving Sepsis Campaign. Crit. Care Med. 2013, 41, 580–637. [Google Scholar] [CrossRef] [PubMed]
- Quinten, V.M.; van Meurs, M.; Wolffensperger, A.E.; ter Maaten, J.C.; Ligtenberg, J.J.M. Sepsis Patients in the Emergency Department. Eur. J. Emerg. Med. 2018, 25, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Komorowski, M.; Celi, L.A.; Badawi, O.; Gordon, A.C.; Faisal, A.A. The Artificial Intelligence Clinician Learns Optimal Treatment Strategies for Sepsis in Intensive Care. Nat. Med. 2018, 24, 1716–1720. [Google Scholar] [CrossRef] [PubMed]
- Davoudi, A.; Malhotra, K.R.; Shickel, B.; Siegel, S.; Williams, S.; Ruppert, M.; Bihorac, E.; Ozrazgat-Baslanti, T.; Tighe, P.J.; Bihorac, A.; et al. Intelligent ICU for Autonomous Patient Monitoring Using Pervasive Sensing and Deep Learning. Sci. Rep. 2019, 9, 8020. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Cao, Z.; Duan, L.; Yang, G.; Yue, T.; Chen, Q. An Experimental Study on Breast Lesion Detection and Classification from Ultrasound Images Using Deep Learning Architectures. BMC Med. Imaging 2019, 19, 51. [Google Scholar] [CrossRef]
- Wang, K.; Lu, X.; Zhou, H.; Gao, Y.; Zheng, J.; Tong, M.; Wu, C.; Liu, C.; Huang, L.; Jiang, T.; et al. Deep Learning Radiomics of Shear Wave Elastography Significantly Improved Diagnostic Performance for Assessing Liver Fibrosis in Chronic Hepatitis B: A Prospective Multicentre Study. Gut 2018, 68, 729–741. [Google Scholar] [CrossRef]
- Lee, J.H.; Joo, I.; Kang, T.W.; Paik, Y.H.; Sinn, D.H.; Ha, S.Y.; Kim, K.; Choi, C.; Lee, G.; Yi, J.; et al. Deep Learning with Ultrasonography: Automated Classification of Liver Fibrosis Using a Deep Convolutional Neural Network. Eur. Radiol. 2019, 30, 1264–1273. [Google Scholar] [CrossRef]
- Ta, C.N.; Kono, Y.; Eghtedari, M.; Oh, Y.T.; Robbin, M.L.; Barr, R.G.; Kummel, A.C.; Mattrey, R.F. Focal Liver Lesions: Computer-Aided Diagnosis by Using Contrast-Enhanced US Cine Recordings. Radiology 2018, 286, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, R.J.; Loram, I.D. Estimation of Absolute States of Human Skeletal Muscle via Standard B-Mode Ultrasound Imaging and Deep Convolutional Neural Networks. J. R. Soc. Interface 2020, 17, 20190715. [Google Scholar] [CrossRef] [PubMed]
- Noort, F.; Vaart, C.H.; Grob, A.T.M.; Waarsenburg, M.K.; Slump, C.H.; Stralen, M. Deep Learning Enables Automatic Quantitative Assessment of Puborectalis Muscle and Urogenital Hiatus in Plane of Minimal Hiatal Dimensions. Ultrasound Obstet. Gynecol. 2019, 54, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Goergen, S.K.; Frazer, H.M.; Reddy, S. Quality Use of Artificial Intelligence in Medical Imaging: What Do Radiologists Need to Know? J. Med. Imaging Radiat. Oncol. 2022, 66, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Fayad, L.M.; Parekh, V.S.; de Castro Luna, R.; Ko, C.C.; Tank, D.; Fritz, J.; Ahlawat, S.; Jacobs, M.A. A Deep Learning System for Synthetic Knee Magnetic Resonance Imaging. Investig. Radiol. 2020, 56, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-J.; Jung, Y.-H.; Cho, B.-H.; Heo, M.-S. An Overview of Deep Learning in the Field of Dentistry. Imaging Sci. Dent. 2019, 49, 1. [Google Scholar] [CrossRef]
- Costa, P.; Galdran, A.; Meyer, M.I.; Niemeijer, M.; Abramoff, M.; Mendonca, A.M.; Campilho, A. End-to-End Adversarial Retinal Image Synthesis. IEEE Trans. Med. Imaging 2018, 37, 781–791. [Google Scholar] [CrossRef]
- Tuzoff, D.V.; Tuzova, L.N.; Bornstein, M.M.; Krasnov, A.S.; Kharchenko, M.A.; Nikolenko, S.I.; Sveshnikov, M.M.; Bednenko, G.B. Tooth Detection and Numbering in Panoramic Radiographs Using Convolutional Neural Networks. Dentomaxillofac. Radiol. 2019, 48, 20180051. [Google Scholar] [CrossRef]
- Kumar, S.; Patnaik, S.; Dixit, A. Predictive Models for Stage and Risk Classification in Head and Neck Squamous Cell Carcinoma (HNSCC). PeerJ 2020, 8, e9656. [Google Scholar] [CrossRef]
