Abstract
In the recent past, microbiome manipulation has emerged as a promising approach to improve plant growth performance by exploring the deep insight of plant–microbe interactions. The exploration of a plant microbiome either present on an ectosphere or endosphere can provide a far better understanding about the potential application of plant-associated microbes for the improvement of plant growth, protection from pathogen invasion, and tolerance to environmental stresses of a diverse nature. In this context, next-generation sequencing methods, omics approaches, and synthetic biology have made significant progress in plant microbiome research and are being frequently used to explore the intriguing role of plant-associated microorganisms. Despite the successfulness of conventional approaches, the incorporation of CRISPR/Cas9, RNA interference technology, rhizosphere engineering, microbiome engineering, and other manipulation techniques appear to be a promising approach to enhancing plant performance, and tolerance against biotic and abiotic stress factors. The present review presents the significance of plant microbe interaction, vital functional aspects, collaborative action, potential constraints, and finally the latest developments in bioengineering approaches destined for microbiome modulation with an objective to improve the performance of a host plant challenged with environmental stressors.
1. Introduction
Currently, agricultural productivity is facing challenges to meet the escalating food demand of a continuously rising global population not only because of declining cultivable land areas, but also due to unfavorable climate change. According to an estimate, agricultural productivity needs to be enhanced by 60% by the year 2050 [1]. Noteworthy is that climate change and a limited availability of natural resources such as agricultural land and water availability limits agricultural productivity [2]. During the past two decades, a substantial progress in agricultural practices has been marked; nevertheless, abiotic environmental stressors such as drought, salinity, flood, nutrient deficiency, soil degradation, indiscriminate application of hazardous agrochemicals, and biotic stress factors including resistance resurgence in pathogens have resulted in compromised crop productivity, suggesting the development of environmentally sound strategies to fulfil the food demand.
Plants and microbes are considered to be continuously co-evolved by forming a complex ecosystem, thereby considerably influencing the growth as well as the survival of each other. The concept of “holobiont” has come into existence to reveal the interaction between the host and associated microorganisms in a given environmental complex with plausible implications in biology, ecology, and the evolutionary process of the host plant. Furthermore, the “holobiont” framework comprising of the host and associated microorganisms is suggested to evolve synergistically for mutual cross-talk and co-signaling [3,4]. The diverse microbiome existing in the rhizosphere, phyllosphere, and endophytically in different plant parts with the characteristics ability to augment host plant growth are referred to as “plant-growth promoting microorganisms” (PGPMs) [5,6]. The PGPMs comprising of different bacteria, fungi, etc. are acknowledged to enhance agricultural productivity [1,7,8,9].
Recently, microbiome manipulation has been carried out to improve agricultural productivity or maintain the plant health via different processes such as tillage practices, synthetic microbial consortia, microbial inoculation, rhizosphere engineering, and microbiome engineering [10,11] (Figure 1). Although these approaches are dependent upon various factors such as the host plant, associated microbial communities, soil, and other environmental factors, manipulation of these factors directly or indirectly influence plant growth or developments.
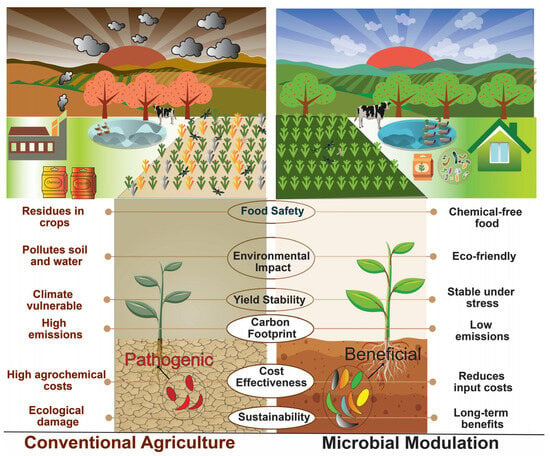
Figure 1.
An overview of microbial modulation or manipulation techniques in sustainable agriculture practices. This figure illustrates the contrasting ecological impacts of microbial manipulation/modulation versus conventional cropping on various environmental components such as soil, air, and water. Microbial modulation promotes eco-friendly practices, enhances soil health, and reduces pollution in air and water. It supports long-term sustainability, stable yields under stress, and chemical-free food production. In contrast, conventional cropping contributes to soil degradation, water contamination, and air pollution, with high emissions, pesticide residues, and vulnerability to climate change, while relying on costly and harmful agrochemical inputs.
Therefore, there is a need to explore various aspects of plant microbe research, microbiome functioning, and how the microbial communities influence plant growth [12,13]. This review aims to provide an overview of the latest developments in microbiome manipulation. Additionally, various aspects of plant–microbe interactions, traditional approaches of microbiome manipulation, and how plant performance can be improved using the latest emerging approaches such as CRISPER/Cas9, RNAi, or microbiome engineering, etc. are discussed.
During the review process, we carried out a bibliometric data analysis to evaluate the significance of topic microbiome manipulation in improving plant performance. We retrieved 68,401 articles published between 2000 and 2025, from the PubMed database by searching for the keywords “Microbiome Manipulation” OR “Microbial Modulation” And “Plant health” OR “Plant performance” OR “Microbiome engineering” OR “Rhizosphere engineering” OR “CRISPER/Cas” or “RNAi” OR “Bioengineering approaches” OR “Holobionts” OR “omics” OR “crop rotation” OR “plant—microbe interactions” OR “tillage intensity” OR “Conventional Agriculture “ OR “Biotic stress” OR “abiotic stress ” (data accessed on 15 February 2025). Out of 15,462 keywords, 277 were selected with a cutoff of 15 occurrences and we downloaded 10,000 papers in a tab-delimited format and created bibliometric analysis (Figure 2) using VOS viewer processing software (v1.6.9). The different colors present in the network show linked terms; however, the size of the labels showing the number of publications and the distances present between two nodes represent the degree through which they are associated.
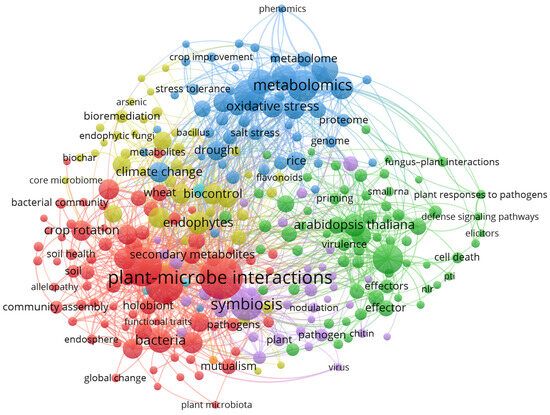
Figure 2.
Network visualization map displaying the key term present in the title and abstract of the published article related to the plant–microbe interactions in abiotic stress management. Nodes of the same color show a cluster of the interconnected phrases; however, the circle size indicates the number of publications, with the larger size indicating a large number of publications. The network has been created using the software VOS viewer processing software (v1.6.9.).
2. Plant–Microbes Interactions and Their Significance
Plants interact with a plethora of microbial species, in which some of them directly or indirectly play a significant role in plant growth and developments [13]. The plant rhizosphere is considered as a hotspot of microbial communities and is defined as a thin layer of soil that surrounds and is influenced by plant roots. The plant root secretes an array of exudates constituted of amino acids, flavonoids, lipids, organic acids, and other secondary metabolites, and these biochemical components act as an energy source or signaling molecules for the microbial species [14,15,16,17]. The amount and the nature of exudates varies with internal or external surrounding factors such as host genotype, seasons, temperature, nature of the soil, etc., which also influence the composition of host-associated microbial communities [18,19]. For example, during the seedling stages, the nature and amount of sugar molecules is different than at the harvest stages; similarly, draught or other stress factors also influence the amount and nature of exudates, which ultimately affect the microbial community structures [20].
Each of the plant organs such as the root, stem, leaves, flowers, etc. have some specific microbial composition which may vary with changing environmental conditions, host genotypes, plant developmental stages, seasons, or stress factors [13]. The microbial communities structure of the plants are generally dominated by the bacterial phyla Proteobacteria, Bacteroidetes, Acidobacteria, and Actinobacteria [21,22,23] and by the fungal phyla Ascomycota and Basidiomycota. In the recent past, various authors studied the microbial communities structure of different plant species. For instance, Fu et al. [24] reported Proteobacteria and Actinobacteria as the most prominent bacterial phyla in the semiarid alpine grassland. Nan et al. [25] reported the dominance of bacterial phyla Actinobacteria and Proteobacteria in the rhizosphere of three species of Stipa. Xu et al. [26] in the global study of citrus rhizosphere also reported Proteobacteria, Actinobacteria, and some other bacterial group like Acidobacteria and Bacteroidetes as the prominent one, while Ascomycota and Basidiomycota were the dominant fungal phyla.
It has been observed that the endosphere contains lesser microbial species than the ectosphere or rhizospheric soil. For example, an abundance of bacterial population in the rhizospheric soil was reported in the range of 106–109, while only 104–108 CFU (colony forming unit) per gram of root tissues was observed in the plant endosphere [27,28,29]. In studies of the plant endosphere, a dominance of Proteobacteria was also reported followed by Actinobacteria. Singh et al. [30] also reported an abundance of phyla Firmicutes and Proteobacteria as endophytic in the Momordica charantia L. Goulart et al. [31] observed a higher abundance of Proteobacteria in the vegetative and reproductive stage of Passiflora incarnata. Other bacterial phyla that were dominant in the vegetative growth were Firmicutes followed by Actinobacteria and Bacteroidetes, while Firmicutes (28.5%) was the second dominant group in the reproductive stages.
