Epigenetics in Knee Osteoarthritis: A 2020–2023 Update Systematic Review
Abstract
:1. Introduction
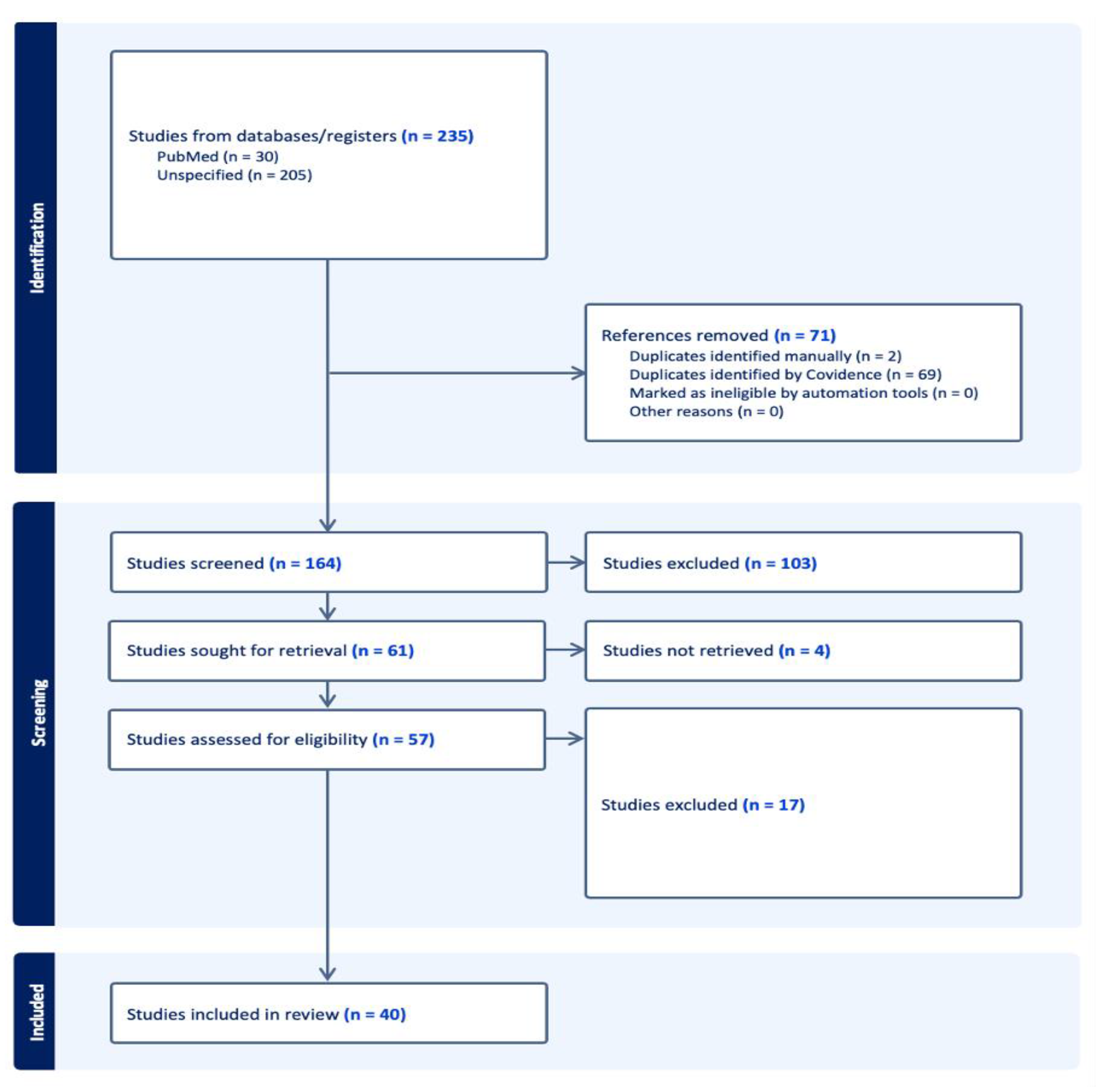
2. Methods
2.1. Review Design
2.2. Search Strategy
2.3. Study Selection
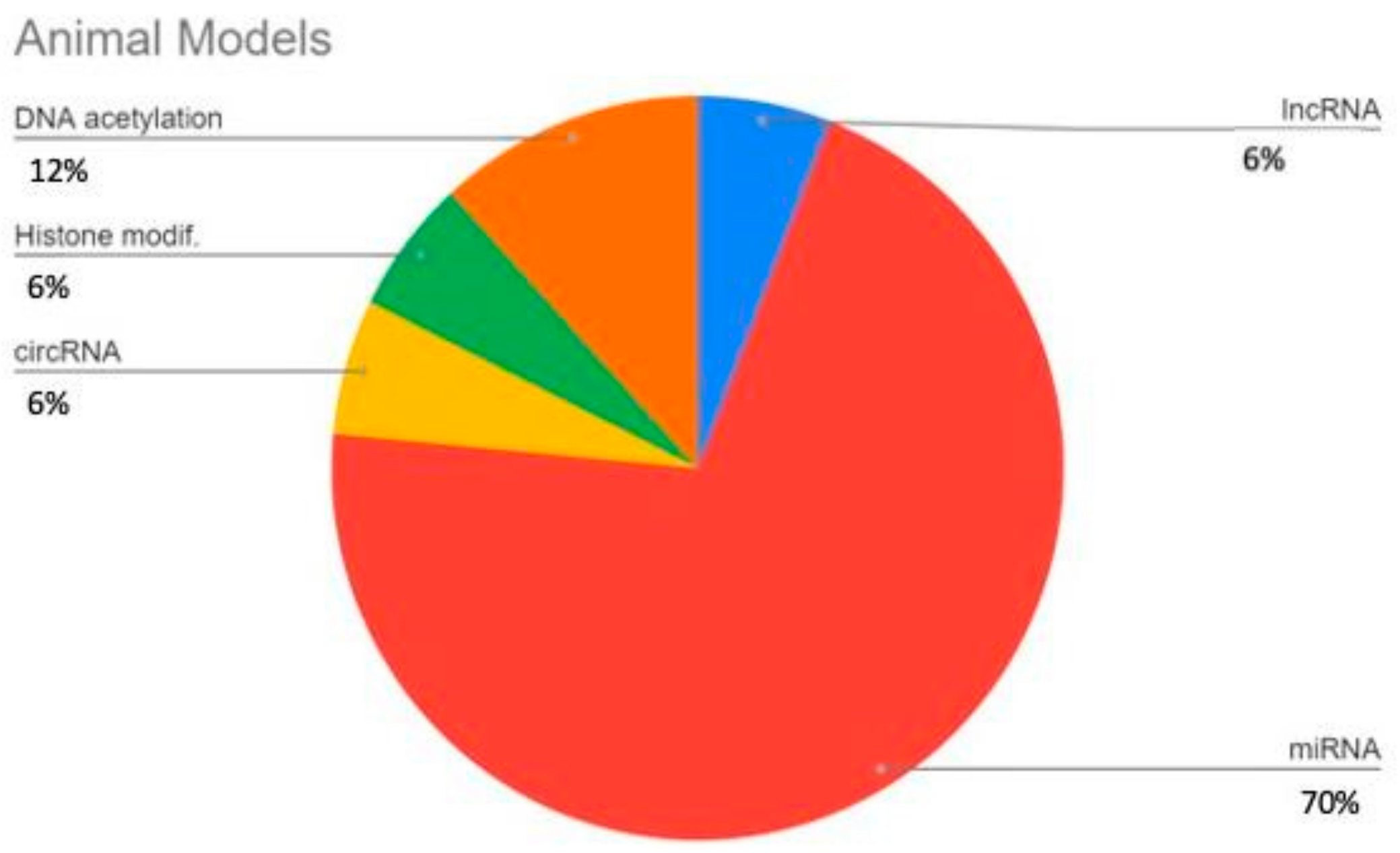
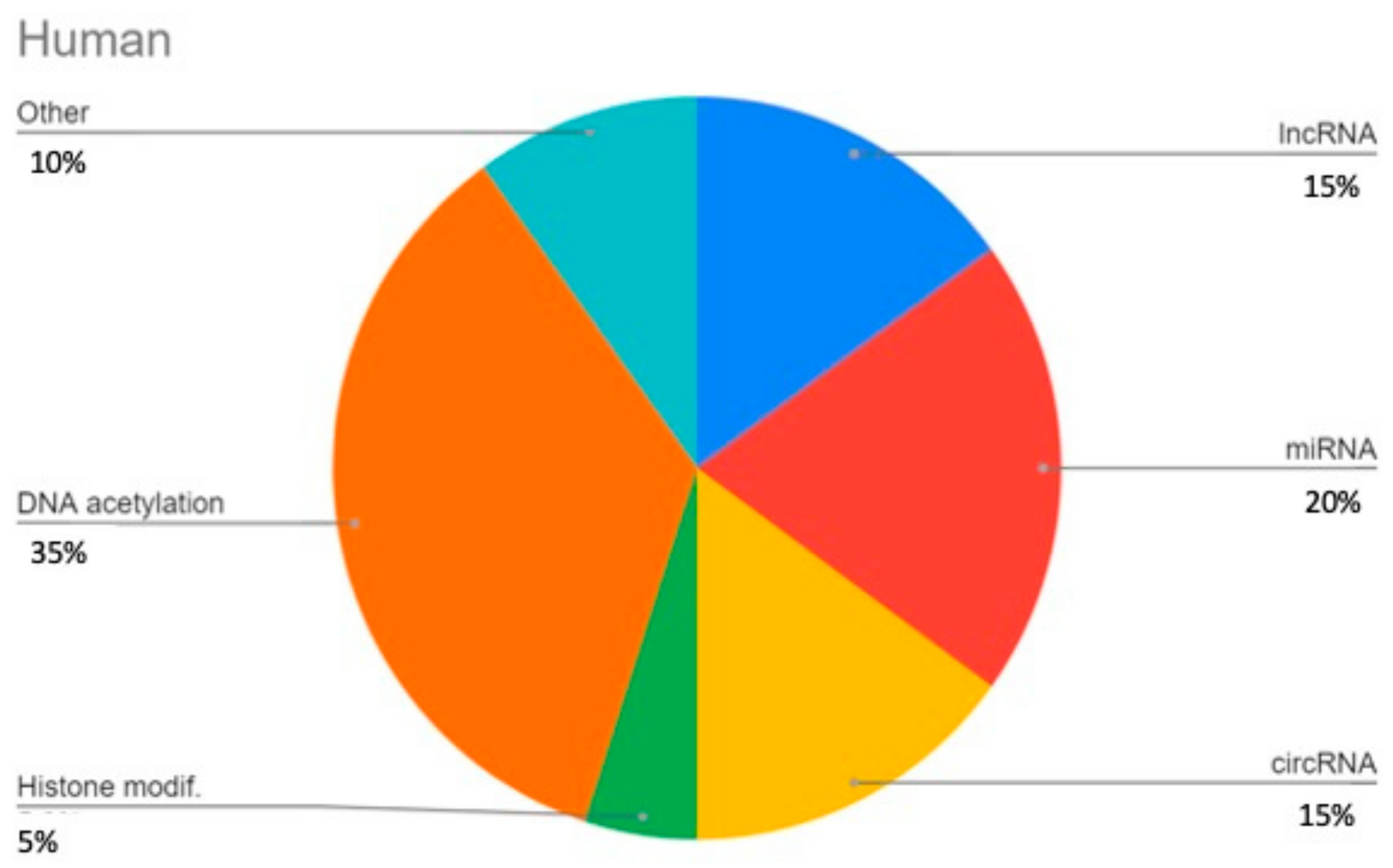
3. Results
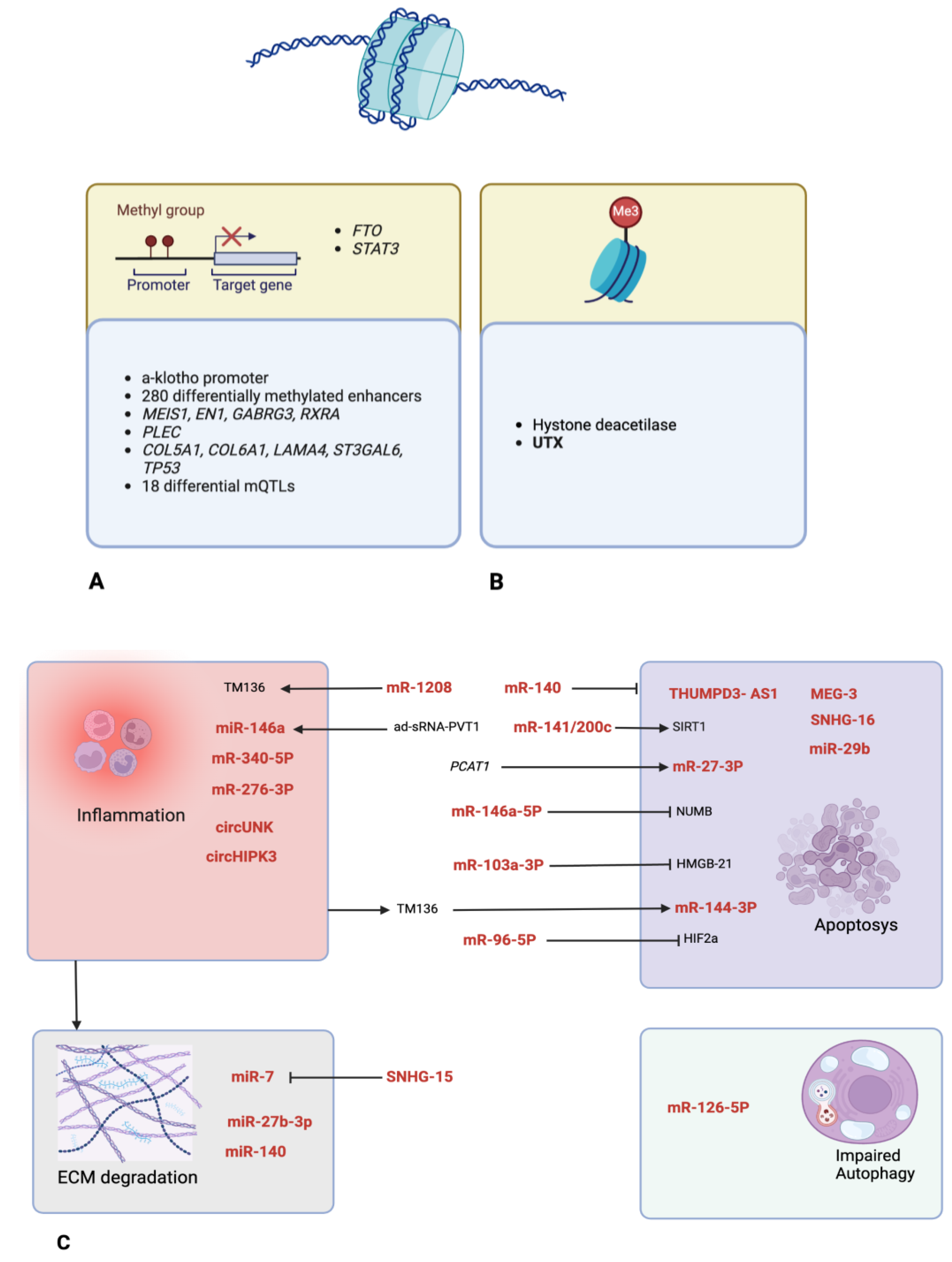
3.1. LncRNAs
3.2. miRNAs
3.3. circRNA
3.4. DNA Methylation
3.5. Histone Modifications
3.6. Others
| Author–Year | Epigenetic Mechanism | Study Design | Main Findings |
|---|---|---|---|
| Wen ZH et al., 2021 [60] | histone modification | Murine animal model of KOA in ACLT mice (4 groups). | The effect of I-A-injected Panobinostat was carried out through MMPs (such as MMP13) activity reduction and less chondral degradation and bone remodeling. This effect was mainly produced via activity modulation of HDAC4 (up-regulated), HDAC6 and HDAC7 (down-regulated) and RUNX2/MMP13 (down-regulated). It also modulated miR-146a, a negative controller of inflammation response in KOA FLSs. |
| Iijima H et al., 2023 [52] | DNA methylation | 4–24-month-old mice (male and female): these ages correspond to ages 20–69 in humans; human articular cartilage tissue from healthy donors. | Increased ECM stiffness drove a-klotho promoter hypermethylation and down-regulated klotho gene expression, accelerating chondrocyte senescence. The inactivation of the a-klotho promoter was attributed to methylation by DNMT1. Exposing the cells to a softer matrix reversed this effect. |
| Cai D et al., 2023 [51] | DNA methylation | KOA cellular model in chondrocytes treated with LPS; KOA in vivo model in rats via MIA I-A injection. | FTO alleviated KKOA (reduces apoptosis, down-regulates IL-6, IL-1β and TNF-α levels and diminishes COX-2 and iNOS expressions) in an m6a-dependent manner via the miR-515-5p/TLR4/MyD88/NF-κB axis. |
| Chen X et al., 2022 [43] | ncRNA | Synovial tissue samples from OA and non-OA pts; animal OA model in DMM mice vs. controls; specific cell culture. | Impaired autophagy occurred in OA FLS, accelerating senescence. Increased m6a modifications were observed, together with up-regulated expression of METTL3. METTL3 plays a key role as autophagy suppressor through m6a modification of ATG7, an autophagy-related mRNA in a YTHDF1-dependent manner. |
| Chen Y et al., 2020 [25] | ncRNA | OA cartilage tissues from 30 patients undergoing TKA vs. 15 normal tissues from amputees; chondrocyte culture; animal murine model (n = 10 OA vs. n = 30 sham) via DMM surgery. | In a KOA murine model, an under-expression of KLF4 and SNHG-15 was detected, with a parallel over-expression of miR-7. SNHG-15 was found to be hypermethylated in OA chondrocytes, leading to its lower effect. SNHG-15 artificial over-expression, on the other hand, inhibited ECM degradation and promoted chondrocyte formation. |
| Cheng M et al., 2020 [34] | ncRNA | Animal model: 12 OA rats (via ani I-A injection) vs. 12 controls; in vitro model: chondrocyte cell culture treated with LPS (4 groups of treatment). | miR-103a-3p is down-regulated in tissues from OA rats and LPS-treated chondrocytes. Up-regulation of miR-103a-3p promotes cell proliferation and reduces inflammation and apoptosis. HMGB1 (OA-related) is a target of miR-103a-3p. Knockdown of HMGB1 mimics the effect of miR-103a-3p. |
| Endisha H et al., 2021 [35] | ncRNA | Joint tissues and plasma from human OA patients and healthy controls; cell culture for human chondrocytes and FLSs; animal OA model in mice via DMM. | Expression of miR-34a-5p was significantly increased in the plasma, cartilage and synovium of patients with late-stage OA and in the cartilage and synovium of mice subjected to DMM. Expression increased in obese patients with OA. miR-34a mimics increased key OA markers. miR-34a KO and injection of the ASO version caused less cartilage degradation in OA development. |
| Fang L et al., 2021 [47] | ncRNA | Cultured chondrocytes OA model; in vivo model in rabbits injected with papain. | Aerobic exercise or OTL treatment—both applied alone—relieved the damage of KOA (cartilage tissue lesions, Mankin score, inflammatory cytokine content, etc.). Combined application showed even better effect than the sum of the two taken alone. Transfection of circUNK attenuated MIA-induced effect on cell viability and apoptosis. |
| Ji M-L et al., 2021 [41] | ncRNA | OA murine model via DMM vs. controls, wt and miR-141/200c KO. | NPs were designed specifically for the purpose of delivering miRNAs and protecting them for a longer period, and their efficacy was positively evaluated. miR-141/200c was then identified and studied as a central actor in KOA development, being up-regulated during OA progression and affecting proliferation, apoptosis and cell growth by targeting SIRT1, which activates IL-6/STAT3 pathway. |
