1. Introduction
The build-up of lipofuscin in the retina has been highlighted as a clinically important feature in the manifestation and progression of the ocular disease geographic atrophy (GA). Lipofuscin is readily highlighted in the retinal fundus autofluorescence (FAF) image as it produces a very distinct emission in the FAF image modality [
1]. The FAF image enables the monitoring of bright
hyperfluorescence patterns in vivo of GA, as areas of increased FAF and are typically observed at the boundaries of GA lesions (which have dark interior areas of hypofluorescence). Previous investigations of the increased presence of FAF and FAF patterns (i.e., ‘hyperfluorescent regions’) in GA patients have revealed the development of new atrophic lesions, suggesting that FAF patterns precede both the development and enlargement of GA [
2]. In particular, the FAF pattern may provide an early indication of the potential rate of future progression of GA.
In a pioneering study, Holz et al. (2007) investigated whether hyperfluorescent patterns around GA are associated with progression of the disease. They divided the increased hyperfluorescent patterns into five
subjective categories: none, focal, banded, patchy, and diffuse [
3]. Diffuse patterns were further divided into five subtypes: reticular, branching, fine granular, and fine granular with peripheral punctate spots (GPS), and trickling. Eyes with no abnormal hyperfluorescence patterns had the slowest growth rates (median: 0.38 mm
2/year; interquartile range [IQR]: 0.13–0.79 mm
2/year), followed by eyes with the focal hyperfluorescent pattern (median: 0.81 mm
2/year; IQR: 0.44–1.07 mm
2/year), then by eyes with the diffuse hyperfluorescent pattern (median: 1.77 mm
2/year; IQR: 0.99–2.58 mm
2/year), and by eyes with the banded hyperfluorescent pattern (median: 1.81 mm
2/year; IQR: 1.41–2.69 mm
2/year). There were three eyes with the patchy hyperfluorescent patterns that had progression rates of 1.37, 1.84, and 2.94 mm
2/year, respectively. However, due to their small frequency, eyes with the patchy hyperfluorescent pattern were not included in further statistical analysis [
4]. This study found a statistically significant difference in atrophy enlargement per year for different abnormal and increased hyperfluorescent patterns between, except for: no abnormal and focal hyperfluorescent pattern (
p = 0.092); and the groups of banded and diffuse hyperfluorescent patterns (
p = 0.510) [
4].
Since the seminal paper by Holz et al. (2007), a number of similar studies have been conducted. For example, Jeong et al. (2014) assessed the association between abnormal hyperfluorescent features on images and GA progression. In this study, abnormal hyperfluorescent patterns in the junctional zone of GA were classified as none or minimal change, focal, patchy, banded, or diffuse; each pattern was evaluated against GA enlargement over time. They found that the mean rate of GA enlargement was the fastest in eyes with the diffuse hyperfluorescent pattern (mean: 1.74 mm
2/year; IQR: 0.82–2.29 mm
2/year), followed by eyes with the banded pattern (mean: 1.69 mm
2/year; IQR: 0.69–2.97 mm
2/year). When assessing diffuse and banded patterns in a logistic regression model, the banded pattern (odds ratio [OR]: 8.33;
p = 0.01) and the diffuse pattern (OR: 4.62;
p = 0.04) showed a statistically significant and high risk association with GA progression when compared with the none or minimal change pattern. Pooling the data for both banded and diffuse patterns showed a highly significant risk factor as well (OR: 3.18;
p = 0.01) [
5].
Batıoğlu et al. (2014) also assessed the role of increased hyperfluorescent patterns surrounding GA lesions in GA progression. This study used none, banded, diffuse nontrickling, diffuse trickling, and focal in their assessment; there was no patchy pattern. Progression rates in eyes with the diffuse trickling pattern (median: 1.42 mm
2/year) were significantly higher than in those with diffuse nontrickling (median: 0.46 mm
2/year) and eyes without hyperfluorescent abnormalities (median: 0.22 mm
2/year). Progression rates in eyes were significantly higher in diffuse trickling pattern as compared to eyes without hyperfluorescent abnormalities (
p = 0.003) and other diffuse patterns (
p = 0.024). No statistically significant difference was found in progression rates between the banded and diffuse trickling (
p = 1.01) or diffuse nontrickling (
p = 1.0). However, eyes with the banded pattern had a significantly higher progression rate than eyes without any hyperfluorescent abnormalities (
p = 0.038) [
6].
Biarnés et al. (2015) also assessed the role of increased hyperfluorescence as a risk factor for GA progression (ClinicalTrials.gov identifier NCT01694095). They found a statistically significant relationship between GA growth and hyperfluorescence patterns, which were categorized as none, focal, banded, diffuse and undetermined. At a median follow-up of 18 months, hyperfluorescence patterns and baseline area of atrophy were strongly associated with GA progression (
p < 0.001). However, they believed that hyperfluorescence patterns were a consequence and not a cause of enlarging atrophy [
7].
