Abstract
Microbiologically influenced corrosion (MIC) is caused by biofilms formed on metal surfaces, and MIC of metal alloys on marine infrastructure leads to severe accidents and great economic losses. Although bacterial community analyses of the biofilms collected from corroded metal have been studied, the analyses of biofilms collected from uncorroded metal are rarely reported. In this study, a biofilm formed on an uncorroded metal joint attached to a metal dock mooring at Akitsu Port was used as a model for bacterial community analysis. The bacterial community was analyzed by high-throughput sequencing of the V3–V4 variable regions of the 16S rRNA gene. Bacterial species contained in the biofilms were identified at the genus level, and Alkanindiges bacteria were the dominant species, which have been not reported as the dominant species in previous research on MIC. The genome sequences of known Alkanindiges bacteria do not have conserved gene clusters required to cause metal corrosion, which suggests that Alkanindiges bacteria do not corrode metals but act on the formation of biofilms. Those findings indicated that the bacterial community may change significantly during the process from biofilm formation to the occurrence of metal corrosion.
1. Introduction
Metal alloys are widely used as raw materials for the construction of vessels, bridges, and factories using established molding technology to create products intended for long-term use in marine environments. In marine environments, biofilms are formed when microorganisms adhere to metal surfaces. Biofilms are complexes of microorganisms with a three-dimensional structure, which is enclosed in an extracellular polymeric substance (EPS) matrix [1]. Inside the biofilm, extracellular matrix proteins and exogenous genes are transported through the EPS matrix, which encourages microbial growth [1]. Moreover, a well-developed biofilm tightly adheres to metal surfaces by extracellular polysaccharides of the EPS matrix, which is difficult to remove completely. The microorganisms inside the biofilm are protected from stresses caused by environmental changes and chemical substances. As such, a favorable environment for microbial growth is formed inside the biofilm, and the microbial community develops to a high density over time. In biofilms formed on metal surfaces, microbiologically influenced corrosion (MIC) is caused by various microbial factors, such as metal oxidation by electron transfer from the metal surface to bacteria [2] and accumulation of corrosive metabolites [3]. MIC causes serious problems for metal infrastructure in the marine environment, and economic losses from MIC were estimated to be $2.5 trillion in 2013, which is equivalent to about 3.4% of gross world product [4]. MIC assessments in the marine environment have been reported for carbon steel [5], stainless steel [6], and low-alloy steel [7]. In those reports, Bacillus, Desulfovibrio, and Desulfobacter have been detected as the dominant bacteria, and the developmental mechanisms of those bacteria are discussed. By contrast, bacterial community analyses of biofilms collected from uncorroded metal have rarely been reported, and our understanding of the community structure is limited. Thus, changes in bacterial community from development of biofilm to occurrence of metal corrosion have not been elucidated in detail. We consider that microbial community analyses of biofilms collected from uncorroded metal in the marine environment are important to understand the mechanism of MIC, and elucidation of the mechanism may lead to the establishment of measures to prevent the occurrence of metal corrosion [8].
High-throughput amplicon sequencing is a powerful method for the identification of target genes in large amounts of unexamined genetic material from the environmental microbiome [9]. In this method, microbial DNA is extracted from the environmental sample, and target genes are then amplified by polymerase chain reaction (PCR) using the extracted DNA as a template and primers designed to bind to the template. Subsequently, the amplified regions (amplicons) are sequenced by a next-generation sequencer. Since hundreds to thousands of different amplicons can be sequenced together by a next-generation sequencer, this method can efficiently identify target genes. In bacterial genomic DNA, several housekeeping genes are conserved to ensure cell maintenance and growth. In particular, the 16S rRNA gene encodes the 30S ribosomal subunit that is essential for the translation of mRNA and protein synthesis, but it has genus-specific variable regions, such as V1–V9 [10]. Almost all bacterial taxa can theoretically be identified based on the 16S rRNA gene sequence. Thus, bacterial community analysis based on high-throughput sequencing of 16S rRNA gene fragments can characterize the bacterial microbiome [10].