| Jin Y et al., 2021 [40] | ncRNA | Animal OA model in mice via DMM + ACLT; cultured human chondrocytes. | BM-MSCs-derived exosomes alleviated cartilage destruction and subchondral bone remodeling in the animal model. In vitro, assays showed that BM-MSCs exosomes could maintain the chondrocyte phenotype. Exosome lncRNA MEG-3 also reduced the senescence and apoptosis of chondrocytes and might partially account for the anti-OA effects of BM-MSC exosomes. |
| Kim D et al., 2021 [33] | ncRNA | Synovial tissue samples from KOA divided in healthy vs. damaged chondral samples; animal KOA model in DMM mice vs. controls; mice cell culture: KOA cases vs. controls. | Loss of PGC1α in chondrocytes due to up-regulation of miR-126-5p during KOA resulted in activation of PRKN-independent mitophagy pathway. Knockdown of PGC1a activated parkin RBR-E3 ubiquitin protein ligase mitochondria autophagy pathway (up-regulation of BCL2 and BNIP3). miR-126-5p is an upstream regulator for PGC1a. |
| Kouroupis D et al., 2023 [44] | ncRNA | IFP–MSC cultures; animal model of fat pad fibrosis/synovitis via MIA injection. | Synoviocytes exposed to sEVs demonstrated reduced proliferation and altered inflammation-related molecular profiles. CD-10-High sEVs treatment indicated chondroprotective effect by inactivating SubstanceP, a pro-inflammatory and pro-fibrosis IFP marker, via CD10 itself. |
| Tavallaee G et al., 2022 [36] | ncRNA | Tissue samples from KOA patients and controls; animal murine KOA model via DMM surgery; FLSs cell culture. | An increased level of miR-27b-3p was observed in synovial fluid and synovia of patients with late-stage KOA. I-A injections in knees of an miR-27b-3p mimic produced in a synovial fibrosis-like phenotype increased inflammation marker production, plus a pro-fibrotic response for FLS. This same injection did not appear to influence any change in the cartilage tissue instead. The axis used for signaling in the synovia was the PPARG/ADAMTS8 in FLSs. |
| Wang YZ et al., 2021 [24] | ncRNA | Animal KOA mice model with diabetes (ACLT, Streptozocin) of 100 mice: 20 controls, 80 KOA cases (20 non-DM, 40 DM, 20 DM treated with pioglitazone). | PVT1 was found to be up-regulated in KOA chondrocytes. PVT1 levels are even higher and miR-146a levels lower in DM KOA knees than in non-DM KOA knees. PVT1 over-expression promoted the apoptosis of chondrocytes and down-regulated miR-146a effect in KOA DM mice. miR-146a-deficient mice can develop early KOA, as it produced a protective effect against KOA. PVT1 activated TGFb/SMAD signal. KOA was found to be histologically worse, too, in KOA diabetic mice. |
| Yi Y et al., 2022 [37] | ncRNA | Cultured FSCs IL-1b treated to simulate KOA (4 groups); animal KOA model in DMM mice (4 groups). | ONECUT2 and SMURF2 genes were found to be significantly up-regulated in the KOA group, and in the FSCs, stimulated with IL-1b in vitro. ONECUT2 was the binding site of miR-144-3p. This miRNA was therefore down-regulated by Il-1b. miR-144-3p was found to be the target of TM1-3p, that is up-regulated by IL-1b in FSCs. Over-expression of TM1-3p acted as a promotion for cell vitality and proliferation, while inhibiting apoptosis, results reversed with a high level of miR-144-3p. |
| Zhang H et al., 2021 [32] | ncRNA | 22 KOA tissue samples vs. 22 healthy controls; animal KOA model in mice through ACLT (4 groups: sham/ACLT/ACLT + NC (negative control for miR-146)/ACLT + AntagomiR); cell culture. | miR-146a-5p was found to be highly expressed in KOA tissue, while NUMB was down-produced and negatively regulated by miR-146a-5p. miR-146a-5p AntagomiR, injected intra-articularly, had a positive effect on KOA progression. |
| Zhou H et al., 2022 [39] | ncRNA | KOA model in mice by DMM + ACLT surgery; human chondrocytes cultured from tissue of patients undergoing TKA. | hucMSCs-EVs slowed down the progression of KOA, promoting chondrocyte activity and inhibiting apoptosis. HucMSCs-EVs could decrease the m6a level of NLRP3 mRNA in macrophages, via miR-1208 binding to METTL3 and thus inactivating it. NLRP3 inflammasome-activated caspase-1, all pro-inflammatory factors. HucMSCs-EVs lost their protective effect in KO mice for NLRP3 (NLRP3−/−). |
| Zhou K et al., 2021 [38] | ncRNA | Animal model in ACLT + DMM mice; murine chondrocytes isolation. | A strongly increased level of SDC-4 was identified in KOA cells. This is a key component implicated in matrix degradation process. Hypoxia can influence production of SDC-4 mRNA and protein. HIF-2a (Hipoxia-Induced Factor 2a) is produced as well in low-oxygen situations and is involved in cartilage degradation. Both SDC-4 and HIF-2a participate in KOA pathogenesis. miR-96-5p was found to be up-regulated post-SDC-4 inhibition. Its up-regulation inhibited HIF-2a activity too. |
