Comparative Analysis of Oral Prevotella intermedia, Tannerella forsythia, Streptococcus sanguinis, and Streptococcus mutans in Patients with Esophageal Squamous Cell Carcinoma and Healthy Controls in Mthatha, South Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Inclusion and Exclusion Criteria
2.3. Data Collection
2.4. Sample Collection
2.5. DNA Extraction and Quantification
2.6. Probe/Primer Sets for Real-Time PCR
2.7. Amplification of DNA Through Real-Time Polymerase Chain Reaction
2.8. Ethical Considerations
2.9. Statistical Analysis
3. Results
3.1. Sociodemographic Parameters of Study Population
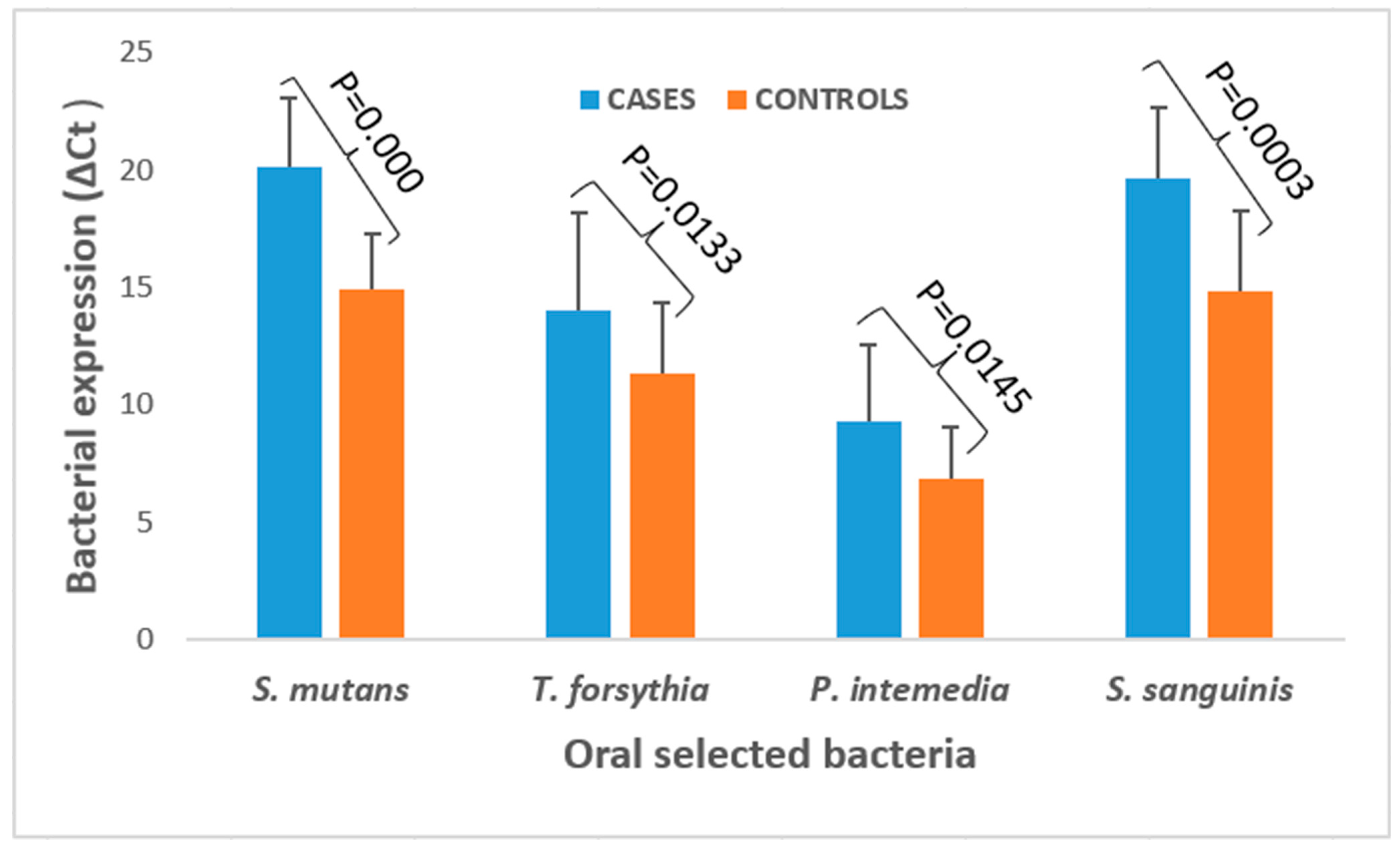
3.2. Bacterial Expression and Quantification in ESCC Cases and Healthy Controls
3.3. Association Between Sociodemographic Characteristics and Bacterial Expression Levels of Selected Oral Bacteria in Healthy Controls and Patients with ESCC
4. Discussion
5. Conclusions
6. Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peters, B.A.; Wu, J.; Pei, Z.; Yang, L.; Purdue, M.P.; Freedman, N.D.; Jacobs, E.J.; Gapstur, S.M.; Hayes, R.B.; Ahn, J. Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Res. 2017, 77, 6777–6787. [Google Scholar] [CrossRef]
- Napier, K.J.; Scheerer, M.; Misra, S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J. Gastrointest. Oncol. 2014, 6, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Kamangar, F.; Malekzadeh, R.; Dawsey, S.M. Esophageal cancer: A global perspective. J. Clin. Gastroenterol. 2019, 53, 461–466. [Google Scholar]
- Drahos, J.; Zhang, Y.; Risch, H.A. Epidemiology of esophageal adenocarcinoma. J. Surg. Oncol. 2020, 121, 257–265. [Google Scholar]
- Alaouna, M.; Hull, R.; Penny, C.; Dlamini, Z. Esophageal cancer genetics in South Africa. Clin. Exp. Gastroenterol. 2019, 12, 157–177. [Google Scholar] [CrossRef] [PubMed]
- Kamsu, G.T.; Ndebia, E.J. Uncovering Risks Associated with Smoking Types and Intensities in Esophageal Cancer within High-Prevalence Regions in Africa: A Comprehensive Meta-analysis. Cancer Epidemiol Biomarkers Prev. 2024, 33, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Ndebia, E.J.; Kamsu, G.T. Drinking patterns, alcoholic beverage types, and esophageal cancer risk in Africa: A comprehensive systematic review and meta-analysis. Front. Oncol. 2023, 13, 1310253. [Google Scholar] [CrossRef] [PubMed]
- di Pietro, M.; Alzoubaidi, D.; Fitzgerald, R.C. Barrett’s esophagus and cancer risk: How research advances can impact clinical practice. Gut Liver 2014, 8, 356–370. [Google Scholar] [CrossRef]
