New Insights on the Glyphosate-Degrading Enzymes C-P Lyase and Glyphosate Oxidoreductase Based on Bioinformatics
Abstract
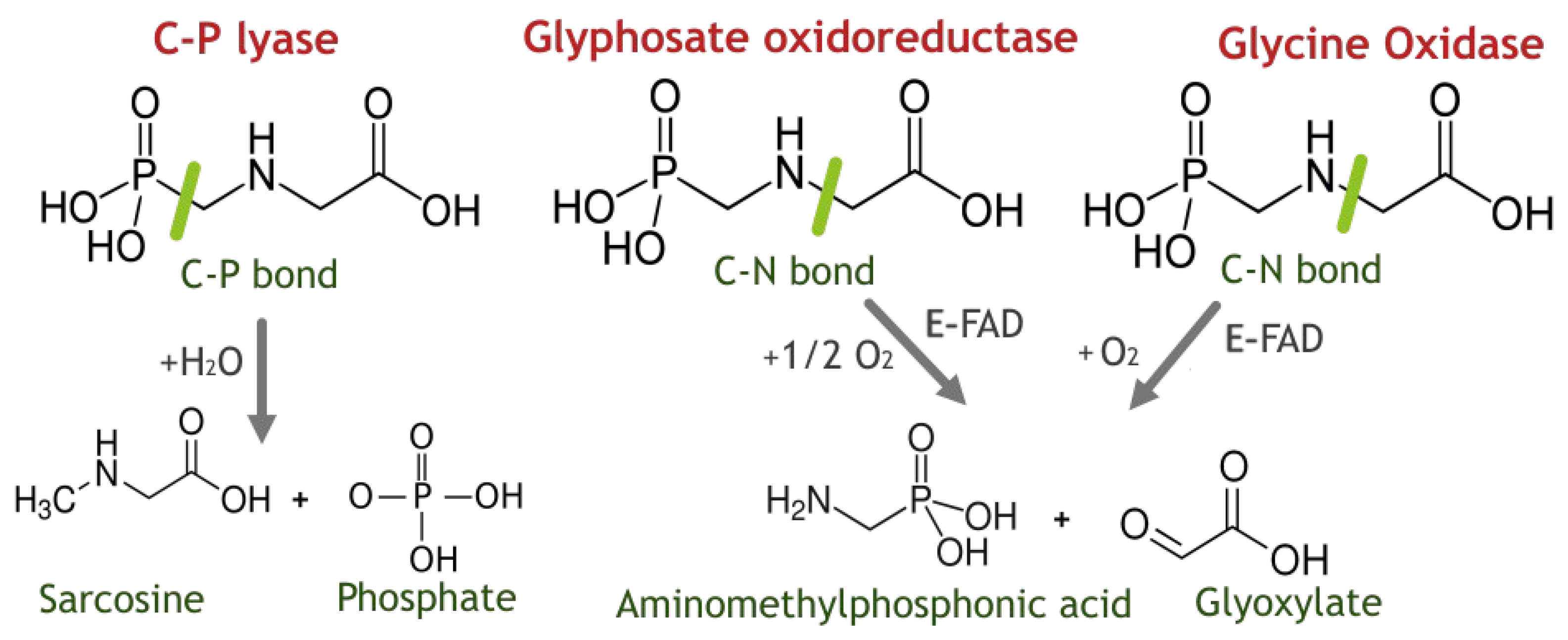
1. Introduction
- CMD: C-terminal mini domain (PhnJ);
- CID: Central insertion domain (PhnJ), which could participate in the binding of PhnK to PhnJ;
- BBD: Beta barrel domain (PhnI);
- NTD: N-terminal domain (PhnI).
2. Materials and Methods
- The nr database (BLASTp_nr), which includes all non-redundant GenBank CDS translations and also entries from PDB, SwissProt, PIR, and PRF, excluding environmental samples from WGS projects.
- RefSeq (BLASTp_ref), which contains only curated entries, i.e., one representative or “Select” transcript for every protein-coding gene, based on specific criteria [55].