| He K et al., 2021 [42] | ncRNA | Rabbit chondrocytes cells cultured; animal KOA model in mice by papain I-A injection in knees. | miR-140 is strongly expressed in normal cartilage, much less in KOA one. Its inhibition induces KOA-like changes in vivo. When up-regulated, it can moderate knee KOA in vivo in mice. A combination of drug therapy (lornoxicam) and gene therapy (miR-140) was then tested for efficacy in cationic liposomes as therapy carrier. The combination of the two treatments together resulted in more effectiveness than the two previous tested alone. |
| Author–Year | Epigenetic Mechanism | Study Design | Main Finding |
|---|---|---|---|
| Lian WS et al., 2022 [61] | hystone modification | RT-PCR, assessment of OA histopathology, immunohistology, chromatin immunoprecipitation (ChIP) sequencing, immunoblotting on 34 patients with radiographic sign of end-stage KOA. | Role of UTX, in concert with PRC2 core components, in controlling H3K27 tri-methylation and articular chondrocyte anabolism and OA development. |
| Lin X et al., 2020 [53] | DNA methylation | Methylation profiles on a public dataset from patients with hip/knee OA (470.870 CpG probes in 108 samples) vs. hip tissue—healthy controls. | 16,816 differentially methylated CpGs, and nearly half (8111) of them were from enhancers, major DNA methylation changes in both types of OA in the enhancer regions. In KOA, 280 differentially methylated enhancer CpGs were identified. |
| Kreitmaier P et al., 2022 [59] | DNA methylation | Epigenome-wide association study of knee cartilage degeneration on 98 OA individuals. | Robustly replicating methylation markers, which reveal an etiologic mechanism linked to the migration of epithelial cells. Created a genome-wide methylation quantitative trait locus (mQTL) map of articular cartilage and synovium and identified 18 disease-grade-specific mQTLs. |
| Wu Z et al., 2022 [54] | DNA methylation | Analysis of the methylation pattern in knee and hip osteoarthritis on 16 OA hip samples, 19 control hip samples and 62 KOA samples. | 12 methylation sites were identified; genes like MEIS1, GABRG3, RXRA and EN1. |
| Sarkar A et al., 2022 [55] | DNA methylation | EWAS, chromatine signature, STAT3 manipulation, unspecified sample size. | STAT3 regulates epigenetic status of cartilage cells, modulating DNA methilation in context-dependent manner. |
| Sorial AK et al., 2020 [56] | DNA methylation | Cartilage, fat pad, synovium and blood samples from TKA patients. A total of 240 samples from 202 patients. | Methylation correlates with PLEC expression. An allele of rs11780978 correlates with reduced PLEC expression and methyl at CpGs in cg19405177 and cg14598846. The rs11780978-PLEC eQTLs, mQTLs and meQTLs that are active in cartilage are also active in synovium (but with differences). PLEC mQTLs across all four tissues. Knocked-down plectin: increased expression of genes for immune response, decreased Wnt signaling (cartilage homeostasis). |
| Zhang Q et al., 2021 [57] | DNA methylation | PCR, Western blot and ELISA determination of epigenetic regulation determining TNF-a expression and its correlation to OA in 50 patients (37 OA + 13 controls). | Significant DNA hypo-methylation was observed in OA cartilage, DNA methylation of the TNF-a promoter suppresses mRNA and protein levels of TNF-a in OA, anacardic acid can inhibit the binding of HAT1 and CBP on the TNF-a promoter and reduce TNF-a secretion. |
| Zhou L et al., 2021 [28] | lncRNA | Analysis of the roles and associations of PCAT-1 and its target miR-27-3p in the pathogenesis OA on 30 articular cartilage samples (15 KOA + 15 healthy). | lncRNA PCAT-1-3 has a pro-apoptotic effect in KOA chondrocytes via regulation through sponging miR-27b-3p in KOA. |
| Fan H et al., 2021 [27] | lncRNA | Analysis on 40 samples (20 KOA + 20 healthy) of the expression of SNHG16 in OA and normal tissues. CHON-001 cells treated with interleukin (IL)-1β serve as an in vitro model of human OA. | lncRNA SNHG16 is up-regulated in KOA. lncRNA SNHG16 promotes the occurrence of KOA by sponging miR-373-3p and its target gene p21. |
| Van Hoolwerff M et al., 2020 [26] | lncRNA | Analysis of differential expression of lncRNAs in macroscopically lesioned and preserved articular cartilage on 98 samples (65 knees, 33 hips). | Antisense lncRNAs play an important role in regulating the pathophysiology of KOA. Antisense lncRNAs can exert their function in cis. |