3. Functional Aspect of Plant Microbiomes
The host plants are considered to offer a habitat for a multitude of microbial communities. The intimate association of microorganisms with plants has been acknowledged to facilitate plant growth [32]. The extensive investigation on the microbiome associated with plants has helped in understanding the intricate communications [33]. The employment of a beneficial microbial population has demonstrated a characteristic role in augmenting crop productivity under changing climatic conditions, diversified stress factors, and a deteriorating soil environment [34,35,36].
Stresses either of biotic and abiotic negatively influence crop productivity worldwide. Noteworthy is that abiotic stress factors have emerged as a major constraint that limits agricultural productivity. The microbial association with the host plant helps alleviate the repercussions of abiotic stressors through the biosynthesis of polymeric substances including exopolysaccharides serving as a protective barrier in the root zone, in addition to the release of osmolytes, including sugar and amino acids as illustrated in the strains of Acinetobacter, Pseudomonas, and Pantoea recovered from drought-resistant and salt-tolerant plants [37]. Additionally, phytohormones and volatiles released by host-associated microbiome triggers induced systemic tolerance (IST). The microbiological biosynthesis of enzyme ACC deaminase lowers the ethylene level. For instance, Achromobacter piechaudii ARV8 equipped with the ability to produce ACC deaminase has been suggested to provide tolerance against drought in Capsicum annuum L. and Solanum lycopersicum L. [38]. Salinity has been envisaged as an important stressor limiting crop productivity substantially. Leveraging salt-tolerant microorganisms could be a sustainable option to mitigate the challenges of agricultural productivity. A recent investigation by Ning et al. [39] explored the feasibility of leveraging halotolerant bacterial strain Enterobacter asburiae D2, endowed with a multitude of growth promotion attributes, to elevate the challenges of salinity stress in Oryza sativa L. Khumairah et al. [40] and Sarker et al. [41] reported on halotolerant rhizobacteria, which after inoculation significantly improved the salinity tolerance and enhanced plant growth [40,41].
PGPMs and their products such as biofilm, exopolysaccharides, and biosurfactants are frequently utilized to mitigate the challenges of heavy metal accumulation in the soil. In addition to mitigation processes of heavy metals, PGPMs also help in growth promotion of plants [42]. For example, treatment with Klebsiella sp. M2 led to an increase in dry weight ranging from 14.3 to 35.9% and the release of metabolic constituents responsible for growth promotion in Triticum aestivum L. Heavy metal–tolerant microbial strains of endophytic nature and those residing in the rhizospheric zone can be exploited for the amelioration of heavy metal toxicity in crop plants. Previous authors reported various different bacterial species such as Enterobacter, Psudomonas, Bacillus, and Klebsiella having the capability to accumulate excess heavy metals [43].
A microbiome can be harnessed for encountering the rising threats of crop damage caused by different phytopathogens. The microbial species having the ability to protect the plants from pathogen invasion or plant diseases are termed as “microbial biocontrol agents.” The microbial biocontrol agents inhibit the phytopathogen growth by secreting antimicrobial metabolites, antibiotics, siderophores, volatile bioactive compounds, etc. or by host immune response or by inducing systemic response [44]. The phenomenon of induced systemic response (ISR) directs the expression of key genes related to defence, such as phenylalanine ammonia lyase (PAL), peroxidase, chitinase, etc. [1]. Also, the release of secondary metabolic products like quorum sensing biomolecules that aggravate the event of ISR. Furthermore, different microbial species can exert various direct as well as indirect effects involving key strategies like enzymes secretion, which causes cell wall degradation, and volatile components restricting the metabolic activities with the resultant inhibition of phytopathogens multiplication [33].
Quorum sensing (QS) is a type of microbial communication mechanism in which bacteria communicate and work together through various chemical cues referred to as “auto-inducers” to govern populations and sustain the equilibrium of a certain process. Additionally, QS modifies the gene expression patterns in response to cell density, which leads to the modification or activation of various transcriptional factors in bacteria [45]. QS also showed direct involvement in the regulation of various functions such as biofilm formation and colonization within the bacterial populations, which plays a significant role in plant growth promotion even under stress conditions [46]. However, the QS molecules like N-acyl homoserine lactones (AHLs) induce the systemic resistance in plants, making them resistant against the pathogens [47].
However, the full potential of a microbiome with reference to sustainable agriculture under diverse climatic conditions has to be explored. The search for newer microbial species, their application as soil or plant inoculants, and the creation of synthetic microbial consortia to control the phytopathogen growth needs to be studied more extensively. The following section deals with the collaborative action of a plant microbiome and strategies being employed for microbiome manipulation using conventional and modern bioengineering approaches.
It is well established that plant-associated microbial species play a significant role in improving plant performance via enhancing plant growth or controlling phytopathogen growth. However, in natural and engineered ecosystems, millions of microbial species reside or co-exist and each possess specific functions and metabolic capabilities [9]. These microbial species perform some specific community’s level performance that contribute a significant role in maintaining plant stability and health during normal or stress conditions [12]. In general, both an energy source and nutrient requirements are fundamental needs for the survival of microorganisms. Although within the microbial community the requirement of an energy resource and the nutrient supplements depends upon the reciprocal exchange of metabolites within the different microorganisms present in the community [12].
4. Dysbiosis in Plant Microbiomes
Dysbiosis signifies an alteration within a microbial community that negatively affects the health of particular plants. Some environmental factors like stress, nutrient content in the soil, and different types of pathogens can throw off the balance within microbes [48]. For example, the increased presence of pathogenic microorganisms within the mycorrhizosphere may suppress the recruitment of good mycorrhizae, thereby reducing plant health and productivity [49]. In addition, certain plants may produce allelochemicals that modify the makeup of their associated microbiomes, resulting in poor interactions with plants, which may lower plant performance [50]. The consequences of dysbiosis are not limited to individual organisms but rather whole ecosystems. The alterations of the diversity of microbes have consequent effects on the microbially mediated processes of nutrient cycling, the health of soils, and the composition of plant communities.
5. Approaches for Microbial Manipulation
In the recent past, a number of studies have suggested different approaches for the manipulation of the microbiome in order to improve plant fitness and agricultural productivity so as to meet the rising food demand for an incessantly growing human population. Some of the commonly used methods such as crop rotation, organic fertilizer input, tillage intensity, and microbiome reorganization are presented in the following sections.
5.1. Crop Rotation
Crop rotation is a common traditional approach practiced to maintain soil productivity and modulate the native microflora of the agricultural soil. It has been observed that repetitive cropping of the same crop can lead to a loss of moisture content, organic matter, and native microbiota [51]. Therefore, maintaining the nutrient status and composition of native microbiota rotation in the cropping system has been preferred for a long time. In a recent study, Yang et al. [52] acquired six years of data and reported a significance of diversified crop rotations and observed that an addition of sweet potato and legumes plants viz. peanut and in the traditional wheat–maize rotation significantly enhanced the soil microbial diversity and agricultural output up to 38%. Additionally, it reduced the emission of N20 by 39% and improved the greenhouse gas balance by 88% in the North China plain. Similar types of observations like growing maize after the cultivation of rice can help with nutrient acquisition and the assembly of beneficial microbial communities studied and reported by various authors in the previous studies [53,54].
5.2. Organic Fertilization
Currently, a huge amount of chemical fertilizers and pesticides are being used to enhance agricultural productivity, but their continuous utilization adversely affects the texture and productivity of plants, native microflora, and human health [6]. Therefore, organic amendments, or bio-organic fertilization, are now being used as efficient methods for maintaining soil productivity or native microflora [55,56].
Soil health and the functioning of an ecosystem largely dependent upon the interactions between the soil organic matter and the native microflora. The presence of a higher content of organic matter positively modulates the growth and metabolism of microbes and also shapes the microbial community structure and diversity [56,57]. The presence of a diverse microbial community not only helps in maintaining soil fertility but also helps in degrading complex environmental contaminants. Delgado-Baquerizo et al. [57] reported that the presence of a diverse microbial community modulates the breakdown of litter and other complex organic matter. Similarly, Sun et al. [58] reported that nutrient-rich leaf litter is more easily decomposed than lignin-rich woody litter, which can constrain microbial growth if not adequately supplied [58].
In conventional practices, nutrient-rich agricultural residues, organic manures, biocomposts, or vermicompost have been used to improve and maintain the soil texture or productivity [59]. Previous authors reported the use of bio-organic fertilization in improving plant growth [60,61,62,63]. For example, Lahbouki et al. [64] used horse manure, and Arbuscular Mycorrhizal Fungi (AMF) to amend the soil and enhance tomato productivity. Li et al. [65] used different bio-organic fertilizers to evaluate the impact on Dendrocalamus farinosus growth and found that the utilization of bioorganic fertilizers enhances the growth via restructuring the soil microbiome and metabolome. Wang et al. [66] reported the impact of bio-organic fertilizers on the yield enhancement of pear, which also shapes the rhizospheric microbiome. Xu et al. [60] reported that organic fertilizers like black soldier fly frass derived from different organic wastes can significantly benefit plant growth by improving soil fertility and microbial diversity. Similarly, a combination of sheep manure and chemical fertilizers showed a beneficial role in plant growth promotion [61]. Furthermore, the application of organic manure in Qingke cultivation has been found to influence the rhizosphere microbiota composition and plant growth [62]. More field studies, however, are required to translate the advantageous actions of the microbiome for augmenting the productivity of a plant challenged with diverse abiotic and biotic factors.