- Ferndale, L.; Aldous, C.; Hift, R.; Thomson, S. Gender Differences in Oesphageal Squamous Cell Carcinoma in a South African Tertiary Hospital. Int. J. Environ. Res. Public Health 2020, 17, 7086. [Google Scholar] [CrossRef]
- Deo, P.N.; Deshmukh, R. Oral microbiome: Unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019, 23, 122–128. [Google Scholar] [CrossRef]
- D’Agostino, S.; Ferrara, E.; Valentini, G.; Stoica, S.A.; Dolci, M. Exploring Oral Microbiome in Healthy Infants and Children: A Systematic Review. Int. J. Environ. Res. Public Health 2022, 19, 11403. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, S.; Valentini, G.; Iarussi, F.; Dolci, M. Effect of Probiotics Lactobacillus rhamnosus and Lactobacillus plantarum on Caries and Periodontal Diseases: A Systematic Review. Dent. J. 2024, 12, 102. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Freedman, N.D.; Alekseyenko, A.V.; Wu, J.; Yang, L.; Pei, Z.; et al. Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome 2018, 24, 59. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xia, L.; Wang, Z.; Zhu, T.; Zhao, L.; Fan, S. A cross-cohort study identifies potential oral microbial markers for esophageal squamous cell carcinoma. iScience 2024, 27, 111453. [Google Scholar] [CrossRef]
- Karpiński, T.M. Role of Oral Microbiota in Cancer Development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef]
- Ahn, H.J.; Lee, D.S. Helicobacter pylori in gastric carcinogenesis. World J. Gastrointest. Oncol. 2015, 7, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhang, Z.; Sun, K.; Li, M.X.; Qi, Y.J. Upper gastrointestinal tract microbiota with oral origin in relation to oesophageal squamous cell carcinoma. Ann. Med. 2023, 55, 2295401. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Gupta, V.; Kwan, B. Second look at Streptococcus sanguinis and the colon. BMJ Case Rep. 2018, 2018, bcr2018224799. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Zhou, Z.; Gao, Q.; Zhu, Z.; Zhu, J.; Lin, J.; Wen, Y.; Qian, F.; Wang, L.; Zhai, Y.; et al. Tumor-derived Prevotella intermedia aggravates gastric cancer by enhancing Perilipin 3 expression. Cancer Sci. 2024, 115, 1141–1153. [Google Scholar] [CrossRef] [PubMed]
- Chukkapalli, S.S.; Rivera-Kweh, M.F.; Velsko, I.M.; Chen, H.; Zheng, D.; Bhattacharyya, I.; Gangula, P.R.; Lucas, A.R.; Kesavalu, L. Chronic oral infection with major periodontal bacteria Tannerella forsythia modulates systemic atherosclerosis risk factors and inflammatory markers. Pathog. Dis. 2015, 73, ftv009. [Google Scholar] [CrossRef]