1.1. Artificial Intelligence
There are no universally accepted
pattern categories that have been defined for the spatial appearance of clusters of hyperfluorescence in FAF images. The clinical description of a pattern is subjective and may be characterized, for example, by the taxonomy originally suggested by Holz et al. [
3]. One aim of the current study was to provide an objective method of pattern classification as opposed to the past subjective approaches.
A recent systematic review revealed that the primary focus of Artificial Intelligence (AI) in GA applications has been the extraction of lesions, with a minor focus on GA progression [
8,
9]. In the case of GA lesions, deep learning has been investigated for lesion segmentation, while information in hyperfluorescent regions appears to have been neglected [
10]. An automated segmentation method with the capability of detecting both GA lesions and hyperfluorescent regions at all stages of the disease would be very valuable in a clinical setting.
Given that hyperfluorescent patterns are complex, ill-defined, and variable, which means that reliable annotations for ground truth for supervised learning models would be very difficult, it is suggested that an unsupervised learning approach would be an appropriate avenue for investigation.
The hypothesis tested was that hyperfluorescent areas can be categorized by their patterns and shapes into respective groups using unsupervised machine learning (ML) algorithms. The different pattern categories could account for variability in GA progression rates from patient to patient, which may not be explained by time-series progression of geometrical area alone. The unsupervised learning approach presented here for pattern classification is referred to as cluster analysis. In this paper, image processing techniques, specifically pseudocoloring (false coloring techniques), together with clustering theory, were used to automate extraction of hyperfluorescent regions.
1.2. Outline of This Paper
This paper describes cluster detection, classification and evaluation based on the following process:
- (i)
Automation of hyperfluorescent region extraction using pseudocoloring techniques.
- (ii)
Identification of major groups of hyperfluorescent regions based on intensity changes in hyperfluorescent areas (i.e., early-stage hyperfluorescent areas, intermediate-stage hyperfluorescent areas, and late-stage hyperfluorescent areas).
- (iii)
Application of an approach for prior assessment of cluster tendency to determine whether the data can be clustered appropriately.
- (iv)
Identification of the optimal number of clusters using unsupervised ML methods.
- (v)
Evaluation of performance using cluster-specific evaluation metrics.
The approach investigated in this study provides an objective assessment of clusters of hyperfluorescence and their association with progression of GA, in contrast to prior work based on subjective assessment of clusters. The approach would augment progression methods based on GA area growth and would also provide a prediction where there is insufficient time-series data.
2. Materials and Methods
2.1. Study Design and Data
This study was approved by the Human Research Ethics Committee of the Royal Victorian Eye and Ear Hospital (RVEEH). This study was conducted at the Centre for Eye Research Australia (CERA; East Melbourne, Australia) and in accordance with the International Conference on Harmonization Guidelines for Good Clinical Practice and tenets of the Declaration of Helsinki. Ethics approval was provided by the Human Research Ethics Committee (HREC: Project No. 95/283H/15) by the RVEEH.
Subjects included in this subanalysis of the case study were age-related macular degeneration (AMD) participants involved in macular natural history studies from CERA and from a private ophthalmology practice [
10]. Cases were referred from a senior medical retinal specialist and graded in the Macular Research Unit grading center. Inclusion criteria included being over the age of 50 years, having a diagnosis of AMD (based on the presence of drusen greater than 125 µm) with progression to GA in either one of both eyes. An atrophic lesion was required to be present in the macular and not extend beyond the limits of the FAF image at the first visit (i.e., baseline). Participants were required to have foveal-centered FAF images and at least three visits recorded over a minimum of 2 years, with FAF imaging of sufficient quality. Good quality images were classified as those having minimal or correctable artefacts (e.g., by correction of illumination with pre-processing techniques), and images should encompass the entire macular area and part (i.e., around half) of the optic disc. No minimum lesion or hyperfluorescent sizes were set, as the objective of this study was to be able to automate these GA features at all stages of the disease process.
Exclusion criteria included participants with neovascular AMD (nAMD) and macular atrophy from causes other than AMD, such as inherited retinal dystrophies, including Stargardt’s disease. These patients were excluded based on past determination by a retinal specialist. Additionally excluded were patients who had undergone any prior treatment or participated in a treatment trial for AMD. Peripapillary atrophy was not included in the analysis and all participants required atrophy in the FAF image to be included. Poor quality images were excluded and were classified as: images that were not salvageable with pre-processing techniques (e.g., excessive blurriness, shadowing, and contrast issues); images where the optic disc was completely absent; and images where the optic disc was in the center of the image, and all information pertaining to GA lesions were completely pushed to the side.