5.3. Tillage Intensity
Tillage intensity refers to the degree and frequency of soil disturbance caused by agricultural practices. It ranges from intensive to no tillage intensity, which involves significant soil disruption, to minimal or no disturbance in soil. In tillage practices, the relocation of crop residues has been carried out, which affects and redistributes moisture, the available carbon, and oxygen availability to the soil [67,68]. Currently, different types of tillage intensity such as conventional tillage, reduced tillage, or no tillage has been practiced that significantly affect soil structure, organic matter distribution, and the soil microbiome [69,70,71]. In previous studies, authors reported on different tillage practices and their impact on maintaining soil or plant health. Dewi et al. [69] reported on long-term no-tillage and integrated the use of biochar to improve soybean production. Hu et al. [70] observed that a reduced tillage system improved carbon, nitrogen, and phosphorous in the soil system.
5.4. Reconstructing the Existing Microbiome by Microbial Inoculation
In sustainable agricultural practices, the screening and characterization of a specific microbial strain is one promising approach [72]. Advancement in the latest next-generation sequencing technology, metagenomics, metabolomics, and transcriptomics study enhanced the understanding of microbial community structure and their functioning. These studies also help in characterizing the microbial strains, having better suitability and capability to use as soil or plant inoculants to enhance agricultural productivity [73]. Although for successful inoculants, the strains must have the capability to survive under field conditions and have the potential to be outcompeted by the native microbiome after the introduction [74]. The transition of microbial strains or consortia from the laboratory to the field condition is one of the most challenging tasks. It has been observed that transition sometimes diminishes the efficacy and survivability of microbial strains [75].
In contrast, in recent times, inoculation of two or more than two microbial species consortia either of the same genera or different genera, such as bacteria–bacteria, bacteria–fungi, bacteria–fungi–yeast, have been formulated to create a diverse functional genetic pool, having the potential to act multidimensionally against environmental challenges or stresses [76]. The synthetic consortia of different microbial strains shows potential to resist against the multiple constraints, where the single strain only has limited options. For instance, under low-phosphorus condition, inoculation of phosphate-solubilizing bacteria along with a synthetic consortia of multiple strains showed a superior effect in the crop growth [74]. These consortia effectively provide sufficient solubilized phosphorus, demonstrating their potential to enhance plant growth in nutrient-deficient soils. Wang et al. [77] identified and characterized 23 strains to create a potential synthetic consortia of different strains to promote rice growth. Manjunath et al. [78] reported two different synthetic consortia, Pseudomonas putida P7 + Paenibacillus favisporus B30 and Pseudomonas putida P45 + Bacillus amyloliquefaciens B17, to successfully enhance the growth of maize plants by 27.78 and 23.21%. Similarly, Olanrewaju and Babalola [79] used different microbial strains singly or in combination to evaluate their potential on maize seed growth and found that a mixture of consortia was more potent than a single inoculation. Similarly, Yin et al. [80] created 10 synthetic consortia of 14 different bacterial strains in which nine syncoms showed an antagonistic role against the wheat pathogen Rhizoctonia solani AG8.
6. Engineering Approach to Improve Plant Performance
Despite the successfulness of conventional approaches suggested for microbiome modulation, the introduction of current bioengineering approaches is considered to have promising potential in improving plant fitness under stressed conditions. Some methods of microbiome manipulation including rhizosphere engineering, deliberated alteration in microbiome characteristics, CRISPR/Cas9, and RNA interference with a profound effect in offering tolerance to host plants under environmental stress are presented in the next section.
6.1. Rhizosphere Engineering
Rhizosphere engineering is an emerging approach through which plant performance has been improved following some key strategies like modification in the root architecture to enhance the root absorption area, modifications in the amount and nature of root exudates to attract some specific microorganisms, plant engineering to overexpress or knockout the genes to attract specific microbial taxa, and soil amendments to promote the growth of some specific plant growth–promoting bacteria [81,82,83].
In previous studies, authors reported on various approaches of rhizosphere engineering. In most of the studies, during root engineering, the primary focus is to knockout the genes that are responsible to check the growth and developments of the root system [84,85] and the functional aspect of the root system can be modulated by modifying the gene expression pattern [86]. Modification in the root architecture is another strategy of rhizosphere engineering to improve the plant performance. In this approach, the architecture of the root has been modified in such a way that the root can hold or absorb a better amount of water and nutrients [84,87,88].
The composition, amount, and the nature of the root exudates affect interactions patterns between the host and microbes and are influenced by the plant development stages, plant genotypes, soil conditions, or the surrounding environmental factors [89]. These root exudates act as chemo attractants or chemo repellents for the microbial species. Modifications in the composition or specific molecules in the root exudates are one of the strategies of rhizosphere engineering, through which specific microbial species enriched in the plant rhizosphere enhance and modulate microbial growth [90].
Inoculation of the soil with beneficial microbial strains is used to improve plant growth and productivity [91,92]. Although environmental conditions and plant species are vital for the success of rhizosphere engineering, which is based on selection and adaptation of microbial strains, recent improvements in high-throughput sequencing and metagenomics have supported the characterization and recognition of beneficial microbial strains from different environmental conditions [91]. For example, unique strains of Bacillus and Paenibacillus were recruited from the rhizosphere of drought-tolerant plants and these were later utilized to improve drought resistance in new crops [92]. Likewise, another suggested track to recover plant performance is the insertion of specific genes into microbial genomes, to progress the microbial performance. For example, Pseudomonas fluorescens was obtained to produce the plant hormone indole-3-acetic acid (IAA), subsequent in progressed root growth and nutrient exploitation in wheat, which demonstrated the microbial functions [93].
6.2. Microbiome Engineering
Microbiome engineering is receiving accelerated attention in view of improving crop productivity and encountering the hurdles of climate change [94,95,96,97]. Although different plant parts are known to harbor specific microbial communities and are endowed with the potential to influence the host biology [98], during the process of microbiome engineering, the microbial communities structure of the plant has been modified or optimized (Figure 3). This involves various processes viz. modification of indigenous microbial strains with the help of genetic engineering approaches like CRISPER/Cas9, RNAi interference, etc.; transplantation of beneficial microbiome from the healthy to the target sites; or the development or inoculation of syncoms. However, the primary goal of microbiome engineering is to improve the functioning of microbes in such a way that shows an improved stress tolerance potential, improved nutrients utilization patterns, etc. [99,100].
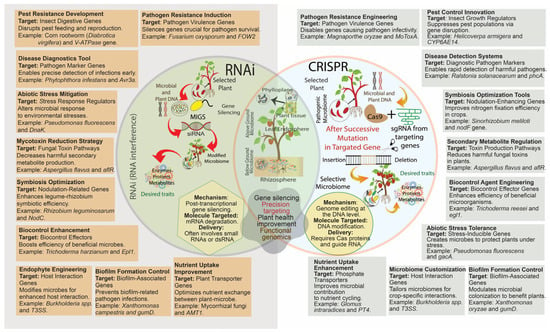
Figure 3.
The interplay between RNAi and CRISPR technologies in plant–microbe interactions, emphasizing their distinct and overlapping roles in microbiome engineering. RNAi (Left Circle) is a post-transcriptional gene-silencing mechanism targeting mRNA degradation, widely used for pest control (e.g., silencing digestive genes in Corn rootworm) and mycotoxin reduction (e.g., Aspergillus flavus toxin pathway). CRISPR (Right Circle) is a genome-editing tool targeting DNA, and facilitates precise modifications for pathogen resistance (e.g., editing Magnaporthe oryzae virulence genes) and nutrient uptake enhancement (e.g., improving phosphate transport in Glomus intraradices). The intersection of RNAi and CRISPR demonstrates shared applications such as gene regulation, precision targeting, and improving plant health. Applications span pathogen resistance, symbiosis optimization, biocontrol enhancement, and abiotic stress tolerance, showcasing the synergistic potential of these tools in advancing sustainable agriculture and functional genomics.
The identification of genes or alleles responsible for the pathogenicity, synergistic, or antagonistic effect is a prime concern in microbiome engineering and their modifications either by enhancing protein expression or by knocking out the gene is a common practice. To complete this process, various techniques such as CRISPR/Cas9, RNA interference have been performed [101,102].
6.2.1. CRISPER/CAS
CRISPR/Cas9 is one of the precise and efficient tools used in gene modification, especially for the knockout or knockdown process of a particular gene [103]. The first-generation CRISPER gene technology has been utilized for the site-specific cleavage of double-standard DNA [104]. The second generation leads to precise conversion of or replacement of single base into another without cleaving double-standard DNA [105].
In the recent past, in the changing climatic conditions eruptions of novel pathogens or virulent strains and their sustainable management is immediate need for optimum agricultural productions [106,107,108]. CRISPR/Cas9 is increasingly recognized as a promising tool for managing plant diseases. Additionally, it has been applied to genetically modify microorganisms to boost their beneficial effects on crops [109,110]. Previously, authors utilized CRISPER/Cas9 technology to control phytopathogens. For example, knockdown of the OsERF922 gene, which is known as an “ethylene responsive factor”, reduced the chance of infection of Magnaporthe oryzae compared to the wild type [111]. Similarly is the knockdown of ERF922 genes, which are upregulated during the M. oryzae infection [111]. The editing in the ERF genes help with resistance in pathogen infections [112]. One study reported a significance of CRISPER/Cas9 technology in generating downy mildew–resistant grapes by editing the MLO-7 gene, which has been upregulated during infection of Erysiphe necator [113]. Paula de Toledo Thomazella et al. [114] reported the significance of CRISPER/Cas9 technology in editing the SlDmr6-1 gene in tomato, which is responsible for providing resistance against different phytopathogens. Further, Wang et al. [115] reported on the knockout of the VvWRKY52 gene through CRISPER/Cas9 technology, which provided resistance against Botrytis cinerea in grapes. Hu et al. [116] reported on the knockout of the CsLOB gene, responsible for bacterial canker diseases in grapefruit. Similarly, knockdown of CsLOB1 in Wanjincheng Orange provided resistance against bacterial canker diseases. Prihatna et al. [117] utilized CRISPER/Cas9 technology to inhibit the Solyc08g075770 (CYCLOPS) gene, which after inhibition enhanced the resistance against the fusarium wilt in tomato. Similarly, Paula de Toledo Thomazella et al. [114] reported broad spectrum disease resistance in tomato via CRISPER-cas9–mediated mutagenesis of DMR6 ortholog.