- Sato, K.; Takahashi, N.; Kato, T. Tannerella forsythia induces inflammation and oxidative stress in human oral epithelial cells. J. Periodontal Res. 2017, 52, 451–460. [Google Scholar]
- Yamamoto, T.; Kato, T.; Takahashi, N. Association between periodontal disease and risk of oesophageal cancer. Cancer Epidemiol. Biomark. Prev. 2012, 21, 2013–2021. [Google Scholar]
- Galvão-Moreira, L.V.; da Cruz, M.C. Oral microbiome, periodontitis and risk of head and neck cancer. Oral Oncol. 2016, 53, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, F.D.; Covenas, R. Biomechanical Mechanisms Associating Alcohol Use Disorders with Cancers. Cancers 2021, 13, 3548. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chu, M.; Huang, Y. Association between oral microbiome and risk of oesophageal cancer. Gut Microbes 2018, 9, 548–558. [Google Scholar]
- May, M.; Abrams, J.A. Emerging Insights into the Esophageal Microbiome. Curr. Treat. Options Gastroenterol. 2018, 16, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Bland, M. An Introduction to Medical Statistics; Oxford University Press: Oxford, UK, 2000; 448p. [Google Scholar]
- Jo, R.; Nishimoto, Y.; Umezawa, K.; Yama, K.; Aita, Y.; Ichiba, Y.; Murakami, S.; Kakizawa, Y.; Kumagai, T.; Yamada, T.; et al. Comparison of oral microbiome profiles in stimulated and unstimulated saliva, tongue, and mouth-rinsed water. Sci. Rep. 2019, 9, 16124. [Google Scholar] [CrossRef] [PubMed]
- Heikrujam, J.; Kishor, R.; Mazumder, P.B. The Chemistry Behind Plant DNA Isolation Protocols. In Biochemical Analysis Tools—Methods for Bio-Molecules Studies; Boldura, O.M., Baltă, C., Awwad, N.S., Eds.; IntechOpen Limited: London, UK, 2020; 206p. [Google Scholar] [CrossRef]
- Barbadoro, P.; Ponzio, E.; Coccia, E.; Prospero, E.; Santarelli, A.; Santarelli, A.; Rappelli, G.G.L.; D’Errico, M.M. Association between hypertension, oral microbiome and salivary nitric oxide: A case-control study. Nitric Oxide Biol. Chem. 2021, 106, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Wroblewski, L.E.; Peek, R.M., Jr.; Wilson, K.T. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin Microbiol Rev. 2010, 23, 713–739. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, B.J.