The FAF images were captured using the Heidelberg HRT-OCT Spectralis instrument (Heidelberg Engineering, Heidelberg, Germany). FAF image files, along with basic demographic data, were retrospectively collected in Tagged Image File Format (i.e., TIFF or TIF) and original sizes of images were either 768 × 768 or 1536 × 1536 pixels with 30° × 30° field-of-view (FOV). As images were collected retrospectively and from real-time clinical settings, automatic real-time tracking (ART) ranged from 5–100.
2.2. Automated Extraction of Hyperfluorescent Regions
The term pseudocoloring refers to application of a colormap to convert the monochrome image into a range of colors. The pseudocoloring process improves the visual perception of image detail and enhances interpretation of image content, due to the fact that the human visual system can discern only a very limited range of grey levels but a much larger range of color differences [
11]. The pseudocoloring procedure developed and applied in this study is summarized as the following algorithm:
2.3. Creation of Foreground Masks for Use in Clustering
The segmentation outputs generated using the automated pseudocoloring methods in the previous section were used to extract the foregrounds (i.e., hyperfluorescent regions) from the backgrounds (i.e., the retina). The objective of this step was to allow the automation method to extract regions of interest from the FAF images so that the FAF image qualities could be used to identify important and clusterable features for hyperfluorescent classification. The following steps were undertaken (
Figure 2):
2.4. Image Feature Extraction from Foreground Masks
Features were extracted from the images using the Visual Graphics Group’s (VGG) transfer learning model—the VGG16 [
15]. The model is complete with pre-trained weights, see Keras (
https://keras.io/ (accessed on 16 April 2021)). The feature extraction process for clustering is described below:
2.5. Cluster Tendency
Testing for cluster tendency is a preliminary step to identify whether the data can form meaningful clusters based on different properties or characteristics [
16]. This process involves the application of statistical tests to evaluate the data structure and assesses whether the data are uniformly distributed. A uniform dataset would suggest there is an absence of structure or pattern and any clustering method tested will be unlikely to yield significant results.
To evaluate cluster tendency, the Hopkins statistical test for spatial randomness of a variable can be used [
16]. The Hopkins test is denoted as [
17]:
where [
refers to a collection of patterns,
, in the dimensional space,
(i.e.,
,
, where
are sampling origins placed at random in space
. In the Hopkins statistic, two distances are used:
refers to the minimum distance from
to its nearest pattern in
, and
is the distance from a randomly selected pattern in
to its nearest neighbor [
17]. The null hypothesis for the Hopkins test is that the data contains no clusters and is uniformly distributed. The alternative hypothesis is that the dataset is not uniformly distributed and contains meaningful clusters. The values for the Hopkins test range from 0 to 1. To reject the null hypothesis, the value of
would be larger than 0.5 and close to 1 for well-defined clusters. For
values less than 0.5, this would suggest the data are regularly spaced and data cannot be clustered. In instances where the space
is uniformly distributed, the distances
and
would be close to each other, whereas if clusters are present, it is anticipated that
distances would be substantially larger than
.
Note on Software: To execute the Hopkins test in the Python language, the
pyclustertend package was used (
https://pypi.org/project/pyclustertend/ (accessed on 16 April 2021)). The package uses the formula
so that clusterability is interpreted as being a value close to 0. For the pyclustertend package, a high score (i.e., 0.3 and above) suggests that the data cannot be clustered.
2.6. Number of Clusters
The number of clusters tested for statistical significance ranged from
k = 2 to
k = 12 clusters. The choice of this range was based on the results reported in a pioneering study by Holz et al. (2007) [
4]. They suggested four primary patterns of hyperfluorescence, with the diffuse category further divided into an additional five subcategories of patterns, making a total of nine hyperfluorescence patterns (not including the “None” category). For completeness, testing from
k = 2 to
k = 12 clusters is a reasonable coverage of potential groupings for both lesions and hyperfluorescence, which is also consistent with conjectures in the literature.
2.7. Unsupervised Clustering Methods
A diverse range of clustering algorithms was investigated as part of this analysis. These included Affinity Propagation, Agglomerative Clustering, Balanced Iterative Reducing and Clustering (BIRCH), Density-Based Spatial Clustering of Applications with Noise (DBSCAN), k-Means, Mini-Batch k-Means, and Spectral Clustering. These approaches are described in more detail as follows.