6.2.2. RNA Interference (RNAi)
RNAi is a gene-silencing approach followed to inhibit or silence the gene regulation by inhibiting the functional aspect of target mRNA. During this process, ds RNA is processed into small interfering RNA (siRNA), which along with some specific proteins bind with the target mRNA and cleaves that specific viral or pathogenic mRNA to control the pathogenicity [118,119,120]. Although for the phytopathogen control, different gene-silencing approaches such as host-induced [121] or spray-induced [122] has been frequently followed.
In the recent past, the scientific community successfully utilized this RNAi technology to inhibit the pathogenicity or protect the crop from phytopathogens including fungi, virus, nematodes, etc. Previous authors have reported on the role of RNAi technology in controlling or inhibiting pathogenicity. Baum et al. [123] reported RNAi in the insect by feeding or injecting ds RNA. However, some of the authors targeted insect genes by expressing dsRNA in the transgenic plants [124,125]. Koch et al. [122] used RNAi technology to control the infection of Fusarium graminearum in barley by spraying CYP3-dsRNA. Mao et al. [126] enhanced the resistance in cotton plant against the cotton bollworm by expressing CYP6AE14 dsRNA. However, utilization of RNAi technology has certain limitations as a gene-silencing approach; for example, the RNAi tool may not be able to target nucleic noncoding RNA or the non-coding RNA present in the nucleus [127]. In addition to this, the RNAi tool may not be able to differentiate the target mRNA or not target the mRNA having very few similarities with the target RNA [128].
7. Future Prospects and Recent Developments
Microbiome engineering is a constantly developing field and is receiving attention from multiple disciplines thereby incorporating novel approaches. However, to engineer the microbiome, multiple hurdles have to be encountered based upon the feasibility and scale-up caliber of the approach. The following section has tried to uncover the newer approaches and their limitations in microbiome engineering.
7.1. Role of Exudate Metabolites
Plants and microbes have co-evolved. The reciprocal benefit to plant and microbes is mediated by means of secondary metabolites. Plants secrete a number of volatile compounds, phytohormones, signaling molecules, extracellular enzymes, and surface receptors in their root exudate, possessing the potential to interfere with their microbial interaction in a positive direction [129]. The potential of root exudates can be harnessed to engineer a rhizospheric microbiome and ultimately increase plant productivity. The selected addition of root exudate can specify the microbiome growth and nutrition acquisition direction [130]. In their study, Jin et al. [131] deciphered that maize seedling exudates promoted growth and reduced oxidative stress in PGPB Pseudomonas fluorescens. However, this approach is associated with certain limitations including difficulty in scale-up and identification of specific metabolites in the root exudate.
7.2. Use of Soil Probiotics
Various research is continuously being conducted to understand the impact of probiotics on the human gut microbiome [132]. Similar studies on the impact of external microbial inoculation on the soil and plant microbiome is warranted. PGPM inoculation is a traditional method to increase plant productivity and control plant pathogens [133]; however, their substantial role in microbiome modulation has not been studied much. Recently, Liu et al. [134] demonstrated that inoculation of Rhodobacter sphaeroides and Bacillus amyloliquefaciens in oilseed rape rhizosphere increased the growth of bacteria belonging to Pseudomonadaceae and Flavobacteriaceae, leading to better soil nutrient cycling. Similarly, the inoculation of Burkholderia vietnamiensis modified soil microbiota and controlled root-knot nematode infestation in watermelon [135]. Nevertheless, this approach is also associated with traditional challenges of competition from the native microbiota. Alternatively, combined approaches of root exudate and PGPR inoculation can be used for a better adaptation to a localized niche.
Increasing agriculture productivity under the pressure of climate change has asked to add another dimension in microbiome modulation. A prompt understanding between synthetic biology, microbiome engineering, and ecology will not only help in climate conservation, but will also increase plant productivity. Harnessing cultured and non-cultured resilient microbes combined with climate smart agriculture can help in combating emerging climatic biotic and abiotic stresses [135]. Epstein et al. [136] explained the role of microbiome modulation in harboring climate resilience in corals, providing some inputs for microbiome engineering in the soil and plant rhizosphere.
7.3. Optimization of Inoculant Size and Growth Conditions Under In Situ Conditions
Microbiome manipulation is directly correlated with the inoculum size and factors encountered in the specialized niche [9]. Whether artificial microbial consortia, traditional soil amendments, microbiome breeding [137] and transplantation of the microbiome, or modification of root architecture and exudates, every approach needs to be optimized for successful application [138,139,140]. Seed bio-priming [141], seed coating, encapsulation, nanoformulations [142], and in situ microbiome engineering are some of the latest approaches used for sustained delivery of microbial formulations [143,144]. Nevertheless, further research is required for homogenous optimization of microbial inoculation.
7.4. Role of NGS, Metagenomics of Uncultured Microbiome, and Use of Genetic Tools
Recent advancements in molecular biology can offer important strategies in microbiome modulation. The firm knowledge of microbiome molecular ecology and interactions is necessary for successful manipulation. Next-generation sequencing (NGS) for studying microbial transcriptome, proteome, and metabolome has revealed dynamic changes during plant–microbe interactions [145,146,147]. High-throughput sequencing of PGPB Pseudomonas putida RA-responsive microRNAs and Bacillus amyloliquefaciens revealed their role in the growth and development of Arabidopsis and the growth of rice under stress [147,148]. Some of the microbes are difficult to culture under laboratory conditions. Metagenomics approaches including high-throughput sequencing, marker gene tags (iTAG), amplicon sequencing, and in situ metagenomics engineering can help in understanding the complexity level and intra-community level interactions [94,149]. Further, genetic editing tools such as miRNA, siRNA-based tools, RNAi, CRISPR/CAS9, and multiomics approaches can be combined with microbiome engineering for increasing plant productivity.
8. Conclusions
In recent decades, to ensure food security for the rising global population, enhancement in agricultural productivity and improved plant performance under stress conditions are some of the main goals. The better understanding of plant–microbe interactions can be a vital option to use the beneficial microbial species as plant or soil inoculants or their use to modulate rhizosphere functioning. Although the isolation, characterization, and selection of beneficial microbial species is complex process, the latest omics and technology make this hurdle easier and help researchers in the identification of microbial communities and their hidden potential. But survivability of the microbial strains in the field trail is one of the challenging tasks. The identification of key genes, microbial engineering, and rhizosphere engineering could be emerging as one of the sustainable solutions to enhance plant performance.
Author Contributions
Conceptualization, A.K. (Ajay Kumar); methodology, M.K.S. and A.K. (Ajay Kumar); software, D.J., A.K. (Amit Kaushik) and V.K.S.; validation, formal analysis and A.K. (Amit Kaushik); investigation, V.K.S. and A.K. (Amit Kaushik); resources, A.K. (Amit Kaushik); writing—original draft preparation, D.J., A.K. (Amit Kaushik), R.K., A.A., G.S., N.K., V.K.S., M.K., N.B. and A.K. (Ajay Kumar); writing—review and editing, D.J., A.K. (Amit Kaushik), R.K., A.A., G.S., N.K., V.K.S., M.K., N.B., M.K.S. and A.K. (Ajay Kumar); visualization, A.K. (Ajay Kumar); supervision, A.K. (Ajay Kumar); funding acquisition, A.K. (Ajay Kumar);. All authors have read and agreed to the published version of the manuscript.
Funding
Author A.K. (Ajay Kumar), thanks to Amity University for funding acquisition.
Data Availability Statement
Data are available within the article.