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Könönen, E.; Fteita, D.; Gursoy, U.K.; Gursoy, M. Prevotella species as oral residents and infectious agents with potential impact on systemic conditions. J. Oral Microbiol. 2022, 14, 2079814. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Macleod, L.C.; Kitten, T.; Xu, P. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future Microbiol. 2018, 13, 915–932. [Google Scholar] [CrossRef]
- Lemos, J.A.; Palmer, S.R.; Zeng, L.; Wen, Z.T.; Kajfasz, J.K.; Freires, I.A.; Abranches, J.; Brady, L.J. The Biology of Streptococcus mutans. Microbiol. Spectr. 2019, 7, GPP3-0051. [Google Scholar] [CrossRef]
- Abranches, J.; Zeng, L.; Kajfasz, J.K.; Palmer, S.R.; Chakraborty, B.; Wen, Z.T.; Richards, V.P.; Brady, L.J.; Lemos, J.A. Biology of Oral Streptococci. Microbiol. Spect. 2018, 6, GPP3-0042. [Google Scholar] [CrossRef]
- Hiam-Galvez, K.J.; Allen, B.M.; Spitzer, M.H. Systemic immunity in cancer. Nat. Rev. Cancer 2021, 21, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Castañeda-Corzo, G.J.; Infante-Rodríguez, L.F.; Villamil-Poveda, J.C.; Bustillo, J.; Cid-Arregui, A.; García-Robayo, D.A. Association of Prevotella intermedia with oropharyngeal cancer: A patient-control study. Heliyon 2023, 9, e14293. [Google Scholar] [CrossRef]
- Yano, Y.; Etemadi, A.; Abnet, C.C. Microbiome and Cancers of the Esophagus: A Review. Microorganisms 2021, 9, 1764. [Google Scholar] [CrossRef]
- Chiang, H.C.; Hughes, M.; Chang, W.L. The role of microbiota in esophageal squamous cell carcinoma: A review of the literature. Thorac. Cancer 2023, 14, 2821–2829. [Google Scholar] [CrossRef]
- Kawasaki, M.; Ikeda, Y.; Ikeda, E.; Takahashi, M.; Tanaka, D.; Nakajima, Y.; Arakawa, S.; Izumi, Y.; Miyake, S. Oral infectious bacteria in dental plaque and saliva as risk factors in patients with esophageal cancer. Cancer 2021, 127, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Lara, E.S.M.; Moyeda, A.L.G.; Ibarra, K.I.J.; Moreno, N.P.F.; Depraect, N.E.Z.; Flores, P.F.; Delgado, K.N.R.; Soto, J.M.S. Tannerella forsythia: A periodontopathic pathogen review. Int. J. Appl. Dent. Sci. 2023, 9, 163–166. [Google Scholar] [CrossRef]
| Bacteria | Primers and Probes |
|---|---|
| Streptococcus sanguinis | 5′-GTGTCAATTCCCAGAAAAG-3′ 54.0 5′-ATTATTGGCTGATGTGGAGTC-3′ 45.3 5′-HEX-AGATGACCACCACCGT-BHQ1-3′ 50.0 |
| Streptococcus mutans | 5′-TCACCAGAAAAGACAAAAGTTAC-3′ 5′-AACTACTAACCAAGCCCAAC-3′ 5′-Cy5-TAGCCGCAGCAATCAATG-BHQ-3′ |