The Affinity Propagation clustering approach identifies samples in the data that are most representative of a cluster, and measures similarities between data samples [
18]. Agglomerative Clustering begins by partitioning the data into single nodes and, step by step, merges paired data which are the closest nodes into a new node, until only a single node is left (i.e., the entire dataset) [
19]. The BIRCH approach takes large datasets and converts them into more compact, summarised versions that retain as much data distribution information as possible, and uses the data summary in lieu of the original dataset [
20]. The DBSCAN approach proposed for spatial databases with noise finds cluster points that are close together within specified distances, and considers points which are far away from the other points as outliers [
21]. The
k-Means approach relies on the distance between points and calculates cluster centers or centroids, and thus partitions that data around these centroids [
22]. The Mini-Batch
-Means approach is similar to the more traditional
-Means approach. However, rather than using an entire dataset, it uses small batches making this a faster algorithm than its traditional counterpart [
23]. The Spectral Clustering approach first constructs a similarity graph of all data points, before using dimensionality reduction and partitioning the data into clusters. Dimensionality reduction refers to the transformation of the data from a high- to low-dimensionality space, with the low-dimensionality space retaining the important properties of the original dataset; this process is also known as spectral embedding [
24].
2.8. Visualization of Clusters
Two types of plots were used in the analysis for graphical visualization of clusters: scatter plots and silhouette plots. Scatterplots are used to visualize the placement of datapoints and the clusters using the first and second order statistical features of the datasets showing lesions and hyperfluorescence. Silhouette plots, on the other hand, show how the data are distributed into the respective clusters (i.e., total number of images that go into each cluster), and they also determine whether each assigned cluster exceeds or falls beneath the average silhouette score.
In addition to the above, following the identification of the optimal number of clusters, the original images (i.e., hyperfluorescence segmentation outputs) are automatically labelled according to the specifications of the clustering model. This enables the assessment of the shapes and patterns within each cluster group, and whether the clustering method accurately groups the shapes and patterns, or whether some are mixed. The objective is not just to identify the optimal number of clusters, but to also ensure the classification of the original images into those clusters is correct.
2.9. Data Transformation
Prior to testing the various clustering algorithms, the following transformations were taken to ensure there was no skewness in the data:
- (1)
Standardization of feature data. This involves removal of the feature mean (average) value and dividing non-constant features by their standard deviation to scale the data, which is equivalent to the statistical z-score [
25]. In Python, this is achieved using the StandardScaler() function in the package sklearn (
https://scikit-learn.org/stable/ (accessed on 20 April 2021)).
- (2)
Reducing the complexity of the data using Principal Component Analysis (PCA), which transforms the collections of correlated features extracted from the images and condenses them into smaller, uncorrelated variables named principal components [
26]. This is achieved using the Python
sklearn function PCA().
2.10. Cluster Evaluation Metrics
There are different metrics which can be used for evaluation of meaningful clusters. The choice is contingent on whether we use an intrinsic (i.e., internal) cluster quality measure or an extrinsic (i.e., external) measure. The analysis presented here is for unsupervised clustering methods where there is no prior knowledge (unlike supervised learning). Therefore, to assess the quality of the clustering method, we adopt an intrinsic approach and evaluate the properties of the data itself. For this analysis, internal validation measures were used, such as the Silhouette Coefficient (SC), the Davies–Bouldin Index (DBI), and the Calinski–Harabasz Index (CHI).
The SC is a proximity measure, assessing both the compactness of clusters as well the separation of clusters from one another. It relies on two features: the partitions obtained from the clustering method, and the proximities of all datapoints. It uses these two features to assess (1) how close each data point is to data points within its own cluster, and (2) how distant each data point is from data points in other clusters. The SC formula is denoted as:
where
is any object in the data (i.e., image features),
is the average dissimilarity of
to all other objects of the cluster
,
is the average dissimilarity of
to all objects of cluster
, and
is smallest number of datapoints which for all clusters where
(i.e.,
= minimum
[
27]. The values for the SC range from −1 to +1. Negative values indicate the assignment of incorrect clusters, while values around 0 may suggest borderline results, and finally positive values indicate good clusters, particularly values which are 0.5 and higher. In fact, the higher the value, the more we can validate that good clustering has been identified (i.e., data points are well matched to their cluster and poorly matched to neighboring clusters).
The DBI is a cluster separation measure and possesses the following qualities: (1) it can be applied to hierarchical datasets, (2) it is computationally feasible for even large datasets, and (3) can yield meaningful results for data of arbitrary dimensionality [
28]. The objective of DBI is to identify whether clusters are well-spaced from each other, and whether each cluster is very dense and likely to be a good cluster. It calculates the ratio of the sum of the average distances between two clusters and between cluster centers [
29]. The formula for the DBI is:
where
is the number of clusters,
is the average distance of all points in cluster
,
is the average distance of all points in cluster
, and
and
are the cluster centroids for clusters
and
, respectively. Conversely to the SC, the rule of thumb for the DBI is that the smaller the value the better the clustering method. The minimum score for this index is 0, and thus ideally anything that is close to the 0 mark is indicative of a good clustering method.