Acknowledgments
Authors are thankful to Amity Institute of Biotechnology, Amity University, Noida, India, for providing lab facilities and no any authors have any conflict of interest.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Arif, I.; Batool, M.; Schenk, P.M. Plant Microbiome Engineering: Expected Benefits for Improved Crop Growth and Resilience. Trends Biotechnol. 2020, 38, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Afridi, M.S.; Ali, S.; Salam, A.; César Terra, W.; Hafeez, A.; Sumaira; Ali, B.; AlTami, M.S.; Ameen, F.; Ercisli, S.; et al. Plant Microbiome Engineering: Hopes or Hypes. Biology 2022, 11, 1782. [Google Scholar] [CrossRef] [PubMed]
- Mesny, F.; Hacquard, S.; Thomma, B.P. Co-Evolution within the Plant Holobiont Drives Host Performance. EMBO Rep. 2023, 24, e57455. [Google Scholar] [CrossRef] [PubMed]
- Lyu, D.; Msimbira, L.A.; Nazari, M.; Antar, M.; Pagé, A.; Shah, A.; Monjezi, N.; Zajonc, J.; Tanney, C.A.; Backer, R.; et al. The Coevolution of Plants and Microbes Underpins Sustainable Agriculture. Microorganisms 2021, 9, 1036. [Google Scholar] [CrossRef]
- Glick, B.R.; Gamalero, E. Recent Developments in the Study of Plant Microbiomes. Microorganisms 2021, 9, 1533. [Google Scholar] [CrossRef]
- Compant, S.; Cassan, F.; Kostić, T.; Johnson, L.; Brader, G.; Trognitz, F.; Sessitsch, A. Harnessing the Plant Microbiome for Sustainable Crop Production. Nat. Rev. Microbiol. 2024, 23, 9–23. [Google Scholar] [CrossRef]
- Solomon, W.; Janda, T.; Molnár, Z. Unveiling the significance of rhizosphere: Implications for plant growth, stress response, and sustainable agriculture. Plant Physiol. Biochem. 2024, 206, 108290. [Google Scholar] [CrossRef]
- Kaul, S.; Choudhary, M.; Gupta, S.; Dhar, M.K. Engineering Host Microbiome for Crop Improvement and Sustainable Agriculture. Front. Microbiol. 2021, 12, 635917. [Google Scholar] [CrossRef]
- Solanki, M.K.; Joshi, N.C.; Singh, P.K.; Singh, S.K.; Santoyo, G.; de Azevedo, L.C.B.; Kumar, A. From Concept to Reality: Transforming Agriculture through Innovative Rhizosphere Engineering for Plant Health and Productivity. Microbiol. Res. 2023, 279, 127553. [Google Scholar] [CrossRef]
- Wang, Z.; Solanki, M.K.; Kumar, A.; Solanki, A.C.; Pang, F.; Ba, Z.X.; Niu, J.Q.; Ren, Z.X. Promoting Plant Resilience Against Stress by Engineering Root Microenvironment with Streptomyces Inoculants. Microbiol. Res. 2023, 277, 127509. [Google Scholar] [CrossRef]
- Lau, S.E.; Teo, W.F.A.; Teoh, E.Y.; Tan, B.C. Microbiome Engineering and Plant Biostimulants for Sustainable Crop Improvement and Mitigation of Biotic and Abiotic Stresses. Discov. Food 2022, 2, 9. [Google Scholar] [CrossRef]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial Interactions within the Plant Holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.; Abdul Rahman, N.S.N. Plant–Microbe Interaction: Aboveground to Belowground, from the Good to the Bad. Int. J. Mol. Sci. 2021, 22, 10388. [Google Scholar] [CrossRef] [PubMed]
- Mommer, L.; Hinsinger, P.; Prigent-Combaret, C.; Visser, E.J. Advances in the Rhizosphere: Stretching the Interface of Life. Plant Soil 2016, 407, 1–8. [Google Scholar] [CrossRef]
- Galindo-Castañeda, T.; Hartmann, M.; Lynch, J.P. Location: Root Architecture Structures Rhizosphere Microbial Associations. J. Exp. Bot. 2024, 75, 594–604. [Google Scholar] [CrossRef]
- Hartman, K.; van der Heijden, M.G.; Roussely-Provent, V.; Walser, J.C.; Schlaeppi, K. Deciphering Composition and Function of the Root Microbiome of a Legume Plant. Microbiome 2017, 5, 2. [Google Scholar] [CrossRef]
- Yuan, T.; Ren, W.; Wang, Z.; Fry, E.L.; Tang, S.; Yin, J.; Zhang, J.; Jia, Z. How Does the Pattern of Root Metabolites Regulating Beneficial Microorganisms Change with Different Grazing Pressures? Front. Plant Sci. 2023, 14, 1180576. [Google Scholar] [CrossRef]
- Kumar, C.; Esposito, A.; Bertani, I.; Musonerimana, S.; Midekssa, M.J.; Tesfaye, K.; Derr, D.C.; Donaldson, L.; Piazza, S.; Bez, C.; et al. Sorghum rhizosphere bacteriome studies and generation of multistrain beneficial bacterial consortia. Microbiol. Res. 2025, 292, 128036. [Google Scholar] [CrossRef]
- Chai, Y.N.; Schachtman, D.P. Root Exudates Impact Plant Performance under Abiotic Stress. Trends Plant Sci. 2022, 27, 80–91. [Google Scholar] [CrossRef]
- Brummell, D.A.; Dal Cin, V.; Crisosto, C.H.; Labavitch, J.M. Cell Wall Metabolism During Maturation, Ripening and Senescence of Peach Fruit. J. Exp. Bot. 2004, 55, 2029–2039. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zuo, S.; Xu, L.; Zou, Y.; Song, W. Study on Diversity of Endophytic Bacterial Communities in Seeds of Hybrid Maize and Their Parental Lines. Arch. Microbiol. 2012, 194, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; Hardoim, C.C.; van Overbeek, L.S.; van Elsas, J.D. Dynamics of Seed-Borne Rice Endophytes on Early Plant Growth Stages. PLoS ONE 2012, 7, e30438. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Yan, Y.; Li, X.; Liu, Y.; Lu, X. Rhizosphere Soil Microbial Community and Its Response to Different Utilization Patterns in the Semi-Arid Alpine Grassland of Northern Tibet. Front. Microbiol. 2022, 13, 931795. [Google Scholar] [CrossRef]
- Nan, J.; Chao, L.; Ma, X.; Xu, D.; Mo, L.; Zhang, X.; Zhao, X.; Bao, Y. Microbial Diversity in the Rhizosphere Soils of Three Stipa Species from the Eastern Inner Mongolian Grasslands. Glob. Ecol. Conserv. 2020, 22, e00992. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Zhang, P.; Trivedi, P.; Riera, N.; Wang, Y.; Liu, X.; Fan, G.; Tang, J.; Coletta-Filho, H.D.; et al. The Structure and Function of the Global Citrus Rhizosphere Microbiome. Nat. Commun. 2018, 9, 4894. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.; Schenk, P.M. Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Sessitsch, A.; Hardoim, P.; Döring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional Characteristics of an Endophyte Community Colonizing Rice Roots as Revealed by Metagenomic Analysis. Mol. Plant Microbe Interact. 2012, 25, 28–36. [Google Scholar] [CrossRef]
- Singh, R.; Pandey, K.D.; Singh, M.; Singh, S.K.; Hashem, A.; Al-Arjani, A.F.; Abd Allah, E.F.; Singh, P.K.; Kumar, A. Isolation and Characterization of Endophytes Bacterial Strains of Momordica charantia L. and Their Possible Approach in Stress Management. Microorganisms 2022, 10, 290. [Google Scholar] [CrossRef]
- Goulart, M.C.; Cueva-Yesquén, L.G.; Hidalgo Martinez, K.J.; Attili-Angelis, D.; Fantinatti-Garboggini, F. Comparison of Specific Endophytic Bacterial Communities in Different Developmental Stages of Passiflora incarnata Using Culture-Dependent and Culture-Independent Analysis. Microbiol. Open 2019, 8, e896. [Google Scholar] [CrossRef] [PubMed]
- Vincze, É.B.; Becze, A.; Laslo, É.; Mara, G. Beneficial Soil Microbiomes and Their Potential Role in Plant Growth and Soil Fertility. Agriculture 2024, 14, 152. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Agunbiade, V.F.; Babalola, O.O. Drought Stress Amelioration Attributes of Plant-Associated Microbiome on Agricultural Plants. Bioinform. Biol. Insights 2024, 18, 11779322241233442. [Google Scholar] [CrossRef]
- Mejia, G.; Jara-Servin, A.; Romero-Chora, L.; Hernandez-Alvarez, C.; Peimbert, M.; Cruz-Ortega, R.; Alcaraz, L.D. Rhizosphere Microbiome Influence on Tomato Growth under Low-Nutrient Settings. bioRxiv 2024. [Google Scholar] [CrossRef]
- Mukherjee, A.; Singh, B.N.; Kaur, S.; Sharma, M.; de Araújo, A.S.F.; de Araujo Pereira, A.P.; Morya, R.; Puopolo, G.; Moënne-Loccoz, Y.; Melo, V.M.M.; et al. Unearthing the Power of Microbes as Plant Microbiome for Sustainable Agriculture. Microbiol. Res. 2024, 286, 127780. [Google Scholar] [CrossRef]
- Leontidou, K.; Genitsaris, S.; Papadopoulou, A.; Kamou, N.; Bosmali, I.; Matsi, T.; Madesis, P.; Vokou, D.; Karamanoli, K.; Mellidou, I. Plant Growth Promoting Rhizobacteria Isolated from Halophytes and Drought-Tolerant Plants: Genomic Characterisation and Exploration of Phyto-Beneficial Traits. Sci. Rep. 2020, 10, 14857. [Google Scholar] [CrossRef]
