Analysis of the Bacterial Community of Metal Scrap Using an Enrichment Culture Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Enrichment Culture
2.2. Genomic DNA Extraction and PCR Conditions
2.3. Sequencing Library Preparation, Sequencing, and Bioinformatics Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Little, B.J.; Lee, J.S. Microbiologically Influenced Corrosion; John Wiley and Sons, Inc.: Hoboken, NJ, USA, 2007; pp. 1–279. [Google Scholar]
- Dai, X.; Wang, H.; Ju, L.K.; Cheng, G.; Cong, H.; Newby, B.Z. Corrosion of aluminum alloy 2024 caused by Aspergillus niger. Int. Biodeterior. Biodegrad. 2016, 115, 1–10. [Google Scholar] [CrossRef]
- Xu, D.; Wen, J.; Fu, W.; Gu, T.; Raad, I.I. D-Amino acids for the enhancement of a binary biocide cocktail consisting of THPS and EDDS against an SRB biofilm. World J. Microbiol. Biotechnol. 2012, 28, 1641–1646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Xu, D.; Li, Y.; Yang, K.; Gu, T. Electron mediators accelerate the microbiologically influenced corrosion of 304 stainless steel by the Desulfovibrio vulgaris biofilm. Bioelectrochemistry 2015, 101, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Yang, C.; Xu, D.; Sun, D.; Nan, L.; Sun, Z.; Li, Q.; Gu, T.; Yang, K. Laboratory investigation of the microbiologically influenced corrosion (MIC) resistance of a novel Cu-bearing 2205 duplex stainless steel in the presence of an aerobic marine Pseudomonas aeruginosa biofilm. Biofouling 2015, 31, 481–492. [Google Scholar] [CrossRef]
- Li, H.; Zhou, E.; Zhang, D.; Xu, D.; Xia, J.; Yang, C.; Feng, H.; Jiang, Z.; Li, X.; Gu, T.; et al. Microbiologically influenced corrosion of 2707 hyper-duplex stainless steel by marine Pseudomonas aeruginosa biofilm. Sci. Rep. 2016, 6, 20190. [Google Scholar] [CrossRef]
- Li, H.; Yang, C.; Zhou, E.; Yang, C.; Feng, H.; Jiang, Z.; Xu, D.; Gu, T.; Yang, K. Microbiologically influenced corrosion behavior of S32654 super austenitic stainless steel in the presence of marine Pseudomonas aeruginosa biofilm. J. Mater. Sci. Technol. 2017, 33, 1596–1603. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Zhang, R.; Guan, F.; Hou, B.; Duan, J. Microbiologically influenced corrosion of marine steels within the interaction between steel and biofilms: A brief view. Appl. Microbiol. Biotechnol. 2020, 104, 515–525. [Google Scholar] [CrossRef]