The CHI, also known as the Variance Ratio Criterion, evaluates the ‘tightness’ of within clusters (i.e., inter-cluster dispersion) as well as between clusters (i.e., between-cluster dispersion) [
29,
30]. The CHI is defined as [
29]:
where
is the number of corresponding clusters,
and is the inter-cluster divergence, and
is the intra-cluster divergence, and
is the total number of samples. A smaller
indicates a good inter-cluster dispersion, while a high
indicates a good between-cluster dispersion. The larger a CHI ratio is the better the clustering effect [
29].
4. Discussion
The automation of hyperfluorescence segmentation is described using a pseudocoloring method employing the application of the JET colormap to capture hyperfluorescent intensity changes. Regions of hyperfluorescence are difficult to annotate due to their spatial distribution, frequency and poor visibility. The pseudocoloring process aided in segmentation of hyperfluorescence flagged in FAF images and also revealed additional areas previously missed. This technique can be used to expedite clinical and research investigations into hyperfluorescence and its association with GA progression. In the segmentation process, three transitionary stages of hyperfluorescence were identified based both on the intensity changes in lipofuscin build-up as well as the proximity of these stages to the GA lesions boundaries. These new transition stages have been labelled as early-, intermediate-, and late-stage hyperfluorescence regions.
The clustering of hyperfluorescence areas is also described in this study, with the newly identified transitionary stages assessed for further analysis. Publications to date have evaluated the categorization of hyperfluorescence patterns, with categories including none, focal, banded, diffuse, and patchy (as described in Holz et al.) [
4]. The diffuse category is further divisible into reticular, branching, fine granular, trickling, and GPS. However, the repeatability and reproducibility of these categories has varied, with associations with these patterns and their influence on GA progression not always showing statistical significance. Furthermore, while Fleckenstein et al. [
33]. showed an illustrative example of how different lesion types correlated with different progression rates, it was an indirect implication—lesion categorization was not fully explored as hyperfluorescence categorization. Therefore, in this study, it was hypothesized that unsupervised clustering algorithms could be used to distinguish the different categories of hyperfluorescence shapes and patterns, and these new cluster patterns could be associated with future GA progression.
The clustering algorithms which were tested included k-Means, Agglomerative, Affinity Propagation, BIRCH, DBSCAN, Mini-Batch k-Means, and Spectral Clustering. A combination of metrics (i.e., SC, DBI, and CHI) were used alongside visualization techniques to determine (1) the optimal number of clusters, and (2) the best unsupervised clustering algorithm for GA data. The number of clusters of interest ranged from k = 2 to k = 12. The choice of this range is in-line with current studies which suggest that regions of hyperfluorescence can be divided into five main categories (i.e., none, focal, diffuse, banded and patchy), and a further five subcategories. Affinity Propagation, DBSCAN, and Spectral Clustering were eliminated early in the evaluation process. Parameter sensitive Affinity Propagation resulted in very large cluster numbers (i.e., over 90 clusters for lesion and hyperfluorescent data). The other parameter-sensitive algorithm, DBSCAN, produced good SC scores, however, less than ideal DB and CHI scores, which resulted in its early elimination. Spectral Clustering was also removed, given that it produced identical results for all clusters, typically after k = 2. The repetitiveness of results with the use of Spectral Clustering can have many factors, but one which may be most probable for this dataset is that the dataset is not balanced (i.e., there is not an equality of cases for each cluster).
Among the remaining algorithms, k-Mean and Agglomerative clustering were the main contenders. They both had favourable SC, DBI, and CHI outcomes, and showed more distinguishable and separable clusters in the scatter plots. However, when it came to seeing how each image was clustered, the shapes and patterns within each cluster as produced by k-Means was much more consistent than Agglomerative, with the latter having the tendency to mix-in different shapes and patterns into one cluster group. As a result, the k-Means, a simple partitioning cluster method, was chosen as the best unsupervised clustering method (in combination with pre-processing techniques, such as scaling and PCA) for GA feature data.
The optimal number of clusters for early-stage hyperfluorescence areas was k = 3, with a SC of 0.597, a DBI of 0.915, and a CHI of 186.99. For intermediate-stage hyperfluorescence, an optimal cluster of k = 2 was identified, with a SC of 0.496, a DBI of 1.282, and a CHI of 144.92. Finally, for the late-stage hyperfluorescence, optimal cluster of k = 3 was identified, with an SC of 0.593, a DBI of 1.013, and a CHI of 217.325. Amongst these results, the one with intermediate-stage hyperfluorescence results present the greatest uncertainty, given it has the lowest value of SC. Scatter plots of lesions demonstrated extremely discernible cluster of three in the scatter plots. While the scatter plots for early-, intermediate and late-stage hyperfluorescence also showed good clustering visually, these clusters were not as distinguishable as those of the lesions cluster plot.