3. Results and Discussion
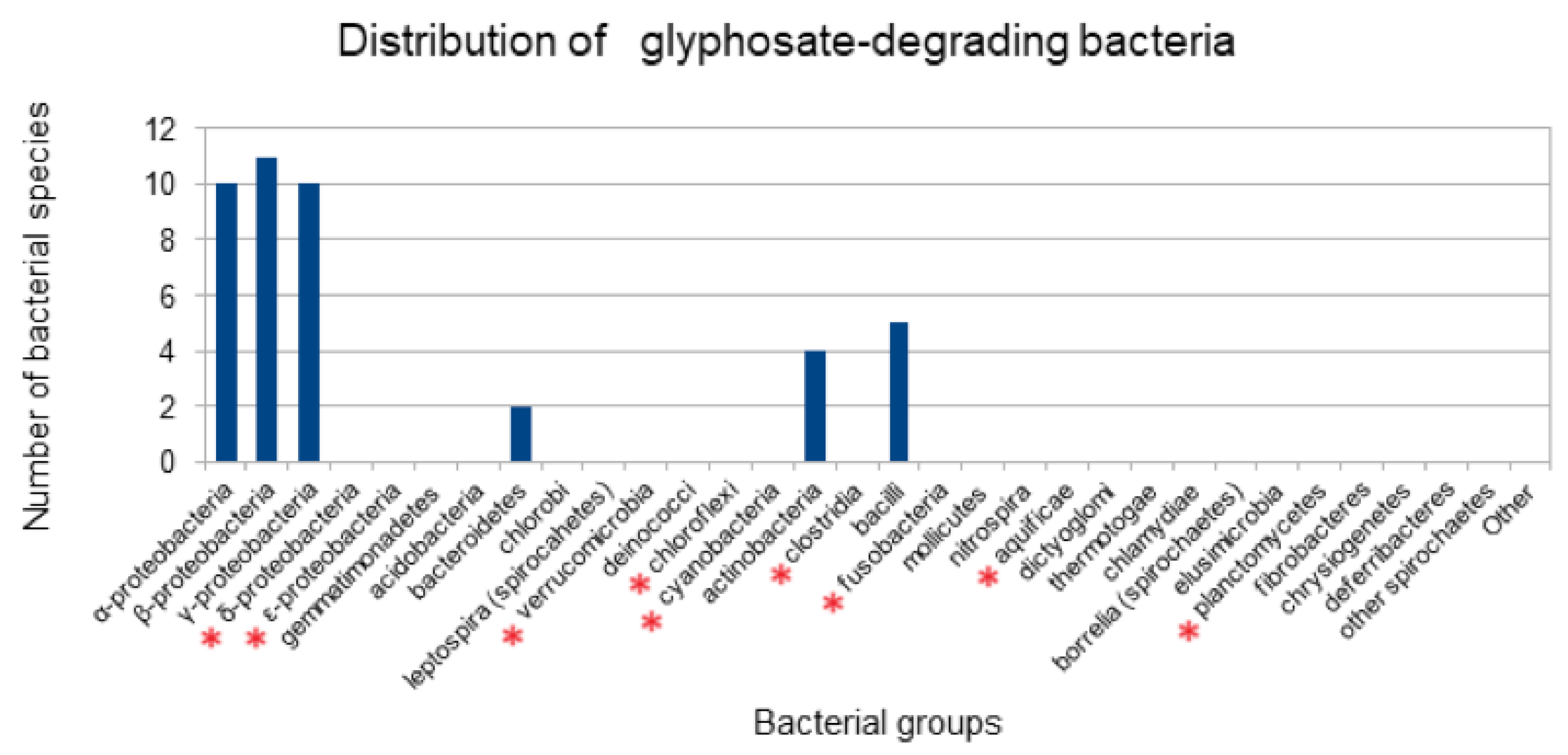
3.1. Distribution of Bacterial Species Previously Reported to Degrade Glyphosate
| Group | Genus, Species | Evidence | Products/Genes Detected | Reference |
|---|---|---|---|---|
| actinobacteria | Arthrobacter sp. GLP-1 | P | Sarcosine | [62] |
| actinobacteria | Streptomycete sp. StC | P | Sarcosine | [63] |
| actinobacteria | Streptomyces sp. StA | P | Sarcosine | [63] |
| actinobacteria | Rhodococcus soli G41 | G | soxB gene | [64] |
| actinomycetes | Arthrobacter atrocyaneus ATCC 13752 | P | AMPA | [65] |
| bacilli | Geobacillus caldoxylosilyticus T20 | P | AMPA | [66] |
| bacilli | Bacillus cereus CB4 | P | AMPA, glyoxylate, sarcosine, formaldehyde | [67] |
| bacilli | Lysinibacillus sphaericus | P | Free phosphorus concentration, glycine | [68] |
| bacilli | Bacillus aryabhattai FACU | G | goxB gene | [69] |
| bacilli | Bacillus. cereus 6 P | - | Conversion of glyphosate to polyphosphate | [70] |
| bacteroidetes | Flavobacterium sp. GD1 | P | AMPA | [33] |
| bacteroidetes | Chryseobacterium sp. Y16C | P | AMPA | [71] |
| αctinobacteria | Arthrobacter atrocyaneus ATCC 13752 | P | AMPA | [65] |
| α-proteobacteria | Agrobacterium radiobacter | P | AMPA | [33] |
| α-proteobacteria | Ochrobactrum anthropi GPK 3 | P | AMPA | [72] |
| α-proteobacteria | Ochrobactrum sp. GDOS | P | AMPA | [73] |
| α-proteobacteria | Ochrobactrum sp. G1 | G | gox gene | [42] |
| α-proteobacteria | Agrobacterium radiobacter SW9 | P | AMPA | [74] |
| α-proteobacteria | Rhizobiaceae meliloti 1021 | G | Homology to part of the phn gene cluster of E. coli | [75] |
| α-proteobacteria | Ochrobactrum sp. GDOS | P | AMPA | [73] |
| α-proteobacteria | Agrobacterium tumefaciens CHLDO | G | phn gene cluster | [76] |
| α-proteobacteria | Ochrobactrum haematophilum SR | - | - | |
| α-proteobacteria | Ochrobactrum intermedium Sq20 | G, P | Sarcosine, glycine, aroA gene (class II EPSPS) | [61] |