- Näslund, J.; Johnsson, J.I. Environmental enrichment for fish in captive environments: Effects of physical structures and substrates. Fish Fish. 2016, 17, 1–30. [Google Scholar] [CrossRef]
- Arechavala-Lopez, P.; Cabrera-Álvarez, M.J.; Maia, C.M.; Saraiva, J.L. Environmental enrichment in fish aquaculture: A review of fundamental and practical aspects. Rev. Aquacult. 2022, 14, 704–728. [Google Scholar] [CrossRef]
- Zhang, Z.; Gao, L.; Zhang, X. Environmental enrichment increases aquatic animal welfare: A systematic review and meta-analysis. Rev. Aquacult. 2022, 14, 1120–1135. [Google Scholar] [CrossRef]
- Guangfeng, X.; Xiaodong, Z.; Shuai, W.; Jie, Y.; Jie, S.; Zhongyi, A.; Yan, L.; Xinlei, Q. Synergistic effect between sulfate-reducing bacteria and Pseudomonas aeruginosa on corrosion behavior of Q235 steel. Int. J. Electrochem. Sci. 2020, 15, 361–370. [Google Scholar]
- Tran, T.T.T.; Kannoorpatti, K.; Padovan, A.; Thennadil, S. A study of bacteria adhesion and microbial corrosion on different stainless steels in environment containing Desulfovibrio vulgaris. R. Soc. Open Sci. 2021, 8, 201577. [Google Scholar] [CrossRef]
- Prasanna, J.; Rosalie, C.; Sun, S.; Martin, S.; Florentin, C.; Sudesh, W.L.; Dominique, T.; Daniel, B.J.; Diane, M.; Scott, R.A.; et al. Onset of microbial influenced corrosion (MIC) in stainless steel exposed to mixed species biofilms from equatorial seawater. J. Electrochem. Soc. 2017, 164, C532–C538. [Google Scholar]
- Bloomfield, S.F.; Stewart, G.S.A.B.; Dodd, C.E.R.; Booth, I.R.; Power, E.G.M. The viable but non-culturable phenomenon explained? Microbiology 1998, 144, 1–3. [Google Scholar] [CrossRef]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1679. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 15 December 2021).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Akita, H.; Shinto, Y.; Kimura, Z.-I. Bacterial Community Analysis of Biofilm Formed on Metal Joint. Appl. Biosci. 2022, 1, 221–228. [Google Scholar] [CrossRef]