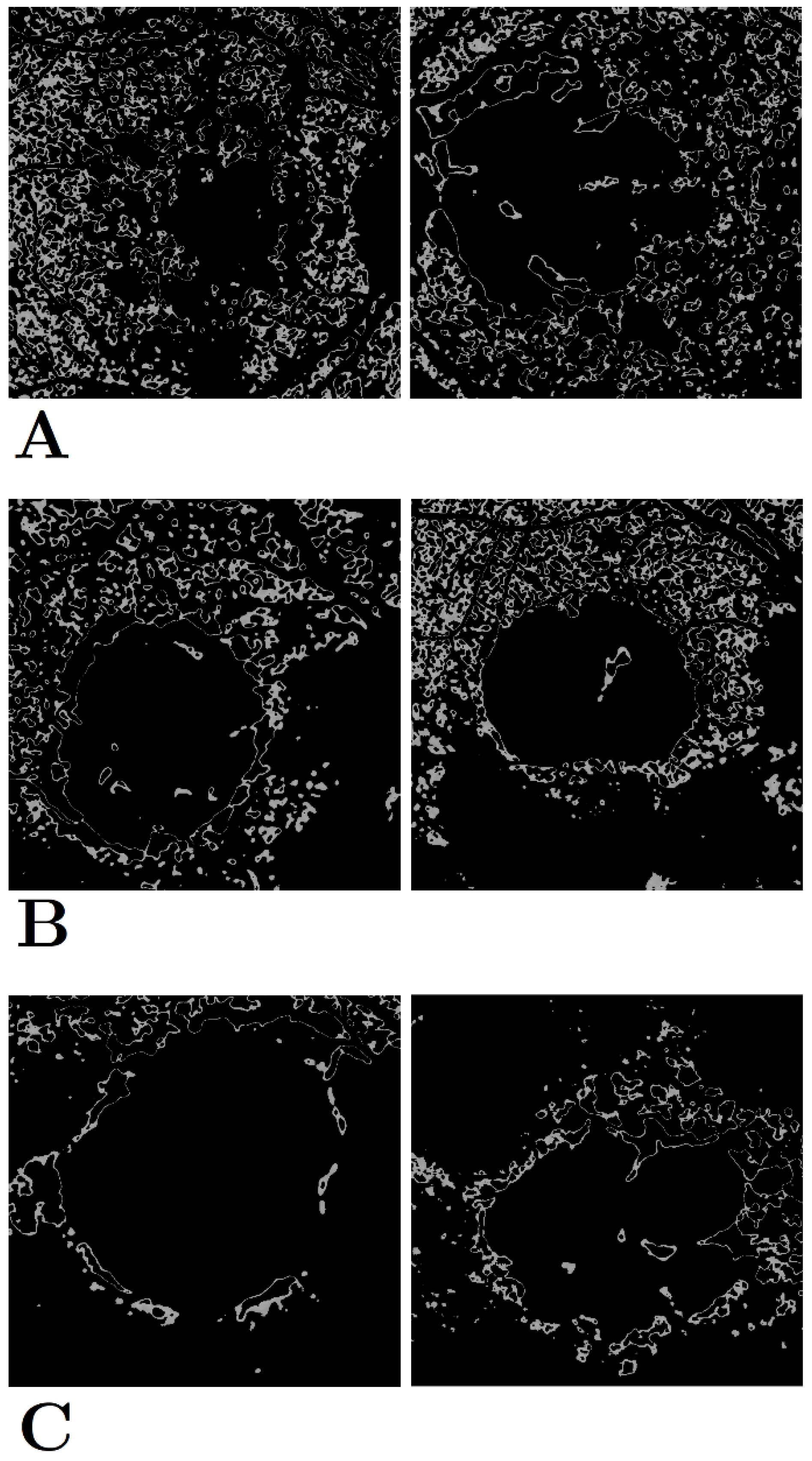
The three categories of early-stage hyperfluorescence clustering are presented in
Figure 5. The cluster groups appear to represent the level of coverage of early-stage hyperfluorescence across the retina. The first pattern involves the coverage of the entire retina, while the second pattern covers the retina partially. Finally, the third pattern covers the perimeter of the lesion. These patterns have been named as (1) early-stage hyperfluorescence complete coverage (EHCC), (2) early-stage hyperfluorescence partial coverage (EHPaC), and (3) early-stage hyperfluorescence proximal coverage (EHPrC).