| β-proteobacteria | Comamonas odontotermitis P2 | G | gox and phnJ genes | [77] |
| β-proteobacteria | Achromobacter sp. strain MPK 7A | P | Sarcosine | [72] |
| β-proteobacteria | Achromobacter sp. Kg 16 | P | AcGP(N-acetylglyphosate), possibly AMPA | [72] |
| β-proteobacteria | Achromobacter Group V D (Agrobacterium sp. LW9) | P | AMPA | [74] |
| β-proteobacteria | Achromobacter sp. MPS 12A | P | Methane, sarcosine, glycine | [41] |
| β-proteobacteria | Alcaligenes sp. GL | P | Sarcosine | [78] |
| β-proteobacteria | Burkholderia vietnamiensis strain AQ5-12 | - | - | [79] |
| β-proteobacteria | Achromobacter denitrificans SOS5 | - | - | [76] |
| β-proteobacteria | Achromobacter insolitus SOR2 | - | - | [76] |
| β-proteobacteria | Achromobacter xylosoxidans SOS3 | - | - | [76] |
| β-proteobacteria | Achromobacter insolitus str Kg 19 (VKM B-3295) | G | phnJ gene | [80] |
| γ-proteobacteria | Pseudomonas sp. LBr | P | AMPA, a very small amount of glycine 5% | [81] |
| γ-proteobacteria | Providencia rettgeri GDB 1 | P | AMPA | [82] |
| γ-proteobacteria | Pseudomonas sp. 4ASW | P | Sarcosine | [83] |
| γ-proteobacteria | Enterobacter cloacae K7 | P | Sarcosine, glycine | [84] |
| γ-proteobacteria | Enterobacter sp. Bisph2 | - | - | [85] |
| γ-proteobacteria | Pseudomonas pseudomallei 22 | G, P | Phosphotransferase genes glpA and glpB | [86] |
| γ-proteobacteria | Pseudomonas sp. GLC11 | - | - | [87] |
| γ-proteobacteria | Pseudomonas sp. PG2982 | P | Sarcosine | [88] |
| γ-proteobacteria | Pseudomonas sp. SG-1 | Glyphosate concentration decreased | [89] | |
| γ-proteobacteria | Pseudomonas nitroreducens TR3 | - | - | [76] |
3.2. Protein Conservation across Prokaryotes
3.3. Conserved Residues in C-P Lyase
- cyb—Synechococcus sp. JA-2-3B’a(2-13) (bacteroidota)—Met269;
- muh—Mucilaginibacter celer HYN0043 (bacteroidota)—Leu276;
- shv—Salinicoccus halodurans H3B36 (bacilli)—Met269;
- crn—Carnobacterium sp. 17-4 (bacilli)—Met27.