In this study, to investigate the bacterial community that contributes to MIC, a biofilm formed on an uncorroded metal joint attached to a metal dock mooring was used as a model. Using bacterial genomic DNA prepared from the biofilm, the variable region V3–V4 of the 16S rRNA gene was amplified by PCR, and high-throughput amplicon sequencing was carried out. The bacterial community was identified at the genus level based on the operation taxonomic units (OTUs).
2. Materials and Methods
2.1. Sampling
A biofilm that formed on an uncorroded metal joint attached to a metal dock mooring was collected in Akitsu Port in Hiroshima prefecture, Japan (Figure 1). After collection, the samples were put into sterile tubes and immediately placed in a cool box at 4 °C, then transported to the lab within a few hours.

Figure 1.
Image of the sampling point. The sampling point for the collection of the biofilm is indicated by a black arrow.
2.2. Genomic DNA Extraction and PCR Conditions
Genomic DNA was extracted from the biofilm using an illustra™ bacteria genomicPrep Mini Spin Kit (GE Healthcare, Chicago, IL, USA) according to the manufacturer’s instructions, and the concentration and purity were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Using the genomic DNA as a template, the V3–V4 variable regions of the 16S rRNA genes were amplified by KOD -Plus- (TOYOBO, Osaka, Japan) using bacterial domain-specific primers 341F (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′) [11] and 805R (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′) [12]. For amplification of the 16S rRNA genes, samples were subjected to initial denaturation at 94 °C for 2 min, followed by 35 cycles of denaturation at 98 °C for 10 s, annealing at 55 °C for 30 s, and extension at 68 °C for 30 s, and the final extension at 68 °C for 5 min.
2.3. Sequencing Library Preparation, Sequencing, and Bioinformatics Analysis
The length of the PCR products was confirmed using 1.0% agarose gel electrophoresis, and the samples (approximately 450 bp) were then purified using a Wizard SV Gel and PCR Clean-up System (Promega, Madison, WI, USA).
The microbiome sequence libraries were prepared using a Nextera XT DNA Library Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The concentrations of the sequence libraries were measured using a Quanti Fluor™ dsDNASystem (Promega). The libraries were sequenced using a MiSeq sequencer (Illumina) with a MiSeq Reagent Kit v3 (Illumina).
The bacterial community was analyzed using the QIIME 2 microbiome bioinformatics platform [13]. After the quality of the sequence data was confirmed using a FastQC ver.0.11.9 [14], the amplicon sequence variants (ASVs) were prepared based on the filtered data using the DADA2 algorithm [15]. The ASVs were grouped into OTUs at 99% sequence identity using the taxonomic databases of Sliva ver. 138 [16,17]. To construct a phylogenetic tree based on the variable regions of 16S rRNA gene sequences, the OTU dataset was also subjected to phylogenetic analysis with MEGA 11 software [18]. The consensus bootstrap phylogenetic tree was constructed using the neighbor-joining method.
3. Results and Discussion
Among the OTUs, a total of 18 genera of bacteria were observed, and the proportion of unclassified bacteria was less than 0.1% (Figure 2). Alkanindiges bacteria were observed as the dominant species, and their relative abundance was more than 95%. Apart from Alkanindiges, several bacteria such as Sphingomonas and Pseudomonas were also observed as OTUs, but the relative abundances of those bacteria were less than 3%. To confirm the phylogenetic relationship of the bacteria comprising more than 1% of the OTUs, a neighbor-joining tree based on the V3–V4 variable regions of the 16S rRNA gene was constructed. In the neighbor-joining tree, nine kinds of Alkanindiges bacteria formed a cluster with A. hongkongensis HKU9T, A. illinoisensis MVAB Hex1T, and A. hydrocarboniclasticus H1T (Figure 3). Alkanindiges bacteria belong to a class of Gammaproteobacteria and can grow using several kinds of hydrocarbons degraded from petroleum [19], so Alkanindiges bacteria are a major contributor to the bioremediation of petroleum-contaminated soil [20,21,22]. To our knowledge, there is a report of Gammaproteobacteria being observed as more than 90% of OTUs in a biofilm collected from a metal surface [6], but there are no reports of Alkanindiges bacteria accounting for more than 90% of the total.