- Bukin, Y.S.; Galachyants, Y.P.; Morozov, I.V.; Bukin, S.V.; Zakharenko, A.S.; Zemskaya, T.I. The effect of 16S rRNA region choice on bacterial community metabarcoding results. Sci. Data 2019, 6, 190007. [Google Scholar] [CrossRef]
- Fadeev, E.; Cardozo-Mino, M.G.; Rapp, J.Z.; Bienhold, C.; Salter, I.; Salman-Carvalho, V.; Molari, M.; Tegetmeyer, H.E.; Buttigieg, P.L.; Boetius, A. Comparison of two 16S rRNA primers (V3–V4 and V4–V5) for studies of arctic microbial communities. Front. Microbiol. 2021, 12, 637526. [Google Scholar] [CrossRef]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The genus Sphingomonas: Physiology and ecology. Curr. Opin. Biotechnol. 1996, 7, 301–306. [Google Scholar] [CrossRef]
- Waigi, M.G.; Sun, K.; Gao, Y. Sphingomonads in microbe-assisted phytoremediation: Tackling soil pollution. Trends Biotechnol. 2017, 35, 883–899. [Google Scholar] [CrossRef]
- Venugopalan, V.P.; Kuehn, M.; Hausner, M.; Springael, D.; Wilderer, P.A.; Wuertz, S. Architecture of a nascent Sphingomonas sp. biofilm under varied hydrodynamic conditions. Appl. Environ. Microbiol. 2005, 71, 2677–2686. [Google Scholar] [CrossRef]
- Beale, D.J.; Morrison, P.D.; Key, C.; Palombo, E.A. Metabolic profiling of biofilm bacteria known to cause microbial influenced corrosion. Water Sci. Technol. 2014, 69, 1–8. [Google Scholar] [CrossRef]
- Meiying, L.; Min, D. A review: Microbiologically influenced corrosion and the effect of cathodic polarization on typical bacteria. Rev. Environ. Sci. Biotechnol. 2018, 17, 431–446. [Google Scholar]
- Kaewyaia, J.; Noophana, P.; Wantawina, C.; Munakata-Marrb, J. Microbiologically influenced corrosion and current mitigation strategies: A state of the art review. Int. Biodeterior. Biodegrad. 2019, 137, 42–58. [Google Scholar]
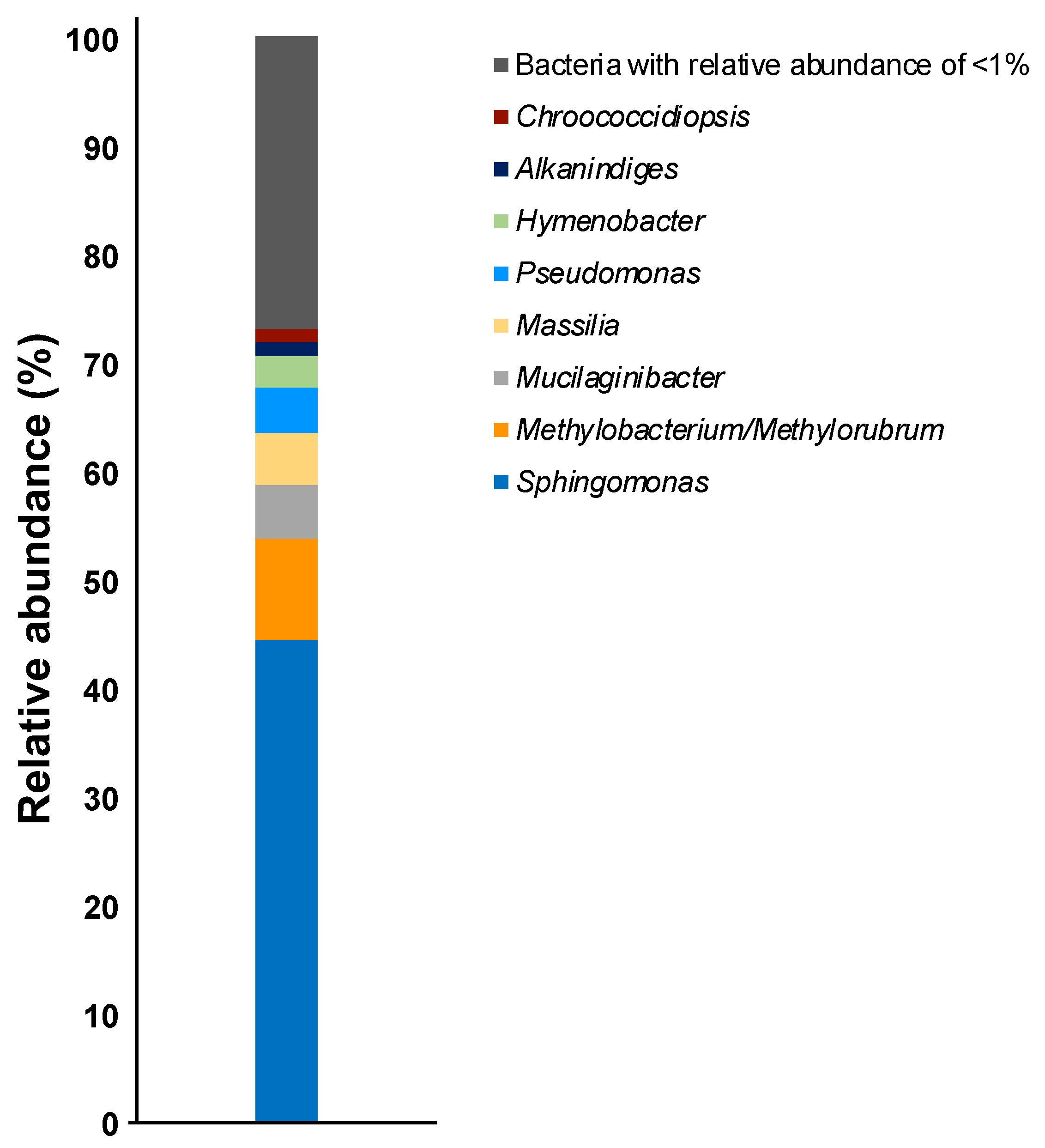
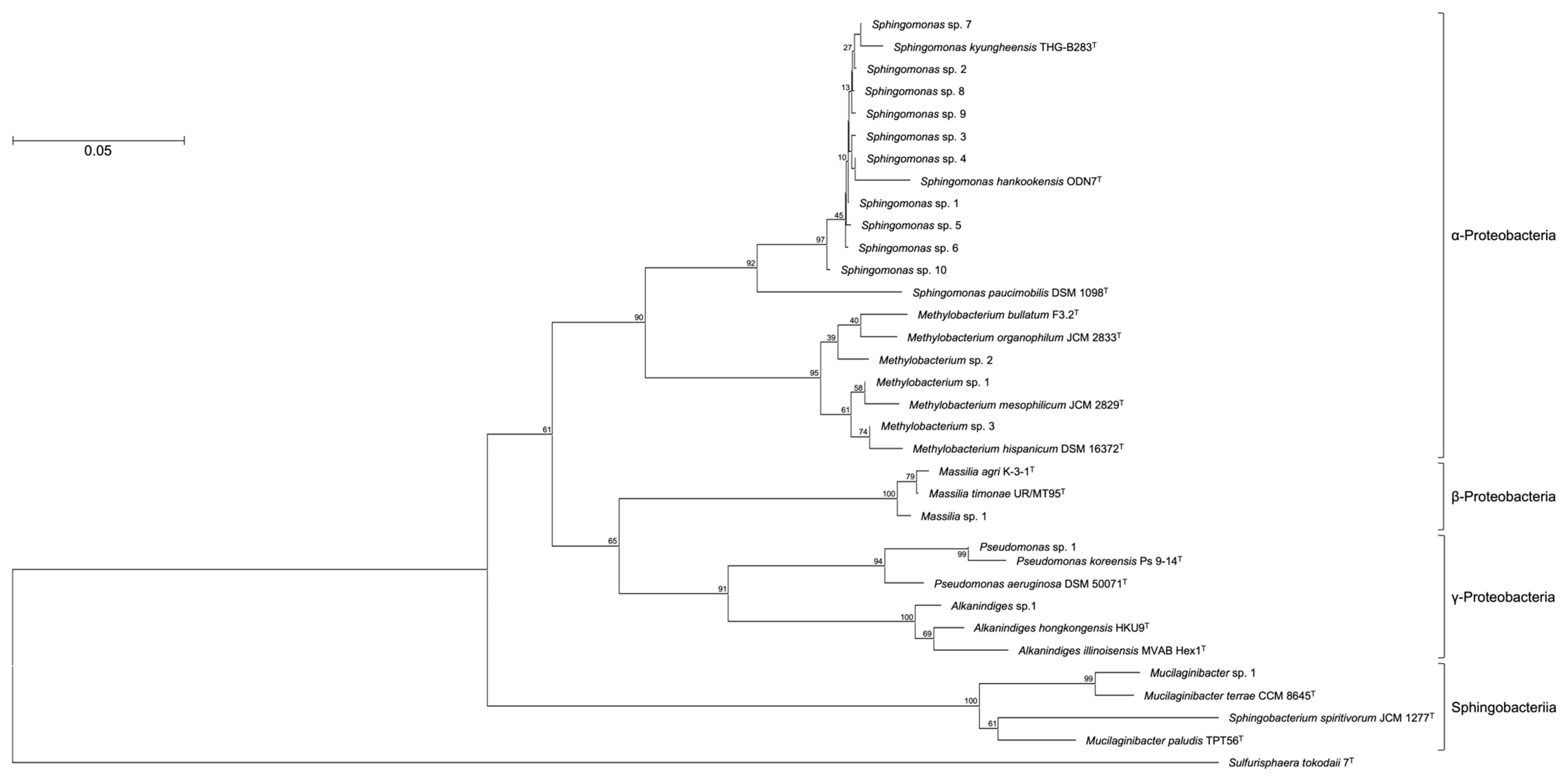
| OTU | Closest Relative | Accession No. | Identity (%) | Relative Abundance (%) | |
|---|---|---|---|---|---|
| Class | Species | ||||
| Sphingomonas sp. 1 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 99.1 | 7.1 |
| Sphingomonas sp. 2 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 99.1 | 5.6 |
| Sphingomonas sp. 3 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 99.1 | 4.8 |
| Sphingomonas sp. 4 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 99.3 | 4.6 |