- Yang, J.; Kloepper, J.W.; Ryu, C.M. Rhizosphere Bacteria Help Plants Tolerate Abiotic Stress. Trends Plant Sci. 2009, 14, 1–4. [Google Scholar] [CrossRef]
- Ning, Z.; Lin, K.; Gao, M.; Han, X.; Guan, Q.; Ji, X.; Yu, S.; Lu, L. Mitigation of Salt Stress in Rice by the Halotolerant Plant Growth-Promoting Bacterium Enterobacter asburiae D2. J. Xenobiotics 2024, 14, 333–349. [Google Scholar] [CrossRef]
- Khumairah, F.H.; Setiawati, M.R.; Fitriatin, B.N.; Simarmata, T.; Alfaraj, S.; Ansari, M.J.; Enshasy, H.A.E.; Sayyed, R.Z.; Najafi, S. Halotolerant Plant Growth-Promoting Rhizobacteria Isolated from Saline Soil Improve Nitrogen Fixation and Alleviate Salt Stress in Rice Plants. Front. Microbiol. 2022, 13, 905210. [Google Scholar] [CrossRef]
- Sarker, P.K.; Karmoker, D.; Shohan, M.U.S.; Saha, A.K.; Rima, F.S.; Begum, R.A.; Islam, M.R.; Seraj, Z.I. Effects of Multiple Halotolerant Rhizobacteria on the Tolerance, Growth, and Yield of Rice Plants under Salt Stress. Folia Microbiol. 2023, 68, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Kan, D.; Tian, M.; Ruan, Y. Phosphate-Solubilizing Bacteria Reshaped the Rhizosphere Microbiome and Metabolic Profile of Wheat to Inhibit Cd Absorption. Environ. Exp. Bot. 2024, 226, 105929. [Google Scholar] [CrossRef]
- Kour, D.; Sharma, B.; Negi, R.; Kumar, S.; Kaur, S.; Kaur, T.; Khan, S.S.; Kour, H.; Ramniwas, S.; Rustegi, S.; et al. Microbial Amelioration of Heavy Metal Toxicity in Plants for Agro-Environmental Sustainability. Water Air Soil Pollut. 2024, 235, 431. [Google Scholar] [CrossRef]
- Samaras, A.; Roumeliotis, E.; Ntasiou, P.; Karaoglanidis, G. Bacillus subtilis MBI600 Promotes Growth of Tomato Plants and Induces Systemic Resistance Contributing to the Control of Soilborne Pathogens. Plants 2021, 10, 1113. [Google Scholar] [CrossRef] [PubMed]
- Kamath, A.; Shukla, A.; Patel, D. Quorum Sensing and Quorum Quenching: Two Sides of the Same Coin. Physiol. Mol. Plant Pathol. 2023, 123, 101927. [Google Scholar] [CrossRef]
- Shreshtha, A.; Schikora, A. AHL-Priming for Enhanced Resistance as a Tool in Sustainable Agriculture. FEMS Microbiol. Ecol. 2020, 96, fiaa226. [Google Scholar]
- Vesuna, A.P.; Nerurkar, A.S. Biocontrol Impact of AHL-Degrading Actinobacteria on Quorum Sensing-Regulated Virulence of Phytopathogen Pectobacterium carotovorum subsp. carotovorum BR1. Plant Soil 2020, 453, 371–388. [Google Scholar] [CrossRef]
- Ketehouli, T.; Pasche, J.; Buttrós, V.H.; Goss, E.M.; Martins, S.J. The Underground World of Plant Disease: Rhizosphere Dysbiosis Reduces Above-Ground Plant Resistance to Bacterial Leaf Spot and Alters Plant Transcriptome. Environ. Microbiol. 2024, 26, e16676. [Google Scholar] [CrossRef]
- Wahab, A.; Muhammad, M.; Munir, A.; Abdi, G.; Zaman, W.; Ayaz, A.; Khizar, C.; Reddy, S.P.P. Role of Arbuscular Mycorrhizal fungi in Regulating Growth, Enhancing Productivity, and Potentially Influencing Ecosystems under Abiotic and Biotic Stresses. Plants 2023, 12, 3102. [Google Scholar] [CrossRef]
- Revillini, D.; David, A.S.; Reyes, A.L.; Knecht, L.D.; Vigo, C.; Allen, P.; Searcy, C.A.; Afkhami, M.E. Allelopathy-Selected Microbiomes Mitigate Chemical Inhibition of Plant Performance. New Phytol. 2023, 240, 2007–2019. [Google Scholar] [CrossRef]
- Yang, L.; Wang, L.; Chu, J.; Zhao, H.; Zhao, J.; Zang, H.; Yang, Y.; Zeng, Z. Improving Soil Quality and Wheat Yield through Diversified Crop Rotations in the North China Plain. Soil Tillage Res. 2024, 244, 106231. [Google Scholar] [CrossRef]
- Gan, Y.; Hamel, C.; O’Donovan, J.T.; Cutforth, H.; Zentner, R.P.; Campbell, C.A.; Niu, Y.; Poppy, L. Diversifying Crop Rotations with Pulses Enhances System Productivity. Sci. Rep. 2015, 5, 14625. [Google Scholar] [CrossRef] [PubMed]
- Venter, Z.S.; Jacobs, K.; Hawkins, H.J. The Impact of Crop Rotation on Soil Microbial Diversity: A Meta-Analysis. Pedobiologia 2016, 59, 215–223. [Google Scholar] [CrossRef]
- Shu, X.; He, J.; Zhou, Z.; Xia, L.; Hu, Y.; Zhang, Y.; Zhang, Y.; Luo, Y.; Chu, H.; Liu, W.; et al. Organic Amendments Enhance Soil Microbial Diversity, Microbial Functionality and Crop Yields: A Meta-Analysis. Sci. Total Environ. 2022, 829, 154627. [Google Scholar] [CrossRef]
- Purohit, H.J.; Pandit, P.; Pal, R.; Warke, R.; Warke, G.M. Soil Microbiome: An Intrinsic Driver for Climate Smart Agriculture. J. Agric. Food Res. 2024, 18, 101433. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial Diversity Drives Multifunctionality in Terrestrial Ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef]
- Sun, X.L.; Zhao, J.; You, Y.M.; Sun, O.J. Soil Microbial Responses to Forest Floor Litter Manipulation and Nitrogen Addition in a Mixed-Wood Forest of Northern China. Sci. Rep. 2016, 6, 19536. [Google Scholar] [CrossRef]
- Rodríguez-Berbel, N.; Soria, R.; Ortega, R.; Lucas-Borja, M.E.; Miralles, I. Benefits of Applying Organic Amendments from Recycled Wastes for Fungal Community Growth in Restored Soils of a Limestone Quarry in a Semiarid Environment. Sci. Total Environ. 2022, 806, 151226. [Google Scholar] [CrossRef]
- Xu, S.; Yuan, M.; Chapman, S.J.; Zheng, N.; Yao, H.; Kuzyakov, Y. Bio-Converted Organic Wastes Shape Microbiota in Maize Rhizosphere: Localization and Identification in Enzyme Hotspots. Soil Biol. Biochem. 2023, 184, 109105. [Google Scholar] [CrossRef]
- Shi, C.H.; Wang, X.Q.; Jiang, S.; Zhang, L.Q.; Luo, J. Revealing the Role of the Rhizosphere Microbiota in Reproductive Growth for Fruit Productivity when Inorganic Fertilizer is Partially Replaced by Organic Fertilizer in Pear Orchard Fields. Microb. Biotechnol. 2023, 16, 1373–1392. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.; Liu, M.; Xu, J.; Bian, H.; Chen, T.; You, E.; Deng, C.; Wei, Y.; Yang, T.; et al. Effects of Different Fertilization Conditions and Different Geographical Locations on the Diversity and Composition of the Rhizosphere Microbiota of Qingke (Hordeum vulgare L.) Plants in Different Growth Stages. Front. Microbiol. 2023, 14, 1094034. [Google Scholar]
- Ayangbenro, A.S.; Chukwuneme, C.F.; Ayilara, M.S.; Kutu, F.R.; Khantsi, M.; Adeleke, B.S.; Glick, B.R.; Babalola, O.O. Harnessing the Rhizosphere Soil Microbiome of Organically Amended Soil for Plant Productivity. Agronomy 2022, 12, 3179. [Google Scholar] [CrossRef]
- Lahbouki, S.; Hashem, A.; Kumar, A.; Abd_Allah, E.F.; Meddich, A. Integration of Horse Manure Vermicompost Doses and Arbuscular Mycorrhizal Fungi to Improve Fruit Quality and Soil Fertility in Tomato Field Facing Drought Stress. Plants 2024, 13, 1449. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fan, W.; Xu, G.; Cao, Y.; Zhao, X.; Hao, S.; Deng, B.; Ren, S.; Hu, S. Bio-Organic Fertilizers Improve Dendrocalamus farinosus Growth by Remolding the Soil Microbiome and Metabolome. Front. Microbiol. 2023, 14, 1117355. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, T.; Mei, X.; Wang, N.; Li, X.; Yang, Q.; Dong, C.; Jiang, G.; Lin, J.; Xu, Y.; et al. Bio-Organic Fertilizer Promotes Pear Yield by Shaping the Rhizosphere Microbiome Composition and Functions. Microbiol. Spectr. 2022, 10, e03572-22. [Google Scholar] [CrossRef]
- Govaerts, B.; Mezzalama, M.; Sayre, K.D.; Crossa, J.; Lichter, K.; Troch, V.; Vanherck, K.; De Corte, P.; Deckers, J. Long-Term Consequences of Tillage, Residue Management, and Crop Rotation on Selected Soil Micro-Flora Groups in the Subtropical Highlands. Appl. Soil Ecol. 2008, 38, 197–210. [Google Scholar] [CrossRef]
- Shanmugam, G.S.; Buehring, N.W.; Prevost, J.D.; Kingery, W.L. Soil Bacterial Community Diversity and Composition as Affected by Tillage Intensity Treatments in Corn-Soybean Production Systems. Microbiol. Res. 2021, 12, 157–172. [Google Scholar] [CrossRef]
- Dewi, R.K.; Huang, Q.; Hashimi, R.; Komatsuzaki, M. Enhancing agroecosystem sustainability: Integrative soil health strategies in regenerative organic soybean production on Andosol in Japan. Geoderma Reg. 2025, 40, e00910. [Google Scholar] [CrossRef]