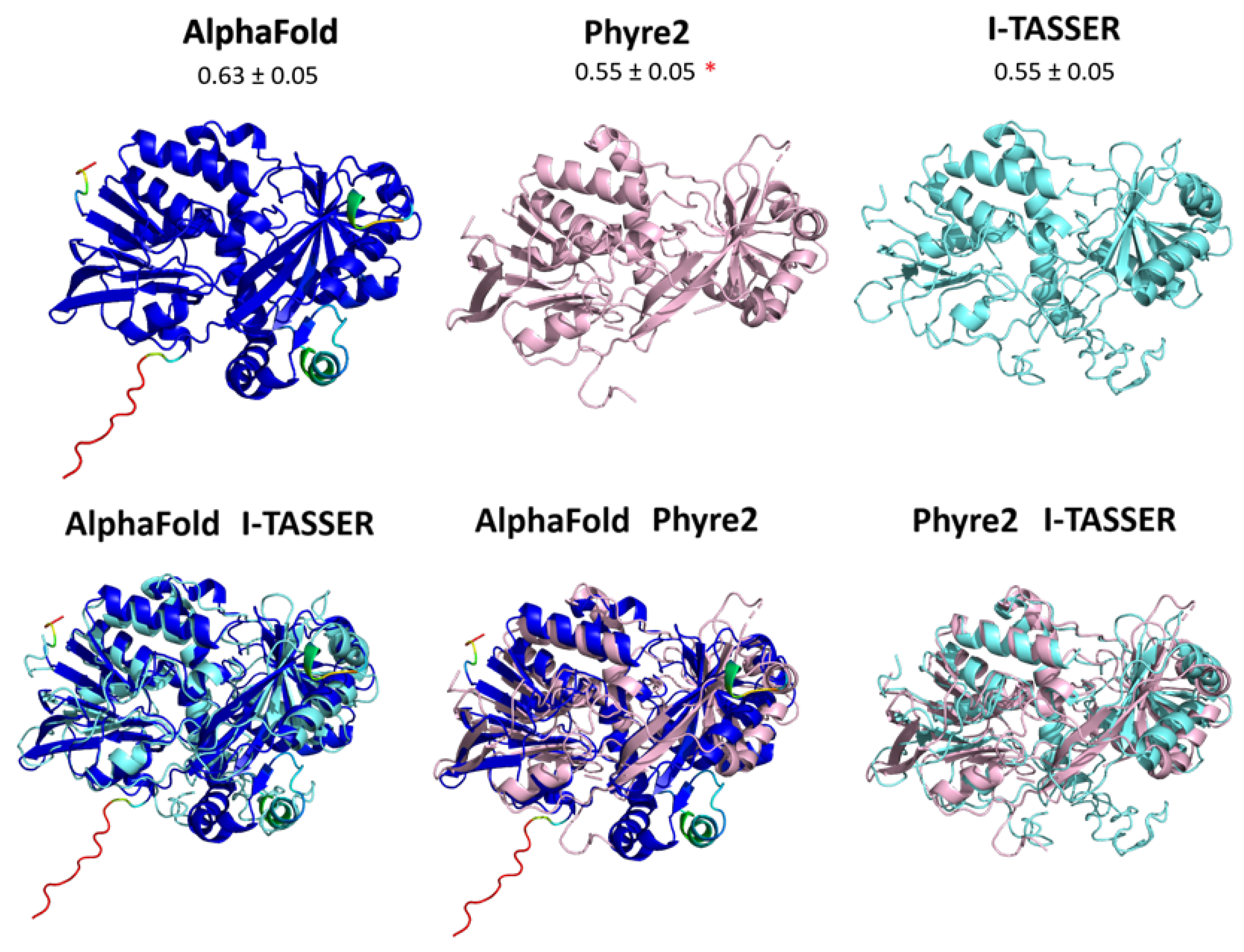
3.4. Structure of Gox
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- PubChem PubChem Compound Summary for CID 3496, Glyphosate. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/3496 (accessed on 10 June 2022).
- Pedotti, M.; Rosini, E.; Molla, G.; Moschetti, T.; Savino, C.; Vallone, B.; Pollegioni, L. Glyphosate Resistance by Engineering the Flavoenzyme Glycine Oxidase. J. Biol. Chem. 2009, 284, 36415–36423. [Google Scholar] [CrossRef] [PubMed]
- Kanissery, R.; Gairhe, B.; Kadyampakeni, D.; Batuman, O.; Alferez, F. Glyphosate: Its Environmental Persistence and Impact on Crop Health and Nutrition. Plants 2019, 8, 499. [Google Scholar] [CrossRef] [PubMed]
- EFSA EFSA Explains the Scientific Assessment of Glyphosate. Available online: https://www.efsa.europa.eu/en/factsheets/efsa-explains-scientific-assessment-glyphosate (accessed on 15 February 2024).