| Li X et al., 2021 [30] | lncRNA | Analysis of existing bulk RNA sequencing (bulk RNA-seq) and single-cell sequencing (scRNA-seq) data for chondrocytes in OA knees, for identification of key transcription factors and lncRNAs on unspecified sample size. | 271 key genes are suggested as involved in the pattern of progression of OA. For these genes, 14 transcription factors, among which TWIST2, MYBL2, RELA, JUN, KLF4 and PTTG1 are mentioned as the key TFs. A total of 8 lncRNAs among the 271 genes and the lncRNA regulation between CYTOR and NRP1 are shown to contribute to the pain and vascularization of cartilage in KOA. |
| Wang Y et al., 2021 [29] | lncRNA | Analysis of the role of lncRNA THUMPD3-AS1 in OA biology on 10 KOA. | lncRNA THUMPD3-AS1 is down-regulated in KOA cartilage tissues and IL-1b-stimulated chondrocytes. Over-expression of lncRNA THUMPD3-AS1 alleviates cell apoptosis and facilitates inflammatory responses. THUMPD3-AS1 increases the levels of inflammatory markers. NF-κB p65 and MAPk p38 are identified target proteins of phosphorylation. |
| Wang G et al., 2022 [31] | lncRNA, miRNA | Analysis of differential expression of lncRNAs and miRNAs in normal chondrocytes and in 10 SDF-1-induced models of chondrocyte degeneration. | 186 lncRNAs have significant expression (88 up-regulated, 98 down-regulated). A total of 684 miRNAs have significant expression. Indication of 10 top core genes: CXCL10, ISG15, MYC, MX1, KOASL, IFIT1, RSAD2, MX2, IFI44L and BST2 associated with chondrocyte degeneration, by inducing expression of N-acetyl-b-d-glucosidase (NAG) and MMP. |
| Tavallaee G et al., 2022 [36] | miRNA | Human: synovial tissue samples from OA patients. Mouse: (1) knee joints collected DMM or sham surgery. For the injection experiments in mice, (2a) miRNA 27b-3p mimic within knees; knees retrieved 5 weeks after; (2b) miRNA 27b-3p inhibitor 1 and 3 weeks after DMM/sham surgery; knees retrieved 5 weeks after. Human: unspecified; mice: (1) 6–10 knees for each group (DMM and sham); (2a) 6 knees per group; and (2b) 10 cases vs. 9 controls. | Association between miRNA 27b-3p expression and OA severity. Transfection with the miR-27b-3p mimic induces pro-fibrotic responses, migration and expression of key ECM genes. RNA sequencing identification of a PPARG/ADAMTS8 signaling axis regulated by miR-27b-3p in OA FLS. |
| Liu Y et al., 2022 [45] | miRNA | Administration of exosomes enriched with miR-140. | Superiority of hUSC exosomes over-expressing miR-140-5p for treating OA. |
| Dou P et al., 2021 [46] | miRNA | Analysis of the relationship between DNMT3B and miR-29b and their implications in humans, 46 OA + 46 normal, and 48 DMM mice. | DNMT3B exerts an anti-apoptotic function by reducing RUNX2. DNMT3B inhibits the expression of miR-29b via DNA methylation. The inhibition of miR-29b increases PTHLH expression. PTHLH impedes the apoptosis of chondrocytes by elevating CDK4. Up-regulation of CDK4 by DNMT3B induces the ubiquitination of RUNX2 protein. |
| Liu P et al., 2022 [48] | circRNA | Real-time PCR identification of circular RNA (circRNA) expression profile in 5 OA and 5 controls. | hsa_circ_0072697 role in the pathogenesis of OA. |
| Wang Y et al., 2020 [49] | circRNA | qRT-PCR detection of circRNA in peripheral blood of OA patients (the Western Ontario and McMaster Index of stiffness and pain). | 1627 differentially expressed circRNAs. hsa_circRNA_0020014 results to be the most significantly differentially expressed and may be identified as a new potential biomarker. |
| Wu Q et al., 2020 [50] | circRNA | RT-qPCR detection of circRNA HIPK3 on OA and its regulatory mechanism in human OA tissue (American College of Rheumatology) on 36 OA patients. | CircHIPK3 is highly expressed, and miR-124 is down-regulated in OA tissues, as they are negatively correlated. CircHIPK3 suppresses apoptosis of OA chondrocytes by the miR-124/SOX8 pathway. |
| Duran-Sotuela A et al., 2023 [62] | mtRNA | Identification of mitochondrial DNA genetic variants associated with the risk of rapid progression of KOA. Characterization of their functional significance with a cell model. OAI cohort (1095), hip (373) and knee (326). | mtDNA variant m.16519C is over-represented in rapid progressors. Cybrids with this variant show increased mtDNA copy number and decreased mitochondrial biosynthesis. Higher amount of ROS and less resistance to oxidative stress. Impairment of autophagic flux. Modulates the transcriptome of cybrids. |
| Park H et al., 2022 [63] | siRNA | Effects of p16INK4a si-RNA NPs (PLGA); unspecified sample size. | Expression of p16INK4a is increased in the synovium and articular cartilage from OA patients. “p16 si_NPs” reduced the levels of TNF-α, IL-1β and IL-6 in FLSs and MMP-13 in chondrocytes. p16 si_NP injection in the model alleviated pain-associated behavior and reduced cartilage damage. |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Allen, K.D.; Choong, P.F.; Davis, A.M.; Dowsey, M.M.; Dziedzic, K.S.; Emery, C.; Hunter, D.J.; Losina, E.; Page, A.E.; Roos, E.M.; et al. Osteoarthritis: Models for appropriate care across the disease continuum. Best Pr. Res. Clin. Rheumatol. 2016, 30, 503–535. [Google Scholar] [CrossRef]