- Hu, X.; Liu, J.; Liang, A.; Gu, H.; Liu, Z.; Jin, J.; Wang, G. Soil metagenomics reveals reduced tillage improves soil functional profiles of carbon, nitrogen, and phosphorus cycling in bulk and rhizosphere soils. Agric. Ecosyst. Environ. 2025, 379, 109371. [Google Scholar] [CrossRef]
- Yang, D.; Liu, Y.; Wang, Y.; Gao, F.; Zhao, J.; Li, Y.; Li, X. Effects of soil tillage, management practices, and mulching film application on soil health and peanut yield in a continuous cropping system. Front. Microbiol. 2020, 11, 570924. [Google Scholar] [CrossRef]
- Sessitsch, A.; Wakelin, S.; Schloter, M.; Maguin, E.; Cernava, T.; Champomier-Verges, M.C.; Charles, T.C.; Cotter, P.D.; Ferrocino, I.; Kriaa, A.; et al. Microbiome Interconnectedness Throughout Environments with Major Consequences for Healthy People and a Healthy Planet. Microbiol. Mol. Biol. Rev. 2023, 87, e00212-22. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, M.; Ballard, R.A.; Wright, D. Soil Microbial Inoculants for Sustainable Agriculture: Limitations and Opportunities. Soil Use Manag. 2022, 38, 1340–1369. [Google Scholar] [CrossRef]
- Beltran-Medina, I.; Romero-Perdomo, F.; Molano-Chavez Gutiérrez, A.Y.; Silva, A.M.; Estrada-Bonilla, G. Inoculation of phosphate-solubilizing bacteria improves soil phosphorus mobilization and maize productivity. Nutr. Cycl. Agroecosyst. 2023, 126, 21–34. [Google Scholar] [CrossRef]
- Nunes, P.S.; Junior, G.V.L.; Mascarin, G.M.; Guimarães, R.A.; Medeiros, F.H.; Arthurs, S.; Bettiol, W. Microbial Consortium of Biological Products: Do They Have a Future? Biol. Control 2024, 188, 105439. [Google Scholar] [CrossRef]
- Cao, Z.; Yan, W.; Ding, M.; Yuan, Y. Construction of Microbial Consortia for Microbial Degradation of Complex Compounds. Front. Bioeng. Biotechnol. 2022, 10, 1051233. [Google Scholar] [CrossRef]
- Wang, Y.; Dall’Agnol, R.F.; Bertani, I.; Bez, C.; Venturi, V. Identification of Synthetic Consortia from a Set of Plant-Beneficial Bacteria. Microb. Biotechnol. 2024, 17, e14330. [Google Scholar] [CrossRef]
- Manjunath, M.; Khokhar, A.; Chary, G.R.; Singh, M.; Yadav, S.K.; Gopinath, K.A.; Jyothilakshmi, N.; Srinivas, K.; Prabhakar, M.; Singh, V.K. Microbial Consortia Enhance the Yield of Maize under Sub-Humid Rainfed Production System of India. Front. Sustain. Food Syst. 2023, 7, 1108492. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Babalola, O.O. Bacterial Consortium for Improved Maize (Zea mays L.) Production. Microorganisms 2019, 7, 519. [Google Scholar] [CrossRef]
- Yin, C.; Hagerty, C.H.; Paulitz, T.C. Synthetic Microbial Consortia Derived from Rhizosphere Soil Protect Wheat Against a Soilborne Fungal Pathogen. Front. Microbiol. 2022, 13, 908981. [Google Scholar] [CrossRef]
- Dessaux, Y.; Grandclément, C.; Faure, D. Engineering the Rhizosphere. Trends Plant Sci. 2016, 21, 266–278. [Google Scholar] [CrossRef]
- Pantigoso, H.A.; Newberger, D.; Vivanco, J.M. The Rhizosphere Microbiome: Plant-Microbial Interactions for Resource Acquisition. J. Appl. Microbiol. 2022, 133, 2864–2876. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P. Harnessing root architecture to address global challenges. Plant J. 2022, 109, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Zhong, V.; Archibald, B.N.; Brophy, J.A.N. Transcriptional and post-transcriptional controls for tuning gene expression in plants. Curr. Opin. Plant Biol. 2023, 71, 102315. [Google Scholar] [CrossRef] [PubMed]
- Yaschenko, A.E.; Fenech, M.; Mazzoni-Putman, S.; Alonso, J.M.; Stepanova, A.N. Deciphering the molecular basis of tissue-specific gene expression in plants: Can synthetic biology help? Curr. Opin. Plant Biol. 2022, 68, 102241. [Google Scholar] [CrossRef]
- Lee, J.Y.; Colinas, J.; Wang, J.Y.; Mace, D.; Ohler, U.; Benfey, P.N. Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2006, 103, 6055–6060. [Google Scholar] [CrossRef]
- Postma, J.A.; Dathe, A.; Lynch, J.P. The optimal lateral root branching density for maize depends on nitrogen and phosphorus availability. Plant Physiol. 2014, 166, 590–602. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Shen, Q.; Huang, X.-F.; Vivanco, J.M.; Reardon, K.F.; Zhang, R. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar]
- Cao, Y.; Zhang, Z.; Ling, N.; Yuan, Y.; Zheng, X.; Shen, B.; Shen, Q. Bacillus subtilis SQR 9 can control Fusarium wilt in cucumber by colonizing plant roots. Biol. Fertil. Soils 2011, 47, 495–506. [Google Scholar] [CrossRef]
- Kumar, A.; Dubey, A. Rhizosphere Microbiome: Engineering Bacterial Competitiveness for Enhancing Crop Production. J. Adv. Res. 2020, 24, 337–352. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev. 2019, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhuang, J.; Zhou, C.; Li, H.; Liu, C.; Liu, C.; Liu, W. CRISPR/Cas9-Mediated Genome Editing of the Filamentous Fungus Neurospora crassa. Methods Mol. Biol. 2019, 1620, 145–159. [Google Scholar]
- Bano, S.; Wu, X.; Zhang, X. Towards Sustainable Agriculture: Rhizosphere Microbiome Engineering. Appl. Microbiol. Biotechnol. 2021, 105, 7141–7160. [Google Scholar] [CrossRef]
- Phour, M.; Sindhu, S.S. Mitigating Abiotic Stress: Microbiome Engineering for Improving Agricultural Production and Environmental Sustainability. Planta 2022, 256, 85. [Google Scholar] [CrossRef]
- Albright, M.B.; Louca, S.; Winkler, D.E.; Feeser, K.L.; Haig, S.J.; Whiteson, K.L.; Emerson, J.B.; Dunbar, J. Solutions in Microbiome Engineering: Prioritizing Barriers to Organism Establishment. ISME J. 2022, 16, 331–338. [Google Scholar] [CrossRef]
- Mahmud, K.; Missaoui, A.; Lee, K.; Ghimire, B.; Presley, H.W.; Makaju, S. Rhizosphere Microbiome Manipulation for Sustainable Crop Production. Curr. Plant Biol. 2021, 27, 100210. [Google Scholar] [CrossRef]
- Ke, J.; Wang, B.; Yoshikuni, Y. Microbiome engineering: Synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2021, 39, 244–261. [Google Scholar] [CrossRef]
- Chesneau, G.; Herpell, J.; Garrido-Oter, R.; Hacquard, S. From synthetic communities to synthetic ecosystems: Exploring causalities in plant–microbe–environment interactions. New Phytol. 2025, 245, 496–502. [Google Scholar] [CrossRef]
- Shelake, R.M.; Pramanik, D.; Kim, J.Y. Exploration of plant-microbe interactions for sustainable agriculture in CRISPR era. Microorganisms 2019, 7, 269. [Google Scholar] [CrossRef]
- Prabhukarthikeyan, S.R.; Parameswaran, C.; Keerthana, U.; Teli, B.; Jagannadham, P.T.; Cayalvizhi, B.; Panneerselvam, P.; Senapati, A.; Nagendran, K.; Kumari, S.; et al. Understanding the plant-microbe interactions in CRISPR/Cas9 era: Indeed a sprinting start in marathon. Curr. Genom. 2020, 21, 429–443. [Google Scholar] [CrossRef] [PubMed]
- Brandt, K.; Barrangou, R. Applications of CRISPR technologies across the food supply chain. Annu. Rev. Food Sci. Technol. 2019, 10, 133–150. [Google Scholar] [CrossRef]
- Hendel, A.; Bak, R.O.; Clark, J.T.; Kennedy, A.B.; Ryan, D.E.; Roy, S.; Steinfeld, I.; Lunstad, B.D.; Kaiser, R.J.; Wilkens, A.B.; et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Giraud, T.; Gladieux, P.; Gavrilets, S. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol. Evol. 2010, 25, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Davierwala, A.P.; Reddy, A.P.K.; Lagu, M.D.; Ranjekar, P.K.; Gupta, V.S. Marker-assisted selection of bacterial blight resistance genes in rice. Biochem. Genet. 2001, 39, 261–278. [Google Scholar] [CrossRef]
- Muñoz, I.V.; Sarrocco, S.; Malfatti, L.; Baroncelli, R.; Vannacci, G. CRISPR-Cas for fungal genome editing: A new tool for the management of plant diseases. Front. Plant Sci. 2019, 10, 135. [Google Scholar] [CrossRef]
- Glandorf, D.C. Re-evaluation of biosafety questions on genetically modified biocontrol bacteria. Eur. J. Plant Pathol. 2019, 154, 43–51. [Google Scholar] [CrossRef]
- Susan, A.; Yadav, M.K.; Kar, S.; Aravindan, S.; Ngangkham, U.; Raghu, S.; Prabhukarthikeyan, S.R.; Keerthana, U.; Mukherjee, S.C.; Salam, J.L.; et al. Molecular identification of blast resistance genes in rice landraces from northeastern India. Plant Pathol. 2019, 68, 537–546. [Google Scholar] [CrossRef]