- Silva, V.; Mol, H.G.J.; Zomer, P.; Tienstra, M.; Ritsema, C.J.; Geissen, V. Pesticide Residues in European Agricultural Soils—A Hidden Reality Unfolded. Sci. Total Environ. 2019, 653, 1532–1545. [Google Scholar] [CrossRef]
- Tauchnitz, N.; Kurzius, F.; Rupp, H.; Schmidt, G.; Hauser, B.; Schrödter, M.; Meissner, R. Assessment of Pesticide Inputs into Surface Waters by Agricultural and Urban Sources—A Case Study in the Querne/Weida Catchment, Central Germany. Environ. Pollut. 2020, 267, 115186. [Google Scholar] [CrossRef]
- Bento, C.P.M.; van der Hoeven, S.; Yang, X.; Riksen, M.M.J.P.M.; Mol, H.G.J.; Ritsema, C.J.; Geissen, V. Dynamics of Glyphosate and AMPA in the Soil Surface Layer of Glyphosate-Resistant Crop Cultivations in the Loess Pampas of Argentina. Environ. Pollut. 2019, 244, 323–331. [Google Scholar] [CrossRef]
- Pelosi, C.; Bertrand, C.; Bretagnolle, V.; Coeurdassier, M.; Delhomme, O.; Deschamps, M.; Gaba, S.; Millet, M.; Nélieu, S.; Fritsch, C. Glyphosate, AMPA and Glufosinate in Soils and Earthworms in a French Arable Landscape. Chemosphere 2022, 301, 134672. [Google Scholar] [CrossRef]
- Baek, Y.; Bobadilla, L.K.; Giacomini, D.A.; Montgomery, J.S.; Murphy, B.P.; Tranel, P.J. Evolution of Glyphosate-Resistant Weeds. Rev. Environ. Contam. Toxicol. 2021, 255, 93–128. [Google Scholar] [CrossRef]
- Fuchs, B.; Saikkonen, K.; Damerau, A.; Yang, B.; Helander, M. Herbicide Residues in Soil Decrease Microbe-Mediated Plant Protection. Plant Biol. 2023, 25, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Kováčik, J.; Novotný, V.; Bujdoš, M.; Dresler, S.; Hladký, J.; Babula, P. Glyphosate Does Not Show Higher Phytotoxicity than Cadmium: Cross Talk and Metabolic Changes in Common Herb. J. Hazard. Mater. 2020, 383, 121250. [Google Scholar] [CrossRef]
- Mertens, M.; Höss, S.; Neumann, G.; Afzal, J.; Reichenbecher, W. Glyphosate, a Chelating Agent—Relevant for Ecological Risk Assessment? Environ. Sci. Pollut. Res. 2018, 25, 5298–5317. [Google Scholar] [CrossRef]
- Smith, D.F.Q.; Camacho, E.; Thakur, R.; Barron, A.J.; Dong, Y.; Dimopoulos, G.; Broderick, N.A.; Casadevall, A. Glyphosate Inhibits Melanization and Increases Susceptibility to Infection in Insects. PLoS Biol. 2021, 19, e3001182. [Google Scholar] [CrossRef] [PubMed]
- Motta, E.V.S.; Raymann, K.; Moran, N.A. Glyphosate Perturbs the Gut Microbiota of Honey Bees. Proc. Natl. Acad. Sci. USA 2018, 115, 10305–10310. [Google Scholar] [CrossRef] [PubMed]
- Motta, E.V.S.; Mak, M.; De Jong, T.K.; Powell, J.E.; O’Donnell, A.; Suhr, K.J.; Riddington, I.M.; Moran, N.A. Oral or Topical Exposure to Glyphosate in Herbicide Formulation Impacts the Gut Microbiota and Survival Rates of Honey Bees. Appl. Environ. Microbiol. 2020, 86, e01150-20. [Google Scholar] [CrossRef] [PubMed]
- Blot, N.; Veillat, L.; Rouzé, R.; Delatte, H. Glyphosate, but Not Its Metabolite AMPA, Alters the Honeybee Gut Microbiota. PLoS ONE 2019, 14, e0215466. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.J.; Stout, J.C.; Stanley, D.A. Contrasting Effects of Fungicide and Herbicide Active Ingredients and Their Formulations on Bumblebee Learning and Behaviour. J. Exp. Biol. 2023, 226, jeb245180. [Google Scholar] [CrossRef]
- Nouvian, M.; Foster, J.J.; Weidenmüller, A. Glyphosate Impairs Aversive Learning in Bumblebees. Sci. Total Environ. 2023, 898, 165527. [Google Scholar] [CrossRef]
- Liu, Z.; Shangguan, Y.; Zhu, P.; Sultan, Y.; Feng, Y.; Li, X.; Ma, J. Developmental Toxicity of Glyphosate on Embryo-Larval Zebrafish (Danio Rerio). Ecotoxicol. Environ. Saf. 2022, 236, 113493. [Google Scholar] [CrossRef]
- Lu, J.; Wang, W.; Zhang, C.; Xu, W.; Chen, W.; Tao, L.; Li, Z.; Cheng, J.; Zhang, Y. Characterization of Glyphosate-Induced Cardiovascular Toxicity and Apoptosis in Zebrafish. Sci. Total Environ. 2022, 851, 158308. [Google Scholar] [CrossRef]