- Dieppe, P.A.; Lohmander, L.S. Pathogenesis and management of pain in osteoarthritis. Lancet 2005, 365, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Yau, M.S.; Yerges-Armstrong, L.M.; Liu, Y.; Lewis, C.E.; Duggan, D.J.; Renner, J.B.; Torner, J.; Felson, D.T.; McCulloch, C.E.; Kwoh, C.K.; et al. Genome-Wide Association Study of Radiographic Knee Osteoarthritis in North American Caucasians. Arthritis Rheumatol. 2016, 69, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liang, Y.; Li, H.; Li, H.; He, Q.; Xue, Y.; Shen, C.; Zhang, C.; Xiang, J.; Ding, J.; et al. ingle Nucleotide Polymorphisms and Osteoarthritis: An Overview and a Meta-Analysis. Medicine 2016, 95, e2811. [Google Scholar] [CrossRef]
- Valdes, A.M.; Spector, T.D. Genetic epidemiology of hip and knee osteoarthritis. Nat. Rev. Rheumatol. 2010, 7, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Styrkarsdottir, U.; Lund, S.H.; Thorleifsson, G.; Zink, F.; Stefansson, O.A.; Sigurdsson, J.K.; Juliusson, K.; Bjarnadottir, K.; Sigurbjornsdottir, S.; Jonsson, S.; et al. Meta-analysis of Icelandic and UK data sets identifies missense variants in SMO, IL11, COL11A1 and 13 more new loci associated with osteoarthritis. Nat. Genet. 2018, 50, 1681–1687. [Google Scholar] [CrossRef]
- Zengini, E.; Hatzikotoulas, K.; Tachmazidou, I.; Steinberg, J.; Hartwig, F.P.; Southam, L.; Hackinger, S.; Boer, C.G.; Styrkarsdottir, U.; Gilly, A.; et al. Genome-wide analyses using UK Biobank data provide insights into the genetic architecture of osteoarthritis. Nat. Genet. 2018, 50, 549–558. [Google Scholar] [CrossRef]
- Styrkarsdottir, U.; Stefansson, O.A.; Gunnarsdottir, K.; Thorleifsson, G.; Lund, S.H.; Stefansdottir, L.; Juliusson, K.; Agustsdottir, A.B.; Zink, F.; Halldorsson, G.H.; et al. GWAS of bone size yields twelve loci that also affect height, BMD, osteoarthritis or fractures. Nat. Commun. 2019, 10, 2054. [Google Scholar] [CrossRef]
- Tachmazidou, I.; Hatzikotoulas, K.; Southam, L.; Esparza-Gordillo, J.; Haberland, V.; Zheng, J.; Johnson, T.; Koprulu, M.; Zengini, E.; Steinberg, J.; et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of UK Biobank data. Nat. Genet. 2019, 51, 230–236. [Google Scholar] [CrossRef]
- Safiri, S.; Kolahi, A.-A.; Smith, E.; Hill, C.; Bettampadi, D.; Mansournia, M.A.; Hoy, D.; Ashrafi-Asgarabad, A.; Sepidarkish, M.; Almasi-Hashiani, A.; et al. Global, regional and national burden of osteoarthritis 1990-2017: A systematic analysis of the Global Burden of Disease Study 2017. Ann. Rheum. Dis. 2020, 79, 819–828. [Google Scholar] [CrossRef]
- Cai, Z.; Long, T.; Zhao, Y.; Lin, R.; Wang, Y. Epigenetic Regulation in Knee Osteoarthritis. Front. Genet. 2022, 13, 942982. [Google Scholar] [CrossRef]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef]
- Ramos, Y.F.; Meulenbelt, I. The role of epigenetics in osteoarthritis: Current perspective. Curr. Opin. Rheumatol. 2017, 29, 119–129. [Google Scholar] [CrossRef]
- Simon, T.C.; Jeffries, M.A. The Epigenomic Landscape in Osteoarthritis. Curr. Rheumatol. Rep. 2017, 19, 30. [Google Scholar] [CrossRef]
- De Almeida, R.C.; Ramos, Y.F.; Meulenbelt, I. Involvement of epigenetics in osteoarthritis. Best Pr. Res. Clin. Rheumatol. 2017, 31, 634–648. [Google Scholar] [CrossRef]
- Messier, S.P.; Beavers, D.P.; Queen, K.; Mihalko, S.L.; Miller, G.D.; Losina, E.; Katz, J.N.; Loeser, R.F.; DeVita, P.; Hunter, D.J.; et al. Effect of Diet and Exercise on Knee Pain in Patients With Osteoarthritis and Overweight or Obesity: A Randomized Clinical Trial. JAMA 2022, 328, 2242–2251. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Ji, L.; He, Z.; Zhang, W.; Tong, Y.; Luo, J.; Hong, Z.; Zhang, Y.; Yu, D.; Zhang, Q.; et al. Association of smoking and osteoarthritis in US (NHANES 1999–2018). Sci. Rep. 2023, 13, 3911. [Google Scholar] [CrossRef] [PubMed]
- Krakowski, P.; Karpiński, R.; Jojczuk, M.; Nogalska, A.; Jonak, J. Knee MRI Underestimates the Grade of Cartilage Lesions. Appl. Sci. 2021, 11, 1552. [Google Scholar] [CrossRef]
- Figueroa, D.; Calvo, R.; Vaisman, A.; Carrasco, M.A.; Moraga, C.; Delgado, I. Knee Chondral Lesions: Incidence and Correlation Between Arthroscopic and Magnetic Resonance Findings. Arthrosc. J. Arthrosc. Relat. Surg. Off. Publ. Arthrosc. Assoc. N. Am. Int. Arthrosc. Assoc. 2007, 23, 312–315. [Google Scholar] [CrossRef] [PubMed]