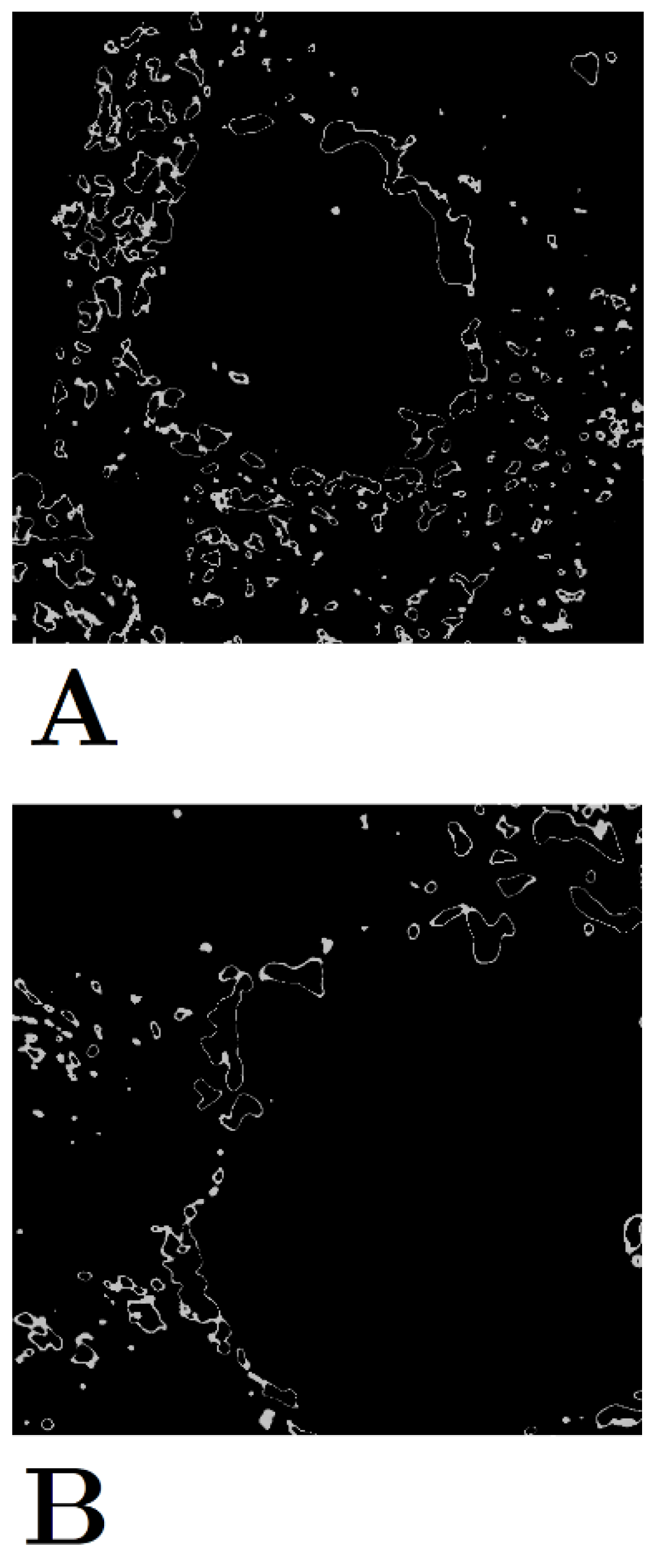
The two clusters of intermediate-stage hyperfluorescence are not as clear-cut in their patterns as the lesions and early-stage hyperfluorescence (
Figure 6), thus explaining the lower SC scores. No names have been given to these yet, given that intermediate-stage clustering may simply represent a transitional stage between the early and late stages of hyperfluorescence.
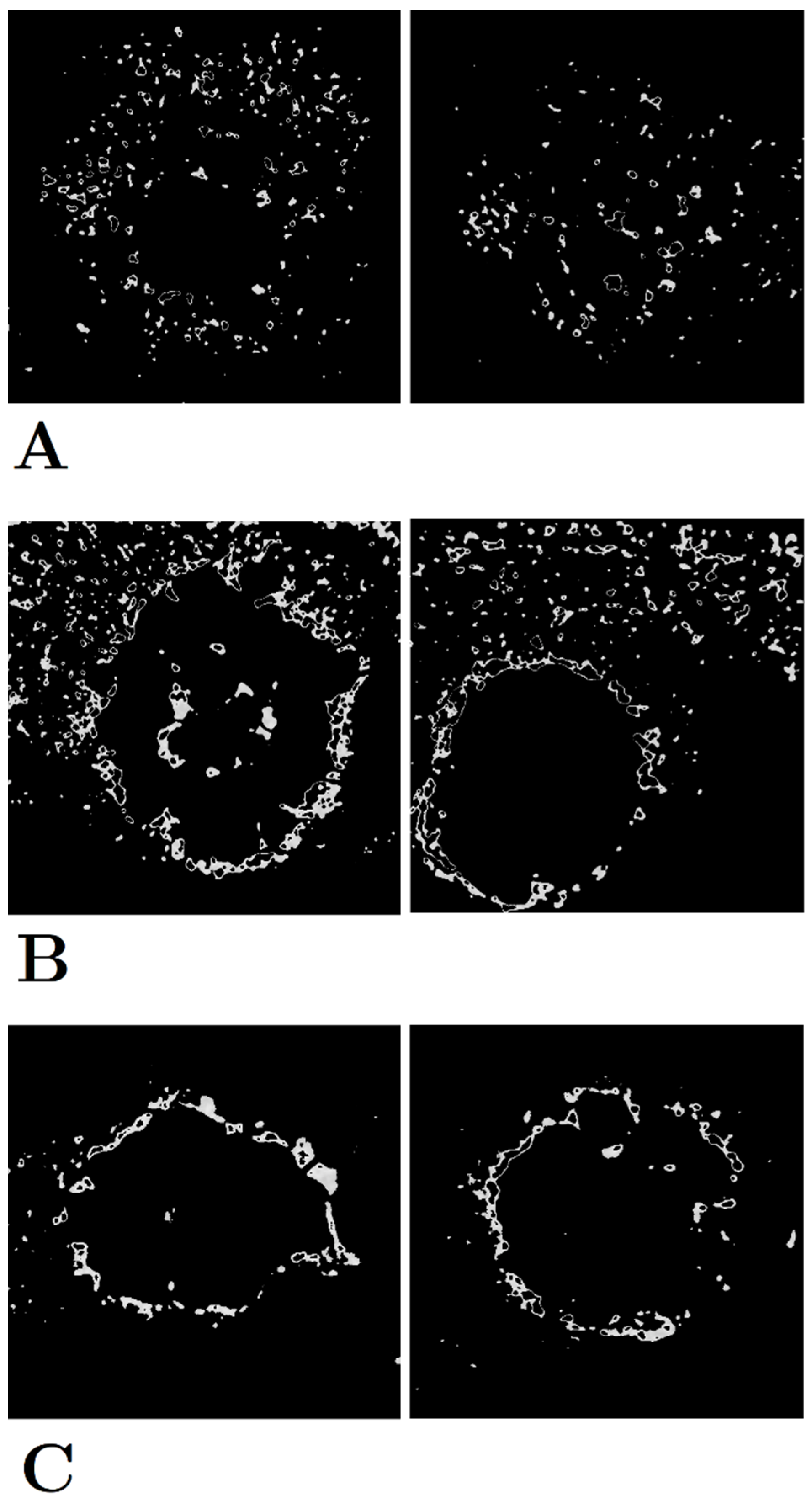
Finally, the three stages of late-stage hyperfluorescence clustering are presented in
Figure 7. The first category of late-stage patterns shows a small, circle scattering of hyperfluorescence droplets, while the second category combines these droplet scatters with a halo surrounding the lesion. Lastly, the third pattern shows a very distinct and dense halo directly surrounding the lesion, that most likely illustrates peak lipofuscin intensity which precedes lesion formation. These regions have been therefore named as (1) late-stage hyperfluorescence droplet scatter (LHDS), (2) late-stage hyperfluorescence halo scatter (LHHS), and (3) late-stage hyperfluorescence halo (LHH). All newly identified GA feature categories can be found in
Table 9.
One of the potential limitations of cluster analysis is the requirement for parameter tuning for some clustering methods, such as Affinity Propagation and DBSCAN. Even though attempts were made to automate the selection of the epsilon parameters for DBSCAN, the results can change given their sensitivity to parameter changes. Proper settings can only be established by trial-and-error, and the analysis presented here was no exception [
28]. From establishing the most appropriate data transformations, to testing the different algorithms, and the selection of parameters, there is a still a component of human error in unsupervised clustering that needs to be addressed. Another potential limitation exists with the cases available in this cohort. The clustering was based on the shapes and patterns of the data in the trial. However, this dataset may not be representative of all GA cases and future research could investigate further applications to new data sets.
Future research could replicate these findings in larger and more diverse cohorts, further establishing the validity of the hyperfluorescent classifications identified in this study. Furthermore, the application of these hyperfluorescent classes in disease progression modelling can be used to validate the influence of the classification on predictive capabilities. In line with previous research, establishing correlations between the identified hyperfluorescent categories and disease progression rates could shed additional light on the significance of hyperfluorescence monitoring in GA progression. Additionally, epidemiological investigations might illuminate how the predictive performance of hyperfluorescent patterns vary across different cohorts. Complementing such research, genetic association studies could provide valuable insights, revealing inheritable patterns that might explain the differing appearances and spatial distributions of hyperfluorescent regions within the retina.
These classifications could lead to the development of more robust prediction models that could be used in a real-time clinical setting to predict progression severity and rate. The developed models, along with their incorporated features, could foster a personalized approach to diagnosis and patient care, leading to the potential formulation of tailored therapies. In a clinical setting, healthcare providers could leverage hyperfluorescence readings to forecast patient prognosis right from the baseline, specifically identifying those at higher risk of rapid progression. This practice could empower clinicians to better manage patients’ well-being, leading to timely and effective interventions.