Lichen is a complex organism of algae, fungi, and bacteria that grows on a wide range of substrates, from natural surfaces to artificial materials such as metal, glass, rubber, and plastic [23]. For example, Stereocaulon japonicum and Cladonia humilis grow in Cu-polluted sites around Cu-roofed temple buildings by their Cu-accumulation capacity [24]. Fernández-Brime et al. reported the bacterial communities associated with a lichen symbiosis, and more than 20 orders of bacteria were observed in several lichenized samples [25]. Moreover, 11 genera of bacteria observed in this study, such as Alkanindiges, Sphingomonas, Pseudomonas, Roseateles, Allorhizobium/Neorhizobium/Pararhizobium/Rhizobium, Brevundimonas, Reyranella, Massilia, Pantoea, Mucilaginibacter, Asinibacterium, Roseomonas, Hymenobacter, Paracoccus, and Bradyrhizobium were also observed in the report by Fernández-Brime et al. [25]. Thus, the occurrence of metal corrosion may be induced by bacteria adhered to metal surfaces via lichen. To improve the accuracy of the measures to prevent the occurrence of metal corrosion, it is necessary to consider bacterial community analysis including lichen in the future.
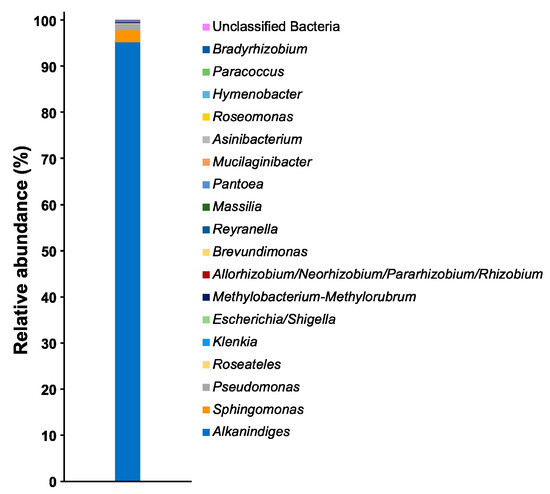
Figure 2.
Relative abundances of the bacterial community in the biofilm at the genus level.
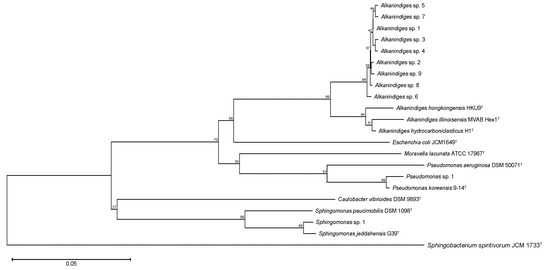
Figure 3.
Consensus bootstrap phylogenetic tree of OTUs comprising more than 1% of the total 16S rRNA gene sequences. Each OTU is denoted by its representative strain type. The numbers at the nodes are percentages indicating the levels of bootstrap support based on neighbor-joining analysis of 1000 resampled datasets. The tree was rooted using Sphingobacterium spiritivorum JCM 1733T as the outgroup. The bar indicates a 0.05% nucleotide substitution rate.
The detailed molecular mechanisms of MIC are still unclear, but electron transfer from the metal surface to MIC-causing bacteria has been demonstrated in a few reports. For example, Pseudomonas aeruginosa MCCC 1A00099 produces water-soluble electron transfer mediators, such as phenazine-1-carboxamide and pyocyanin, by phenazine-modifying enzyme, methyl transferase, and flavin-containing monooxygenase, which are encoded by phzH, phzM, and phzS genes. Those mediators activate the extracellular electron transfer pathway [26,27]. In the genome of Methanococcus maripaludis OS7, the genes that encode the small and large subunits of a [NiFe] hydrogenase (MMOS7_11590 and MMOS7_11600), the membrane proteins for secretion of the [NiFe] hydrogenase (MMOS7_11620 and MMOS7_11630), and a hydrogenase maturation protease (MMOS7_11610) are clustered [28]. Although M. maripaludis OS7 promotes MIC of iron, a deletion mutant strain lacking the gene cluster does not corrode iron. In Alkanindiges bacteria, the genome has been sequenced for two species, including A. hydrocarboniclasticus H1T and A. illinoisensis DSM 15370T. In those genome sequences, with the exception of MMOS7_11620 and MMOS7_11630, genes related to MIC are not present. This suggests that Alkanindiges bacteria do not cause MIC but act in the formation of biofilms before MIC occurrence.