- Faria, M.; Bedrossiantz, J.; Ramírez, J.R.R.; Mayol, M.; García, G.H.; Bellot, M.; Prats, E.; Garcia-Reyero, N.; Gómez-Canela, C.; Gómez-Oliván, L.M.; et al. Glyphosate Targets Fish Monoaminergic Systems Leading to Oxidative Stress and Anxiety. Environ. Int. 2021, 146, 106253. [Google Scholar] [CrossRef]
- Abdelmagid, A.D.; Said, A.M.; Abd El-Gawad, E.A.; Shalaby, S.A.; Dawood, M.A.O. Glyphosate-Induced Liver and Kidney Dysfunction, Oxidative Stress, Immunosuppression in Nile Tilapia, but Ginger Showed a Protection Role. Vet. Res. Commun. 2023, 47, 445–455. [Google Scholar] [CrossRef]
- Sabio y García, C.A.; Schiaffino, M.R.; Lozano, V.L.; Vera, M.S.; Ferraro, M.; Izaguirre, I.; Pizarro, H. New Findings on the Effect of Glyphosate on Autotrophic and Heterotrophic Picoplankton Structure: A Microcosm Approach. Aquat. Toxicol. 2020, 222, 105463. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lin, X.; Li, L.; Lin, S. Differential Growth Responses of Marine Phytoplankton to Herbicide Glyphosate. PLoS ONE 2016, 11, e0151633. [Google Scholar] [CrossRef]
- Duforestel, M.; Nadaradjane, A.; Bougras-Cartron, G.; Briand, J.; Olivier, C.; Frenel, J.S.; Vallette, F.M.; Lelièvre, S.A.; Cartron, P.F. Glyphosate Primes Mammary Cells for Tumorigenesis by Reprogramming the Epigenome in a TET3-Dependent Manner. Front. Genet. 2019, 10, 885. [Google Scholar] [CrossRef]
- Woźniak, E.; Reszka, E.; Jabłońska, E.; Balcerczyk, A.; Broncel, M.; Bukowska, B. Glyphosate Affects Methylation in the Promoter Regions of Selected Tumor Suppressors as Well as Expression of Major Cell Cycle and Apoptosis Drivers in PBMCs (in Vitro Study). Toxicol. Vitr. 2020, 63, 104736. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, E.; Reszka, E.; Jabłońska, E.; Michałowicz, J.; Huras, B.; Bukowska, B. Glyphosate and Ampa Induce Alterations in Expression of Genes Involved in Chromatin Architecture in Human Peripheral Blood Mononuclear Cells (In Vitro). Int. J. Mol. Sci. 2021, 22, 2966. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, E.; Sicińska, P.; Michałowicz, J.; Woźniak, K.; Reszka, E.; Huras, B.; Zakrzewski, J.; Bukowska, B. The Mechanism of DNA Damage Induced by Roundup 360 PLUS, Glyphosate and AMPA in Human Peripheral Blood Mononuclear Cells—Genotoxic Risk Assessement. Food Chem. Toxicol. 2018, 120, 510–522. [Google Scholar] [CrossRef]
- Tarboush, N.A.; Almomani, D.H.; Khabour, O.F.; Azzam, M.I. Genotoxicity of Glyphosate on Cultured Human Lymphocytes. Int. J. Toxicol. 2022, 41, 126–131. [Google Scholar] [CrossRef]
- Lucia, R.M.; Huang, W.L.; Pathak, K.V.; McGilvrey, M.; David-Dirgo, V.; Alvarez, A.; Goodman, D.; Masunaka, I.; Odegaard, A.O.; Ziogas, A.; et al. Association of Glyphosate Exposure with Blood DNA Methylation in a Cross-Sectional Study of Postmenopausal Women. Environ. Health Perspect. 2022, 130, 047001. [Google Scholar] [CrossRef]
- Cosemans, C.; Van Larebeke, N.; Janssen, B.G.; Martens, D.S.; Baeyens, W.; Bruckers, L.; Den Hond, E.; Coertjens, D.; Nelen, V.; Schoeters, G.; et al. Glyphosate and AMPA Exposure in Relation to Markers of Biological Aging in an Adult Population-Based Study. Int. J. Hyg. Environ. Health 2022, 240, 113895. [Google Scholar] [CrossRef]
- Buchenauer, L.; Junge, K.M.; Haange, S.B.; Simon, J.C.; von Bergen, M.; Hoh, A.L.; Aust, G.; Zenclussen, A.C.; Stangl, G.I.; Polte, T. Glyphosate Differentially Affects the Allergic Immune Response across Generations in Mice. Sci. Total Environ. 2022, 850, 157973. [Google Scholar] [CrossRef]