- Bredella, M.A.; Tirman, P.F.; Peterfy, C.G.; Zarlingo, M.; Feller, J.F.; Bost, F.W.; Belzer, J.P.; Wischer, T.K.; Genant, H.K.; Bredella, P.F.T.M.A.; et al. Accuracy of T2-weighted fast spin-echo MR imaging with fat saturation in detecting cartilage defects in the knee: Comparison with arthroscopy in 130 patients. Am. J. Roentgenol. 1999, 172, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Karpiński, R.; Krakowski, P.; Jonak, J.; Machrowska, A.; Maciejewski, M.; Nogalski, A. Diagnostics of Articular Cartilage Damage Based on Generated Acoustic Signals Using ANN—Part II: Patellofemoral Joint. Sensors 2022, 22, 3765. [Google Scholar] [CrossRef] [PubMed]
- Karpiński, R. Knee Joint Osteoarthritis Diagnosis Based On Selected Acoustic Signal Discriminants Using Machine Learning. Appl. Comput. Sci. 2022, 18, 71–85. [Google Scholar] [CrossRef]
- Izda, V.; Martin, J.; Sturdy, C.; Jeffries, M.A. DNA methylation and noncoding RNA in OA: Recent findings and methodological advances. Osteoarthr. Cartil. Open 2021, 3, 100208. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Z.; Li, Y.; Liang, S.-K.; Ding, L.-B.; Li, F.; Guan, J.; Wang, H.-J. LncPVT1 promotes cartilage degradation in diabetic OA mice by downregulating miR-146a and activating TGF-β/SMAD4 signaling. J. Bone Miner. Metab. 2021, 39, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guo, H.; Li, L.; Bao, D.; Gao, F.; Li, Q.; Huang, Q.; Duan, X.; Xiang, Z. Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 15 (SNHG15) Alleviates Osteoarthritis Progression by Regulation of Extracellular Matrix Homeostasis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e923868. [Google Scholar] [CrossRef]
- Van Hoolwerff, M.; Metselaar, P.I.; Tuerlings, M.; Suchiman, H.E.D.; Lakenberg, N.; Ramos, Y.F.; Cats, D.; Nelissen, R.G.; Broekhuis, D.; Mei, H.; et al. Elucidating Epigenetic Regulation by Identifying Functional cis-Acting Long Noncoding RNAs and Their Targets in Osteoarthritic Articular Cartilage. Arthritis Rheumatol. 2020, 72, 1845–1854. [Google Scholar] [CrossRef]
- Fan, H.; Ding, L.; Yang, Y. lncRNA SNHG16 promotes the occurrence of osteoarthritis by sponging miR-373-3p. Mol. Med. Rep. 2020, 23, 117. [Google Scholar] [CrossRef]
- Zhou, L.; Gu, M.; Ma, X.; Wen, L.; Zhang, B.; Lin, Y.; Pan, J. Long non-coding RNA PCAT-1 regulates apoptosis of chondrocytes in osteoarthritis by sponging miR-27b-3p. J. Bone Miner. Metab. 2021, 39, 139–147. [Google Scholar] [CrossRef]
- Wang, Y.; Li, T.; Yang, Q.; Feng, B.; Xiang, Y.; Lv, Z.; Weng, X. LncRNA THUMPD3-AS1 enhances the proliferation and inflammatory response of chondrocytes in osteoarthritis. Int. Immunopharmacol. 2021, 100, 108138. [Google Scholar] [CrossRef]
- Li, X.; Liao, Z.; Deng, Z.; Chen, N.; Zhao, L. Combining bulk and single-cell RNA-sequencing data to reveal gene expression pattern of chondrocytes in the osteoarthritic knee. Bioengineered 2021, 12, 997–1007. [Google Scholar] [CrossRef]
- Wang, G.; He, L.; Xiang, Y.; Jia, D.; Li, Y. Long noncoding and micro-RNA expression in a model of articular chondrocyte degeneration induced by stromal cell-derived factor-1. Asian Biomed. Res. Rev. News 2022, 16, 169–179. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, W.; Li, D.; Zheng, J. miR-146a-5p Promotes Chondrocyte Apoptosis and Inhibits Autophagy of Osteoarthritis by Targeting NUMB. Cartilage 2021, 13, 1467S–1477S. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Song, J.; Jin, E.-J. BNIP3-Dependent Mitophagy via PGC1α Promotes Cartilage Degradation. Cells 2021, 10, 1839. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Wang, Y. Downregulation of HMGB1 by miR-103a-3p Promotes Cell Proliferation, Alleviates Apoptosis and in Flammation in a Cell Model of Osteoarthritis. Iran J. Biotechnol. 2020, 18, 24–31. [Google Scholar] [CrossRef]
- Endisha, H.; Datta, P.; Sharma, A.; Nakamura, S.; Rossomacha, E.; Younan, C.; Ali, S.A.; Tavallaee, G.; Lively, S.; Potla, P.; et al. MicroRNA-34a-5p Promotes Joint Destruction During Osteoarthritis. Arthritis Rheumatol. 2020, 73, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Tavallaee, G.; Lively, S.; Rockel, J.S.; Ali, S.A.; Im, M.; Sarda, C.; Mitchell, G.M.; Rossomacha, E.; Nakamura, S.; Potla, P.; et al. Contribution of MicroRNA-27b-3p to Synovial Fibrotic Responses in Knee Osteoarthritis. Arthritis Rheumatol. 2022, 74, 1928–1942. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Yang, N.; Yang, Z.; Tao, X.; Li, Y. LncRNA TM1-3P Regulates Proliferation, Apoptosis and Inflammation of Fibroblasts in Osteoarthritis through miR-144-3p/ONECUT2 Axis. Orthop. Surg. 2022, 14, 3078–3091. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; He, S.; Yu, H.; Pei, F.; Zhou, Z. Inhibition of syndecan-4 reduces cartilage degradation in murine models of osteoarthritis through the downregulation of HIF-2α by miR-96-5p. Lab. Investig. J. Tech. Methods Pathol. 2021, 101, 1060–1070. [Google Scholar] [CrossRef]