In the KEGG PATHWAY Database [29], the biofilm formation pathway is registered for three species of Gammaproteobacteria, including Escherichia coli, P. aeruginosa, and Vibrio cholerae. To investigate the biofilm-forming capacity of Alkanindiges bacteria, the biofilm formation pathways of A. hydrocarboniclasticus H1T and A. illinoisensis DSM 15370T were compared using that of P. aeruginosa, which is the closest of the three species of Gammaproteobacteria to Alkanindiges bacteria (Figure 4). In P. aeruginosa, cAMP/Vfr signaling, Quorum sensing, and Gac/Rsm and c-di-GMP signaling pathways have been identified, and those pathways except the Quorum sensing pathway seem to be conserved in both strains. In particular, the function of c-di-GMP signaling pathway is enhanced by Psl, which is a neutral exopolysaccharide typically comprising D-glucose, D-mannose, and L-rhamnose. This exopolysaccharide is a crucial adhesive component of the biofilm matrix that promotes both cell-to-cell interactions and cell-to-metal surface attachment [30]. Thus, Alkanindiges bacteria may adhere to a metal surface by the function of the c-di-GMP signaling pathway, resulting in the formation of a biofilm. In addition, the development of biofilms can migrate to bacterial communities composed of MIC-causing bacteria and eventually cause MIC on the surface of metal joints. Pseudomonas species have been isolated and identified from various sources, such as soil, fresh water, and metal surfaces, and several species are suggested to be MIC-causing strains because those species can obtain energy by transferring electrons to extracellular solid substances [31]. In fact, we also observed Pseudomonas sp. 1, which shows high identity with the MIC-causing strain P. koreensis Ps 9-14T [32]. Thus, the relative abundance of Pseudomonas sp. 1 in the biofilm may increase as time proceeds, which then encourages MIC on the surface of the metal joint.

Figure 4.
Presence/absence matrix of biofilm formation pathway genes in A. hydrocarboniclasticus H1T and A. illinoisensis DSM 15370T. Strains: 1, A. hydrocarboniclasticus H1T; 2, A. illinoisensis DSM 15370T. Green, orange, and gray squares indicate cases where the genes are conserved, similar genes are conserved, and genes are not conserved, respectively.
4. Conclusions
Bacterial community analyses of metal corrosion-free biofilms have rarely been reported, though analyses within different environments are required to understand the mechanism of MIC. Unlike previous studies on MIC, Alkanindiges bacteria were observed as the dominant species in this study. Moreover, the gene clusters required to cause metal corrosion were not conserved in the genome sequences of known Alkanindiges bacteria. Those findings indicated that the bacterial community changes significantly during the process from development of biofilm to occurrence of metal corrosion. For more detailed investigations, it will be necessary to analyze changes in the bacterial community over time.