| Prevotella intermedia | 5′-TCCACCGATGAATCTTTGGTC-3′ 5′-ATCCAACCTTCCCTCCACTC-3′ 5′-FAM-CGTCAGATGCCATATGTGGACAACATCG-TAMRA-3′ |
| Tannerella forsythia | 5′-AGCGATGGTAGCAATACCTGTC-3′ 5′-TTCGCCGGGTTATCCCTC-3′ 5′-FAM-TGAGTAACGCGTATGTAAACCTGCCCGC-TAMRA-3′ |
| Universal | 5′-TCCTACGGGAGGCAGCAGT-3′ 5′-GGACTACCAGGGTATCTAATCCTGTT-3′ 5′-FAM-CGTATTACCGCGGCTGCTGGCAC-TAMRA-3′ |
| Parameters | Groups | Healthy Controls n = 24(%) | Cases (ESCC) n = 24(%) | p-Value |
|---|---|---|---|---|
| Age | Mean | 51.34 | 66.42 | / |
| Gender | Female | 11 (45.83) | 13 (54.16) | 0.773 |
| Male | 13 (54.16) | 11(45.83) | ||
| Level of education | Primary | 1 (4.16) | 20 (83.33) | 0.000 |
| Secondary | 23 (95.83) | 4 (16.66) | ||
| Marital status | Single | 17 (70.83) | 19 (79.16) | 0.740 |
| Married | 7 (29.16) | 5 (20.83) | ||
| Teeth brushing | Once | 11 (45.83) | 14(58.33) | 0.772 |
| ≥2 | 13 (54.16) | 10 (41.66) | ||
| Missing teeth | 0 | 7 (29.16)) | 4 (16.66) | 0.666 |
| 1–3 | 13 (54.16) | 16 (66.66) | ||
| >3 | 4 (16.66) | 4 (16.66) | ||
| Smoking status | Never | 20 (83.33) | 7 (29.16) | 0.000 |
| Ever | 4 (16.66) | 17 (70.83) | ||
| Alcohol consumption | Never | 12 (50) | 8 (33.33) | 0.380 |
| Ever | 12 (50) | 16 (66.66) |
| Parameters | Groups | S. mutans | S. sanguinis | P. intermedia | T. forsythia | ||||
|---|---|---|---|---|---|---|---|---|---|
| Log (Mean ± SD) | p-Value | Log (Mean ± SD) | p-Value | Log (Mean ± SD) | p-Value | Log (Mean ± SD) | p-Value | ||
| Age | Mean | / | 0.000 | / | 0.009 | / | 0.131 | / | 0.367 |
| Gender | Female | 1.25 ± 0.10 | 0.189 | 1.25 ± 0.13 | 0.076 | 0.89 ± 0.16 | 0.603 | 1.10 ± 0.13 | 0.248 |
| Male | 1.22 ± 0.08 | 1.19 ± 0.12 | 0.86 ± 0.17 | 1.06 ± 0.11 | |||||
| Education level | Primary | 1.30 ± 0.09 | 0.000 | 1.28 ± 0.12 | 0.004 | 0.92 ± 0.18 | 0.069 | 1.13 ± 0.12 | 0.014 |
| Secondary | 1.19 ± 0.07 | 1.18 ± 0.13 | 0.84 ± 0.14 | 1.04 ± 0.10 | |||||
| Marital status | Single | 1.24 ± 0.09 | 0.207 | 1.22 ± 0.13 | 0.825 | 0.88 ± 0.17 | 0.534 | 1.07 ± 0.12 | 0.275 |
| Married | 1.20 ± 0.12 | 1.23 ± 0.14 | 0.85 ± 0.16 | 1.12 ± 0.13 | |||||
| Teeth brushing | Once | 1.25 ± 0.09 | 0.279 | 0.86 ± 0.17 | 0.589 | 0.86 ± 0.17 | 0.589 | 1.10 ± 0.14 | 0.737 |
| ≥2 | 1.22 ± 0.10 | 0.89 ± 0.16 | 0.89 ± 0.16 | 1.07 ± 0.11 | |||||
| Missing teeth | 0 | 1.24 ± 0.08 | 0.692 | 1.24 ± 0.11 | 0.643 | 0.92 ± 0.21 | 0.521 | 1.07 ± 0.11 | 0.777 |
| 1–3 | 1.23 ± 0.10 | 1.22 ± 0.14 | 0.86 ± 0.15 | 1.08 ± 0.13 | |||||
| >3 | 1.21 ± 0.11 | 1.18 ± 0.12 | 0.87 ± 0.15 | 1.11 ± 0.12 | |||||