- Zhou, H.; Shen, X.; Yan, C.; Xiong, W.; Ma, Z.; Tan, Z.; Wang, J.; Li, Y.; Liu, J.; Duan, A.; et al. Extracellular vesicles derived from human umbilical cord mesenchymal stem cells alleviate osteoarthritis of the knee in mice model by interacting with METTL3 to reduce m6A of NLRP3 in macrophage. Stem Cell Res. Ther. 2022, 13, 322. [Google Scholar] [CrossRef]
- Jin, Y.; Xu, M.; Zhu, H.; Dong, C.; Ji, J.; Liu, Y.; Deng, A.; Gu, Z. Therapeutic effects of bone marrow mesenchymal stem cells-derived exosomes on osteoarthritis. J. Cell. Mol. Med. 2021, 25, 9281–9294. [Google Scholar] [CrossRef]
- Ji, M.-L.; Jiang, H.; Wu, F.; Geng, R.; Ya, L.K.; Lin, Y.C.; Xu, J.H.; Wu, X.T.; Lu, J. Precise targeting of miR-141/200c cluster in chondrocytes attenuates osteoarthritis development. Ann. Rheum. Dis. 2020, 80, 356–366. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Huang, X.; Shan, R.; Yang, X.; Song, R.; Xie, F.; Huang, G. Intra-articular Injection of Lornoxicam and MicroRNA-140 Co-loaded Cationic Liposomes Enhanced the Therapeutic Treatment of Experimental Osteoarthritis. AAPS Pharmscitech 2021, 23, 9. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gong, W.; Shao, X.; Shi, T.; Zhang, L.; Dong, J.; Shi, Y.; Shen, S.; Qin, J.; Jiang, Q.; et al. METTL3-mediated m6A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann. Rheum. Dis. 2021, 81, 85–97. [Google Scholar] [CrossRef]
- Kouroupis, D.; Kaplan, L.D.; Huard, J.; Best, T.M. CD10-Bound Human Mesenchymal Stem/Stromal Cell-Derived Small Extracellular Vesicles Possess Immunomodulatory Cargo and Maintain Cartilage Homeostasis under Inflammatory Conditions. Cells 2023, 12, 1824. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zeng, Y.; Si, H.-B.; Tang, L.; Xie, H.-Q.; Shen, B. Exosomes Derived From Human Urine–Derived Stem Cells Overexpressing miR-140-5p Alleviate Knee Osteoarthritis Through Downregulation of VEGFA in a Rat Model. Am. J. Sports Med. 2022, 50, 1088–1105. [Google Scholar] [CrossRef]
- Dou, P.; He, Y.; Yu, B.; Duan, J. Downregulation of microRNA-29b by DNMT3B decelerates chondrocyte apoptosis and the progression of osteoarthritis via PTHLH/CDK4/RUNX2 axis. Aging 2020, 13, 7676–7690. [Google Scholar] [CrossRef]
- Fang, L.; Lin, L.; Lv, Y.; Huang, Z.; Lin, X.; Wang, X.; Chen, B. The mechanism of aerobic exercise combined with glucosamine therapy and circUNK in improving knee osteoarthritis in rabbits. Life Sci. 2021, 275, 119375. [Google Scholar] [CrossRef]
- Liu, P.; Gao, G.; Zhou, X.; Zhang, X.; Cai, Q.; Xiang, Z.; Shen, X.; Wu, X. Circular RNA profiles of osteoarthritic synovium. Mol. Omics 2022, 18, 439–448. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, C.; Zhang, Y.; Yang, Y.; Ren, Z.; Lammi, M.J.; Guo, X. Screening for differentially expressed circRNA between Kashin–Beck disease and osteoarthritis patients based on circRNA chips. Clin. Chim. Acta Int. J. Clin. Chem. 2019, 501, 92–101. [Google Scholar] [CrossRef]
- Wu, Q.; Yuan, Z.-H.; Ma, X.-B.; Tang, X.-H. Low expression of CircRNA HIPK3 promotes osteoarthritis chondrocyte apoptosis by serving as a sponge of miR-124 to regulate SOX8. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7937–7945. [Google Scholar] [CrossRef]
- Cai, D.; Zhang, J.; Yang, J.; Lv, Q.; Zhong, C. Overexpression of FTO alleviates osteoarthritis by regulating the processing of miR-515-5p and the TLR4/MyD88/NF-κB axis. Int. Immunopharmacol. 2023, 114, 109524. [Google Scholar] [CrossRef]
- Iijima, H.; Gilmer, G.; Wang, K.; Bean, A.C.; He, Y.; Lin, H.; Tang, W.-Y.; Lamont, D.; Tai, C.; Ito, A.; et al. Age-related matrix stiffening epigenetically regulates α-Klotho expression and compromises chondrocyte integrity. Nat. Commun. 2023, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, L.; Liu, X.; Tian, J.; Zheng, W.; Li, J.; Wang, L. Genome-wide analysis of aberrant methylation of enhancer DNA in human osteoarthritis. BMC Med. Genom. 2020, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Shou, L.; Wang, J.; Huang, T.; Xu, X. The Methylation Pattern for Knee and Hip Osteoarthritis. Front. Cell Dev. Biol. 2020, 8, 602024. [Google Scholar] [CrossRef]
- Sarkar, A.; Liu, N.Q.; Magallanes, J.; Tassey, J.; Lee, S.; Shkhyan, R.; Lee, Y.; Lu, J.; Ouyang, Y.; Tang, H.; et al. STAT3 promotes a youthful epigenetic state in articular chondrocytes. Aging Cell 2023, 22, e13773. [Google Scholar] [CrossRef]
- Sorial, A.; Hofer, I.; Tselepi, M.; Cheung, K.; Parker, E.; Deehan, D.; Rice, S.; Loughlin, J. Multi-tissue epigenetic analysis of the osteoarthritis susceptibility locus mapping to the plectin gene PLEC. Osteoarthr. Cartil. 2020, 28, 1448–1458. [Google Scholar] [CrossRef]