Author Contributions
Conceptualization, H.A.; methodology, H.A. and Y.S.; validation, H.A. and Y.S.; formal analysis, H.A. and Y.S.; investigation, H.A. and Y.S.; resources, H.A.; data curation, H.A. and Y.S.; writing—original draft preparation, H.A.; writing—review and editing, H.A., Y.S., and Z.-i.K.; visualization, H.A.; supervision, H.A. and Z.-i.K.; project administration, H.A.; funding acquisition, H.A. and Z.-i.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from The Salt Science Research Foundation (No. 2101).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The 16S rRNA gene sequences of A. hongkongensis HKU9T, A. hydrocarboniclasticus H1T, A. illinoisensis MVAB Hex1T, C. vibrioides DSM 9893T, E. coli JCM 1649T, M. lacunata ATCC 17967T, P. aeruginosa DSM 50071T, P. koreensis 9-14T, S. jeddahensis G39T, S. paucimobilis DSM 1098T, and S. spiritivorum JCM 1733T are available in the GenBank/EMBL/DDBJ databases under accession numbers NR_114676, LT827059, NR_025254, NR_037099, NR_024570, NR_036825, AF094713, NR_025228, NR_158139, LN681566, and LC060921, respectively. Draft genome sequences of A. hydrocarboniclasticus H1T and A. illinoisensis DSM 15370T were deposited in DDBJ/EMBL/GenBank databases under accession numbers MLCN01000001–MLCN01000092 and JHUX01000001–JHUX01000012, respectively.
Acknowledgments
We are grateful to all members of the Bio-conversion Research Group at our Institute (Research Institute for Sustainable Chemistry, National Institute of Advanced Industrial Science and Technology (AIST)) for their technical assistance and valuable discussion.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Meiying, L.; Min, D. A review: Microbiologically influenced corrosion and the effect of cathodic polarization on typical bacteria. Rev. Environ. Sci. Biotechnol. 2018, 17, 431–446. [Google Scholar]
- Jia, R.; Unsal, T.; Xu, D.; Lekbach, Y.; Gu, T. Microbiologically influenced corrosion and current mitigation strategies: A state of the art review. Int. Biodeterior. Biodegradation 2019, 137, 42–58. [Google Scholar] [CrossRef]
- Koch, G.; Varney, J.; Thompson, N.; Moghissi, O.; Gould, M.; Payer, J. International Measures of Prevention, Application, and Economics of Corrosion Technologies Study; Jacobson, G., Ed.; NACE International: Houston, TX, USA, 2016; pp. 1–216. [Google Scholar]
- Procópio, L. Microbial community profiles grown on 1020 carbon steel surfaces in seawater-isolated microcosm. Ann. Microbiol. 2020, 70, 13. [Google Scholar] [CrossRef]
- Capão, A.; Moreira-Filho, P.; Garcia, M.; Bitati, S.; Procópio, L. Marine bacterial community analysis on 316L stainless steel coupons by Illumina MiSeq sequencing. Biotechnol. Lett. 2020, 42, 1431–1448. [Google Scholar] [CrossRef]
- Lv, M.; Du, M.; Li, Z. Investigation of mixed species biofilm on corrosion of X65 steel in seawater environment. Bioelectrochemistry 2022, 143, 107951. [Google Scholar] [CrossRef] [PubMed]
- Procópio, L. The role of biofilms in the corrosion of steel in marine environments. World J. Microbiol. Biotechnol. 2019, 35, 73. [Google Scholar] [CrossRef] [PubMed]
- Gołębiewski, M.; Tretyn, A. Generating amplicon reads for microbial community assessment with next-generation sequencing. J. Appl. Microbiol. 2020, 128, 330–354. [Google Scholar] [CrossRef]
- Johnson, J.S.; Spakowicz, D.J.; Hong, B.Y.; Petersen, L.M.; Demkowicz, P.; Chen, L.; Leopold, S.R.; Hanson, B.M.; Agresta, H.O.; Gerstein, M.; et al. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 2019, 10, 5029. [Google Scholar] [CrossRef]
- Muyzer, G.; de Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gel electrophoresis analysis of polymerase chain reaction amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef]
- Herlemann, D.P.; Labrenz, M.; Jürgens, K.; Bertilsson, S.; Waniek, J.J.; Andersson, A.F. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 2011, 5, 1571–1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 30 November 2021).