- la Cecilia, D.; Maggi, F. Analysis of Glyphosate Degradation in a Soil Microcosm. Environ. Pollut. 2018, 233, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, P.; Flores, F.; Mueller, J.F.; Carter, S.; Negri, A.P. Glyphosate Persistence in Seawater. Mar. Pollut. Bull. 2014, 85, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Sviridov, A.V.; Shushkova, T.V.; Ermakova, I.T.; Ivanova, E.V.; Epiktetov, D.O.; Leontievsky, A.A. Microbial Degradation of Glyphosate Herbicides (Review). Appl. Biochem. Microbiol. 2015, 51, 188–195. [Google Scholar] [CrossRef]
- Hove-jensen, B.; Zechel, D.L. Utilization of Glyphosate as Phosphate Source: Biochemistry and Genetics of Bacterial Carbon-Phosphorus Lyase. Biotechnol. Mol. Biol. Rev. 2014, 78, 176–197. [Google Scholar] [CrossRef]
- Seweryn, P.; Van, L.B.; Kjeldgaard, M.; Russo, C.J.; Passmore, L.A.; Hove-Jensen, B.; Jochimsen, B.; Brodersen, D.E. Structural Insights into the Bacterial Carbon-Phosphorus Lyase Machinery. Nature 2015, 525, 68–72. [Google Scholar] [CrossRef]
- Amstrup, S.K.; Ong, S.C.; Sofos, N.; Karlsen, J.L.; Skjerning, R.B.; Boesen, T.; Enghild, J.J.; Hove-Jensen, B.; Brodersen, D.E. Structural Remodelling of the Carbon–Phosphorus Lyase Machinery by a Dual ABC ATPase. Nat. Commun. 2023, 14, 1001. [Google Scholar] [CrossRef]
- Kamat, S.S.; Raushel, F.M. PhnJ—A Novel Radical SAM Enzyme from the C–P Lyase Complex. Perspect Sci. 2015, 4, 32–37. [Google Scholar] [CrossRef]
- Kononova, S.V.; Trutko, S.M.; Laurinavichus, K.S. Detection of C-P-Lyase Activity in a Cell-Free Extract of Escherichia Coli. Appl. Biochem. Microbiol. 2007, 43, 394–398. [Google Scholar] [CrossRef]
- Sviridov, A.V.; Shushkova, T.V.; Zelenkova, N.F.; Vinokurova, N.G.; Morgunov, I.G.; Ermakova, I.T.; Leontievsky, A.A. Distribution of Glyphosate and Methylphosphonate Catabolism Systems in Soil Bacteria Ochrobactrum anthropi and Achromobacter Sp. Appl. Microbiol. Biotechnol. 2012, 93, 787–796. [Google Scholar] [CrossRef]
- Barry, G.F.; Kishore, G.M. Glyphosate Tolerant Plants. US Patent 5463175, 31 October 1995. [Google Scholar]
- Bhatt, P.; Joshi, T.; Bhatt, K.; Zhang, W.; Huang, Y.; Chen, S. Binding Interaction of Glyphosate with Glyphosate Oxidoreductase and C–P Lyase: Molecular Docking and Molecular Dynamics Simulation Studies. J. Hazard. Mater. 2021, 409, 124927. [Google Scholar] [CrossRef]
- Yao, P.; Lin, Y.; Wu, G.; Lu, Y.; Zhan, T.; Kumar, A.; Zhang, L.; Liu, Z. Improvement of Glycine Oxidase by DNA Shuffling, and Site-Saturation Mutagenesis of F247 Residue. Int. J. Biol. Macromol. 2015, 79, 965–970. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Zhang, K.; Chen, Y.; Lin, Y.; Wu, G.; Zhang, L.; Yao, P.; Shao, Z.; Liu, Z. Improving Glyphosate Oxidation Activity of Glycine Oxidase from Bacillus cereus by Directed Evolution. PLoS ONE 2013, 8, e79175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Guo, Y.; Yao, P.; Lin, Y.; Kumar, A.; Liu, Z.; Wu, G.; Zhang, L. Characterization and Directed Evolution of BliGO, a Novel Glycine Oxidase from Bacillus licheniformis. Enzym. Microb. Technol. 2016, 85, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Wu, G.; Guo, Y.; Ke, D.; Yin, J.; Wang, D.; Fan, X.; Liu, Z.; Ruan, L.; Hu, Y. Engineered Glyphosate Oxidase Coupled to Spore-Based Chemiluminescence System for Glyphosate Detection. Anal. Chim. Acta 2020, 1133, 39–47. [Google Scholar] [CrossRef] [PubMed]
- National Library of Medicine; National Institutes of Health; U.S. Department of Health and Human Services PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 12 December 2021).
- Elsevier, B.V. ScienceDirect. Available online: https://www.sciencedirect.com/ (accessed on 10 June 2022).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Kanehisa Laboratories KEGG: Kyoto Encyclopedia of Genes and Genomes. Available online: https://www.kegg.jp/kegg/ (accessed on 15 June 2021).