- Papadopoulos, D.N.; Karalis, V. Variational Autoencoders for Data Augmentation in Clinical Studies. Appl. Sci. 2023, 13, 8793. [Google Scholar] [CrossRef]
- Kumar, P.; Chauhan, S.; Awasthi, L.K. Artificial Intelligence in Healthcare: Review, Ethics, Trust Challenges & Future Research Directions. Eng. Appl. Artif. Intell. 2023, 120, 105894. [Google Scholar]
- Denecke, K.; Gabarron, E. How Artificial Intelligence for Healthcare Look Like in the Future? In Studies in Health Technology and Informatics; IOS Press: Amsterdam, The Netherlands, 2021; Volume 281, pp. 860–864. [Google Scholar]
- Davenport, T.; Kalakota, R. The Potential for Artificial Intelligence in Healthcare. Future Healthc. J. 2019, 6, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Zech, J.R.; Badgeley, M.A.; Liu, M.; Costa, A.B.; Titano, J.J.; Oermann, E.K. Variable Generalization Performance of a Deep Learning Model to Detect Pneumonia in Chest Radiographs: A Cross-Sectional Study. PLoS Med. 2018, 15, e1002683. [Google Scholar] [CrossRef] [PubMed]
- Olsen, H.P.; Slosser, J.L.; Hildebrandt, T.T.; Wiesener, C. What’s in the Box? The Legal Requirement of Explainability in Computationally Aided Decision-Making in Public Administration. SSRN Electron. J. 2019. [Google Scholar] [CrossRef]
- Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I. Explainability for Artificial Intelligence in Healthcare: A Multidisciplinary Perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration. Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SAMD): Discussion Paper and Request for Feedback. 2019. Available online: https://www.fda.gov/files/medical%20devices/published/US-FDA-Artificial-Intelligence-and-Machine-Learning-Discussion-Paper.pdf (accessed on 24 September 2023).
- Chaddad, A.; Peng, J.; Xu, J.; Bouridane, A. Survey of Explainable AI Techniques in Healthcare. Sensors 2023, 23, 634. [Google Scholar] [CrossRef]
- Cohen, I.G. Informed Consent and Medical Artificial Intelligence: What to Tell the Patient? SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Proposal for a Regulation of The European Parliament and of The Council Laying Down Harmonised Rules on Artificial Intelligence (Artificial Intelligence Act) and Amending Certain Union Legislative Acts; European Commission: Brussels, Belgium. 2021. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=celex%3A52021PC0206 (accessed on 24 September 2023).
- US FDA. Artificial Intelligence and Machine Learning in Software as a Medical Device. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-software-medical-device (accessed on 24 September 2023).
- Takshi, S. Unexpected Inequality: Disparate-Impact from Artificial Intelligence in Healthcare Decisions. J. Law Health 2021, 34, 215–251. [Google Scholar]
- Galvani, A.P.; Parpia, A.S.; Foster, E.M.; Singer, B.H.; Fitzpatrick, M.C. Improving the Prognosis of Health Care in the USA. Lancet 2020, 395, 524–533. [Google Scholar] [CrossRef]
- O’Sullivan, S.; Nevejans, N.; Allen, C.; Blyth, A.; Leonard, S.; Pagallo, U.; Holzinger, K.; Holzinger, A.; Sajid, M.I.; Ashrafian, H. Legal, Regulatory, and Ethical Frameworks for Development of Standards in Artificial Intelligence (AI) and Autonomous Robotic Surgery. Int. J. Med. Robot. Comput. Assist. Surg. 2019, 15, e1968. [Google Scholar] [CrossRef]
- Harvey, H.B.; Gowda, V. Clinical Applications of AI in MSK Imaging: A Liability Perspective. Skelet. Radiol. 2022, 51, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Bodaghi, A.; Fattahi, N.; Ramazani, A. Biomarkers: Promising and Valuable Tools towards Diagnosis, Prognosis and Treatment of COVID-19 and Other Diseases. Heliyon 2023, 9, e13323. [Google Scholar] [CrossRef] [PubMed]
- Alber, S.; Kumar, S.; Liu, J.; Huang, Z.-M.; Paez, D.; Hong, J.; Chang, H.-W.; Bhutani, T.; Gensler, L.S.; Liao, W. Single Cell Transcriptome and Surface Epitope Analysis of Ankylosing Spondylitis Facilitates Disease Classification by Machine Learning. Front. Immunol. 2022, 13, 838636. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).