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bogan, B.W.; Sullivan, W.R.; Kayser, K.J.; Derr, K.D.; Aldrich, H.C.; Paterek, J.R. Alkanindiges illinoisensis gen. nov., sp. nov., an obligately hydrocarbonoclastic, aerobic squalane-degrading bacterium isolated from oilfield soils. Int. J. Syst. Evol. Microbiol. 2003, 53, 1389–1395. [Google Scholar] [CrossRef]
- Zheng, J.; Feng, J.Q.; Zhou, L.; Mbadinga, S.M.; Gu, J.D.; Mu, B.Z. Characterization of bacterial composition and diversity in a long-term petroleum contaminated soil and isolation of high-efficiency alkane-degrading strains using an improved medium. World J. Microbiol. Biotechnol. 2018, 34, 34. [Google Scholar] [CrossRef]
- Vergeynst, L.; Greer, C.W.; Mosbech, A.; Gustavson, K.; Meire, L.; Poulsen, K.G.; Christensen, J.H. Biodegradation, photo-oxidation, and dissolution of petroleum compounds in an arctic fjord during summer. Environ. Sci. Technol. 2019, 53, 12197–12206. [Google Scholar] [CrossRef]
- Liang, J.; Gao, S.; Wu, Z.; Rijnaarts, H.H.M.; Grotenhuis, T. DNA-SIP identification of phenanthrene-degrading bacteria undergoing bioaugmentation and natural attenuation in petroleum-contaminated soil. Chemosphere 2021, 266, 128984. [Google Scholar] [CrossRef]
- Fernández-Brime, S.; Muggia, L.; Maier, S.; Grube, M.; Wedin, M. Bacterial communities in an optional lichen symbiosis are determined by substrate, not algal photobionts. FEMS Microbiol. Ecol. 2019, 95, fiz012. [Google Scholar] [CrossRef] [PubMed]
- Sierra, M.A.; Danko, D.C.; Sandoval, T.A.; Pishchany, G.; Moncada, B.; Kolter, R.; Mason, C.E.; Zambrano, M.M. The Microbiomes of seven lichen genera reveal host specificity, a reduced core community and potential as source of antimicrobials. Front. Microbiol. 2020, 11, 398. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, H.; Yamamoto, Y.; Yoshitani, A.; Itoh, K. Effect of metal stress on photosynthetic pigments in the Cu-hyperaccumulating lichens Cladonia humilis and Stereocaulon japonicum growing in Cu-polluted sites in Japan. Ecotoxicol. Environ. Saf. 2013, 97, 154–159. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, E.; Jiang, C.; Jia, R.; Liu, S.; Xu, D.; Gu, T.; Wang, F. Endogenous phenazine-1-carboxamide encoding gene PhzH regulated the extracellular electron transfer in biocorrosion of stainless steel by marine Pseudomonas aeruginosa. Electrochem. Commun. 2018, 94, 9–13. [Google Scholar] [CrossRef]
- Huang, L.; Huang, Y.; Lou, Y.; Qian, H.; Xu, D.; Ma, L.; Jiang, C.; Zhang, D. Pyocyanin-modifying genes phzM and phzS regulated the extracellular T electron transfer in microbiologically-influenced corrosion of X80 carbon steel by Pseudomonas aeruginosa. Corros. Sci. 2020, 164, 108355. [Google Scholar] [CrossRef]
- Tsurumaru, H.; Ito, N.; Mori, K.; Wakai, S.; Uchiyama, T.; Iino, T.; Hosoyama, A.; Ataku, H.; Nishijima, K.; Mise, M.; et al. An extracellular [NiFe] hydrogenase mediating iron corrosion is encoded in a genetically unstable genomic island in Methanococcus maripaludis. Sci. Rep. 2018, 8, 15149. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Chugh, B.; Sheetal; Singh, M.; Thakur, S.; Pani, B.; Singh, A.K.; Sajil, V.S. Extracellular electron transfer by Pseudomonas aeruginosa in biocorrosion: A review. ACS Biomater. Sci. Eng. 2022, 8, 1049–1059. [Google Scholar] [CrossRef]
- Raut, I.; Calin, M.; Oancea, F.; Arsene, M.L.; Jecu, L. Isolation and identification of microbial strains involved in industrial systems materials corrosion in aquatic environment. Rev. Roum. Chim. 2013, 58, 59–64. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).