| Smoking status | Never | 1.19 ± 0.08 | 0.000 | 1.19 ± 0.14 | 0.060 | 0.85 ± 0.17 | 0.317 | 1.05 ± 0.10 | 0.030 |
| Ever | 1.30 ± 0.08 | 1.26 ± 0.11 | 0.90 ± 0.16 | 1.12 ± 0.13 | |||||
| Alcohol consumption | Never | 1.21 ± 0.11 | 0.097 | 1.22 ± 0.15 | 0.974 | 0.88 ± 0.16 | 0.832 | 1.07 ± 0.10 | 0.724 |
| Ever | 1.25 ± 0.08 | 1.22 ± 0.12 | 0.87 ± 0.17 | 1.09 ± 0.13 | |||||
| Tannerella forsythia | Streptococcus mutans | Streptococcus sanguinis | Prevotella intermedia | |||||
|---|---|---|---|---|---|---|---|---|
| Variable | Adjusted Coefficient (95% CI) | p-Value | Adjusted Coefficient (95% CI) | p-Value | Adjusted Coefficient (95% CI) | p-Value | Adjusted Coefficient (95% CI) | p-Value |
| Age | / | / | −0.026 (−0.090–0.038) | 0.417 | −0.010 (−0.120–0.101) | 0.862 | / | / |
| Education Level | −0.049 (−0.163–0.065) | 0.392 | −0.017 (−0.083–0.049) | 0.614 | −0.002 (−0.115–0.111) | 0.970 | / | / |
| Smoking status | 0.044 (−0.038–0.127) | 0.283 | 0.043 (−0.005–0.090) | 0.076 | / | / | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nokamatye, Y.Y.; Kamsu, G.T.; Ndebia, E.J. Comparative Analysis of Oral Prevotella intermedia, Tannerella forsythia, Streptococcus sanguinis, and Streptococcus mutans in Patients with Esophageal Squamous Cell Carcinoma and Healthy Controls in Mthatha, South Africa. Bacteria 2025, 4, 11. https://doi.org/10.3390/bacteria4010011
Nokamatye YY, Kamsu GT, Ndebia EJ. Comparative Analysis of Oral Prevotella intermedia, Tannerella forsythia, Streptococcus sanguinis, and Streptococcus mutans in Patients with Esophageal Squamous Cell Carcinoma and Healthy Controls in Mthatha, South Africa. Bacteria. 2025; 4(1):11. https://doi.org/10.3390/bacteria4010011
Chicago/Turabian StyleNokamatye, Yolanda Yolisa, Gabriel Tchuente Kamsu, and Eugene Jamot Ndebia. 2025. "Comparative Analysis of Oral Prevotella intermedia, Tannerella forsythia, Streptococcus sanguinis, and Streptococcus mutans in Patients with Esophageal Squamous Cell Carcinoma and Healthy Controls in Mthatha, South Africa" Bacteria 4, no. 1: 11. https://doi.org/10.3390/bacteria4010011
APA StyleNokamatye, Y. Y., Kamsu, G. T., & Ndebia, E. J. (2025). Comparative Analysis of Oral Prevotella intermedia, Tannerella forsythia, Streptococcus sanguinis, and Streptococcus mutans in Patients with Esophageal Squamous Cell Carcinoma and Healthy Controls in Mthatha, South Africa. Bacteria, 4(1), 11. https://doi.org/10.3390/bacteria4010011