| Mucilaginibacter sp. 1 | Sphingobacteria | Mucilaginibacter terrae CCM 8645T | NR_158094 | 99.7 | 4.3 |
| Sphingomonas sp. 5 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 97.6 | 4.2 |
| Sphingomonas sp. 6 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 99.1 | 4.0 |
| Methylobacterium sp. 1 | α-Proteobacteria | Methylobacterium mesophilicum JCM 2829T | NR_115550 | 99.1 | 3.7 |
| Pseudomonas sp. 1 | γ-Proteobacteria | Pseudomonas koreensis Ps 9-14T | NR_025228 | 99.1 | 3.5 |
| Sphingomonas sp. 7 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 99.3 | 3.1 |
| Sphingomonas sp. 8 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 99.3 | 2.4 |
| Sphingomonas sp. 9 | α-Proteobacteria | Sphingomonas kyungheensis THG-B283T | NR_118263 | 99.1 | 2.0 |
| Massilia sp. 1 | β-Proteobacteria | Massilia agri K-3-1T | NR_157781 | 98.5 | 1.9 |
| Methylobacterium sp. 2 | α-Proteobacteria | Methylobacterium bullatum F3.2T | NR_116548 | 98.2 | 1.4 |
| Sphingomonas sp. 10 | α-Proteobacteria | Sphingomonas hankookensis ODN7T | NR_116570 | 98.6 | 1.3 |
| Alkanindiges sp. 1 | γ-Proteobacteria | Alkanindiges hongkongensis HKU9T | NR_115178 | 97.4 | 1.2 |
| Methylobacterium sp. 3 | α-Proteobacteria | Methylobacterium hispanicum DSM 16372T | NR_112613 | 99.1 | 1.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akita, H.; Shinto, Y.; Kimura, Z.-i. Analysis of the Bacterial Community of Metal Scrap Using an Enrichment Culture Approach. Appl. Biosci. 2023, 2, 23-30. https://doi.org/10.3390/applbiosci2010004
Akita H, Shinto Y, Kimura Z-i. Analysis of the Bacterial Community of Metal Scrap Using an Enrichment Culture Approach. Applied Biosciences. 2023; 2(1):23-30. https://doi.org/10.3390/applbiosci2010004
Chicago/Turabian StyleAkita, Hironaga, Yoshiki Shinto, and Zen-ichiro Kimura. 2023. "Analysis of the Bacterial Community of Metal Scrap Using an Enrichment Culture Approach" Applied Biosciences 2, no. 1: 23-30. https://doi.org/10.3390/applbiosci2010004
APA StyleAkita, H., Shinto, Y., & Kimura, Z.-i. (2023). Analysis of the Bacterial Community of Metal Scrap Using an Enrichment Culture Approach. Applied Biosciences, 2(1), 23-30. https://doi.org/10.3390/applbiosci2010004