- Zhang, Q.; Ouyang, Z.; Song, X.; Zhu, W.; Tang, X.; Liu, Z.; Chen, X. Epigenetic modifications of tumor necrosis factor-alpha in joint cartilage tissue from osteoarthritis patients—CONSORT. Medicine 2021, 100, e27868. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Ma, J.; Chen, Z.; Wang, F.; Li, Z.; Shang, Z.; Dong, J. Osteoarthritis related epigenetic variations in miRNA expression and DNA methylation. BMC Med. Genom. 2023, 16, 163. [Google Scholar] [CrossRef]
- Kreitmaier, P.; Suderman, M.; Southam, L.; De Almeida, R.C.; Hatzikotoulas, K.; Meulenbelt, I.; Steinberg, J.; Relton, C.L.; Wilkinson, J.M.; Zeggini, E. An epigenome-wide view of osteoarthritis in primary tissues. Am. J. Hum. Genet. 2022, 109, 1255–1271. [Google Scholar] [CrossRef]
- Wen, Z.-H.; Huang, J.-S.; Lin, Y.-Y.; Yao, Z.-K.; Lai, Y.-C.; Chen, W.-F.; Liu, H.-T.; Lin, S.-C.; Tsai, Y.-C.; Tsai, T.-C.; et al. Chondroprotective Effects of a Histone Deacetylase Inhibitor, Panobinostat, on Pain Behavior and Cartilage Degradation in Anterior Cruciate Ligament Transection-Induced Experimental Osteoarthritic Rats. Int. J. Mol. Sci. 2021, 22, 7290. [Google Scholar] [CrossRef]
- Lian, W.-S.; Wu, R.-W.; Ko, J.-Y.; Chen, Y.-S.; Wang, S.-Y.; Yu, C.-P.; Jahr, H.; Wang, F.-S. Histone H3K27 demethylase UTX compromises articular chondrocyte anabolism and aggravates osteoarthritic degeneration. Cell Death Dis. 2022, 13, 538. [Google Scholar] [CrossRef]
- Durán-Sotuela, A.; Fernandez-Moreno, M.; Suárez-Ulloa, V.; Vázquez-García, J.; Relaño, S.; Hermida-Gómez, T.; Balboa-Barreiro, V.; Lourido-Salas, L.; Calamia, V.; Fernandez-Puente, P.; et al. A meta-analysis and a functional study support the influence of mtDNA variant m.16519C on the risk of rapid progression of knee osteoarthritis. Ann. Rheum. Dis. 2023, 82, 974–984. [Google Scholar] [CrossRef]
- Park, H.; Lee, H.-R.; Shin, H.J.; Park, J.A.; Joo, Y.; Kim, S.M.; Beom, J.; Kang, S.W.; Kim, D.W.; Kim, J. p16INK4a-siRNA nanoparticles attenuate cartilage degeneration in osteoarthritis by inhibiting inflammation in fibroblast-like synoviocytes. Biomater. Sci. 2022, 10, 3223–3235. [Google Scholar] [CrossRef]
- Khavari, B.; Cairns, M.J. Epigenomic Dysregulation in Schizophrenia: In Search of Disease Etiology and Biomarkers. Cells 2020, 9, 1837. [Google Scholar] [CrossRef] [PubMed]
- Quilichini, E.; Haumaitre, C. Implication of epigenetics in pancreas development and disease. Best Pr. Res. Clin. Endocrinol. Metab. 2015, 29, 883–898. [Google Scholar] [CrossRef] [PubMed]
- Nacev, B.A.; Jones, K.B.; Intlekofer, A.M.; Yu, J.S.E.; Allis, C.D.; Tap, W.D.; Ladanyi, M.; Nielsen, T.O. The epigenomics of sarcoma. Nat. Rev. Cancer 2020, 20, 608–623. [Google Scholar] [CrossRef] [PubMed]
- D’agnelli, S.; Arendt-Nielsen, L.; Gerra, M.C.; Zatorri, K.; Boggiani, L.; Baciarello, M.; Bignami, E. Fibromyalgia: Genetics and epigenetics insights may provide the basis for the development of diagnostic biomarkers. Mol. Pain 2018, 15, 1744806918819944. [Google Scholar] [CrossRef] [PubMed]
- Berson, A.; Nativio, R.; Berger, S.L.; Bonini, N.M. Epigenetic Regulation in Neurodegenerative Diseases. Trends Neurosci. 2018, 41, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, M. Epigenetic alterations and advancement of treatment in peripheral T-cell lymphoma. Clin. Epigenetics 2020, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, Z.; Mortaz, E.; Adcock, I.; Moin, M. Role of Epigenetics in the Pathogenesis of Asthma. Iran. J. Allergy Asthma Immunol. 2017, 16, 82–91. [Google Scholar] [PubMed]
- Nemoda, Z.; Massart, R.; Suderman, M.; Hallett, M.; Li, T.; Coote, M.; Cody, N.; Sun, Z.S.; Soares, C.N.; Turecki, G.; et al. Maternal depression is associated with DNA methylation changes in cord blood T lymphocytes and adult hippocampi. Transl. Psychiatry 2015, 5, e545. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, K.; Karssiens, T.; Kumar, V.; Pandit, H. Obesity and osteoarthritis. Maturitas 2016, 89, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Carro, C.; Blanco-Blanco, M.; Villagrán-Andrade, K.M.; Blanco, F.J.; De Andrés, M.C. Epigenetics as a Therapeutic Target in Osteoarthritis. Pharmaceuticals 2023, 16, 156. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caldo, D.; Massarini, E.; Rucci, M.; Deaglio, S.; Ferracini, R. Epigenetics in Knee Osteoarthritis: A 2020–2023 Update Systematic Review. Life 2024, 14, 269. https://doi.org/10.3390/life14020269
Caldo D, Massarini E, Rucci M, Deaglio S, Ferracini R. Epigenetics in Knee Osteoarthritis: A 2020–2023 Update Systematic Review. Life. 2024; 14(2):269. https://doi.org/10.3390/life14020269
Chicago/Turabian StyleCaldo, Davide, Eugenia Massarini, Massimiliano Rucci, Silvia Deaglio, and Riccardo Ferracini. 2024. "Epigenetics in Knee Osteoarthritis: A 2020–2023 Update Systematic Review" Life 14, no. 2: 269. https://doi.org/10.3390/life14020269