- Cao, Y.; Wu, Y.; Zheng, Z.; Song, F. Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiol. Mol. Plant Pathol. 2005, 67, 202–211. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-free genetically edited grapevine and apple protoplasts using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef] [PubMed]
- de Toledo Thomazella, D.P.; Brail, Q.; Dahlbeck, D.; Staskawicz, B. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv 2016. [Google Scholar] [CrossRef]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y. Citrus Canker Requires Targeting of a Susceptibility Gene by Specific TAL Effectors Present in Xanthomonas Citri; University of Florida: Gainesville, FL, USA, 2013. [Google Scholar]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef]
- Prihatna, C.; Barbetti, M.J.; Barker, S.J. A novel tomato fusarium wilt tolerance gene. Front. Microbiol. 2018, 9, 1226. [Google Scholar] [CrossRef]
- Niu, D.; Hamby, R.; Sanchez, J.N.; Cai, Q.; Yan, Q.; Jin, H. RNAs—A New Frontier in Crop Protection. Curr. Opin. Biotechnol. 2021, 70, 204–212. [Google Scholar] [CrossRef]
- Mezzetti, B.; Smagghe, G.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Kostov, K.; Sabbadini, S.; Opsahl-Sorteberg, H.G.; Ventura, V.; et al. RNAi: What is its position in agriculture? J. Pest Sci. 2020, 93, 1125–1130. [Google Scholar] [CrossRef]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Zotti, M.; Dos Santos, E.A.; Cagliari, D.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNA interference technology in crop protection against arthropod pests, pathogens, and nematodes. Pest Manag. Sci. 2018, 74, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Di Lelio, I.; Barra, E.; Coppola, M.; Corrado, G.; Rao, R.; Caccia, S. Transgenic plants expressing immunosuppressive dsRNA improve entomopathogen efficacy against Spodoptera littoralis larvae. J. Pest Sci. 2022, 95, 1413–1428. [Google Scholar] [CrossRef]
- Mao, J.; Zeng, F. Plant-mediated RNAi of a gap gene enhances tobacco tolerance against the Myzus persicae. Transgenic Res. 2014, 23, 145–152. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642. [Google Scholar] [CrossRef]
- Quiza, L.; St-Arnaud, M.; Yergeau, E. Harnessing Phytomicrobiome Signaling for Rhizosphere Microbiome Engineering. Front. Plant Sci. 2015, 6, 507. [Google Scholar] [CrossRef]
- Singh, R.K.; Singh, P.; Sharma, A.; Guo, D.J.; Upadhyay, S.K.; Song, Q.Q.; Verma, K.K.; Li, D.P.; Malviya, M.K.; Song, X.P.; et al. Unraveling Nitrogen Fixing Potential of Endophytic Diazotrophs of Different Saccharum Species for Sustainable Sugarcane Growth. Int. J. Mol. Sci. 2022, 23, 6242. [Google Scholar] [CrossRef]
- Jin, J.; Wang, M.; Lu, W.; Zhang, L.; Jiang, Q.; Jin, Y.; Lu, K.; Sun, S.; Cao, Q.; Wang, Y.; et al. Effect of Plants and Their Root Exudate on Bacterial Activities During Rhizobacterium–Plant Remediation of Phenol from Water. Environ. Int. 2019, 127, 114–124. [Google Scholar] [CrossRef]
- Hemarajata, P.; Versalovic, J. Effects of Probiotics on Gut Microbiota: Mechanisms of Intestinal Immunomodulation and Neuromodulation. Ther. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef]
- Mishra, S.; Mishra, A.; Chauhan, P.S.; Mishra, S.K.; Kumari, M.; Niranjan, A.; Nautiyal, C.S. Pseudomonas putida NBRIC19 Dihydrolipoamide Succinyltransferase (SucB) Gene Controls Degradation of Toxic Allelochemicals Produced by Parthenium hysterophorus. J. Appl. Microbiol. 2012, 112, 793–808. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, J.; Bai, Z.; Wu, S.; Li, X.; Wang, N.; Du, X.; Fan, H.; Zhuang, G.; Bohu, T.; et al. Unraveling Mechanisms and Impact of Microbial Recruitment on Oilseed Rape (Brassica napus L.) and the Rhizosphere Mediated by Plant Growth-Promoting Rhizobacteria. Microorganisms 2021, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Philp, J.; Wang, Y.; Hu, J.; Wei, Y.; Li, J.; Ryder, M.; Toh, R.; Zhou, Y.; Denton, M.D.; et al. Plant Growth-Promoting Rhizobacteria Burkholderia vietnamiensis B418 Inhibits Root-Knot Nematode on Watermelon by Modifying the Rhizosphere Microbial Community. Sci. Rep. 2022, 12, 8381. [Google Scholar] [CrossRef] [PubMed]
- Epstein, H.E.; Smith, H.A.; Torda, G.; van Oppen, M.J. Microbiome Engineering: Enhancing Climate Resilience in Corals. Front. Ecol. Environ. 2019, 17, 100–108. [Google Scholar] [CrossRef]
- Knief, C. Analysis of Plant-Microbe Interactions in the Era of Next Generation Sequencing Technologies. Front. Plant Sci. 2014, 5, 216. [Google Scholar] [CrossRef]
- Mueller, U.G.; Linksvayer, T.A. Microbiome breeding: Conceptual and practical issues. Trends Microbiol. 2022, 30, 997–1011. [Google Scholar] [CrossRef]
- Prasannath, K.; Peter Arulraj, S.; Shakthivel, K.; Rakulan, T.; Chandranath Karunarathna, S. Revisiting the ecological significance, composition, and functions of plant microbiome: A review. N. Z. J. Crop Hortic. Sci. 2025, 1–33. [Google Scholar] [CrossRef]
- Misu, I.J.; Kayess, M.O.; Siddiqui, M.N.; Gupta, D.R.; Islam, M.N.; Islam, T. Microbiome engineering for sustainable rice production: Strategies for biofertilization, stress tolerance, and climate resilience. Microorganisms 2025, 13, 233. [Google Scholar] [CrossRef]
- Singh, V.; Upadhyay, R.S.; Sarma, B.K.; Singh, H.B. Seed bio-priming with Trichoderma asperellum effectively modulates plant growth promotion in pea. Int. J. Agric. Environ. Biotechnol. 2016, 9, 361–365. [Google Scholar] [CrossRef]
- Rana, A.; Rani, A.; Nayana, K.R.; Deswal, S.; Singh, A.P.; Rana, S.; Chahar, M.; Singh, N.; Dhaka, R.K. Biotic stress alleviation in plants using rhizobacteria: An overview of mechanism of action, antimicrobial compounds production, (nano) formulations and employment methods. Indian J. Microbiol. 2025, 1–27. [Google Scholar] [CrossRef]
- Qiu, Z.; Egidi, E.; Liu, H.; Kaur, S.; Singh, B.K. New Frontiers in Agriculture Productivity: Optimised Microbial Inoculants and In Situ Microbiome Engineering. Biotechnol. Adv. 2019, 37, 107371. [Google Scholar] [CrossRef] [PubMed]
- Giri, V.P.; Pandey, S.; Kumari, M.; Tripathi, A.; Katiyar, R.; White, J.C.; Mishra, A. Hybridization of Chitosan and Biosynthesized Silver Nanoparticles to Enhance Antimicrobial Activity Against Phytopathogens in Tomato (Solanum lycopersicum). ACS Agric. Sci. Technol. 2022, 2, 719–733. [Google Scholar] [CrossRef]
- Tan, C.; Kalhoro, M.T.; Faqir, Y.; Ma, J.; Osei, M.D.; Khaliq, G. Climate-Resilient Microbial Biotechnology: A Perspective on Sustainable Agriculture. Sustainability 2022, 14, 5574. [Google Scholar] [CrossRef]
- Kumari, M.; Pandey, S.; Mishra, S.K.; Giri, V.P.; Agarwal, L.; Dwivedi, S.; Pandey, A.K.; Nautiyal, C.S.; Mishra, A. Omics-Based Mechanistic Insight into the Role of Bioengineered Nanoparticles for Biotic Stress Amelioration by Modulating Plant Metabolic Pathways. Front. Bioeng. Biotechnol. 2020, 8, 242. [Google Scholar] [CrossRef] [PubMed]
- Jatan, R.; Chauhan, P.S.; Lata, C. High-Throughput Sequencing and Expression Analysis Suggest the Involvement of Pseudomonas putida RA-Responsive miRNAs in Growth and Development of Arabidopsis. Int. J. Mol. Sci. 2020, 21, 5468. [Google Scholar] [CrossRef]
- Chauhan, P.S.; Lata, C.; Tiwari, S.; Chauhan, A.S.; Mishra, S.K.; Agrawal, L.; Chakrabarty, D.; Nautiyal, C.S. Transcriptional Alterations Reveal Bacillus amyloliquefaciens-Rice Cooperation Under Salt Stress. Sci. Rep. 2019, 9, 11912. [Google Scholar] [CrossRef]
- Hakim, S.; Nawaz, M.S.; Siddique, M.J.; Hayat, M.; Gulzar, U.; Imran, A. Metagenomics for Rhizosphere Engineering. In Rhizosphere Engineering; Academic Press: Cambridge, MA, USA, 2022; pp. 395–416. [Google Scholar]
- Lawson, C.E.; Harcombe, W.R.; Hatzenpichler, R.; Lindemann, S.R.; Löffler, F.E.; O’Malley, M.A.; García Martín, H.; Pfleger, B.F.; Raskin, L.; Venturelli, O.S.; et al. Common Principles and Best Practices for Engineering Microbiomes. Nat. Rev. Microbiol. 2019, 17, 725–741. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).