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System, Version 2.5.2; Schrödinger, LLC.: New York, NY, USA, 2021. Available online: https://pymol.org/support.html (accessed on 11 November 2023).
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2-A Multiple Sequence Alignment Editor and Analysis Workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Madden Thomas The BLAST Sequence Analysis Tool In The NCBI Handbook [Internet]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK153387/ (accessed on 20 February 2024).
- Jun, S.R.; Sims, G.E.; Wu, G.A.; Kim, S.H. Whole-Proteome Phylogeny of Prokaryotes by Feature Frequency Profiles: An Alignment-Free Method with Optimal Feature Resolution. Proc. Natl. Acad. Sci. USA 2010, 107, 133–138. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A Unified Platform for Automated Protein Structure and Function Prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Studer, G.; Rempfer, C.; Waterhouse, A.M.; Gumienny, G.; Haas, J.; Schwede, T. QMEANDisCo—Distance Constraints Applied on Model Quality Estimation. Bioinformatics 2020, 36, 1765–1771. [Google Scholar] [CrossRef]
- Firdous, S.; Iqbal, S.; Anwar, S.; Jabeen, H. Identification and Analysis of 5-Enolpyruvylshikimate-3-Phosphate Synthase (EPSPS) Gene from Glyphosate-Resistant Ochrobactrum Intermedium Sq20. Pest. Manag. Sci. 2018, 74, 1184–1196. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Elgorriaga, A.; Amrhein, N. Evidence for Two Distinct Phosphonate-Degrading Enzymes (C-P Lyases) in Arthrobacter Sp. GLP-1. Biodegradation 1991, 2, 53–59. [Google Scholar] [CrossRef]
- Obojska, A.; Lejczak, B.; Kubrak, M. Degradation of Phosphonates by Streptomycete Isolates. Appl. Microbiol. Biotechnol. 1999, 51, 872–876. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.N.; Vo, V.T.; Nguyen, T.H.P.; Kiefer, R. Isolation and Optimization of a Glyphosate-Degrading Rhodococcus soli G41 for Bioremediation. Arch. Microbiol. 2022, 204, 252. [Google Scholar] [CrossRef] [PubMed]
- Pipke, R.; Amrheint, N. Degradation of the Phosphonate Herbicide Glyphosate by Arthrobacter atrocyaneus ATCC 13752. Appl. Environ. Microbiol. 1988, 54, 1293–1296. [Google Scholar] [CrossRef]
- Obojska, A.; Ternan, N.G.; Lejczak, B.; Kafarski, P.; McMullan, G. Organophosphonate Utilization by the Thermophile Geobacillus caldoxylosilyticus T20. Appl. Environ. Microbiol. 2002, 68, 2081–2084. [Google Scholar] [CrossRef]
- Fan, J.; Yang, G.; Zhao, H.; Shi, G.; Geng, Y.; Hou, T.; Tao, K. Isolation, Identification and Characterization of a Glyphosate-Degrading Bacterium, Bacillus cereus CB4, from Soil. J. Gen. Appl. Microbiol. 2012, 58, 263–271. [Google Scholar] [CrossRef]
- González-Valenzuela, L.E.; Dussán, J. Molecular Assessment of Glyphosate-Degradation Pathway via Sarcosine Intermediate in Lysinibacillus sphaericus. Environ. Sci. Pollut. Res. 2018, 25, 22790–22796. [Google Scholar] [CrossRef]
- Elarabi, N.I.; Abdelhadi, A.A.; Ahmed, R.H.; Saleh, I.; Arif, I.A.; Osman, G.; Ahmed, D.S. Bacillus aryabhattai FACU: A Promising Bacterial Strain Capable of Manipulate the Glyphosate Herbicide Residues. Saudi J. Biol. Sci. 2020, 27, 2207–2214. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Cortés, A.G.; Martinez-Ledezma, C.; López-Chuken, U.J.; Kaushik, G.; Nimesh, S.; Villarreal-Chiu, J.F. Polyphosphate Recovery by a Native Bacillus cereus Strain as a Direct Effect of Glyphosate Uptake. ISME J. 2019, 13, 1497–1505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, J.; Zhang, Y.; Wu, X.; Zhou, Z.; Huang, Y.; Zhao, Y.; Mishra, S.; Bhatt, P.; Chen, S. Characterization of a Novel Glyphosate-Degrading Bacterial Species, Chryseobacterium Sp. Y16C, and Evaluation of Its Effects on Microbial Communities in Glyphosate-Contaminated Soil. J. Hazard. Mater. 2022, 432, 128689. [Google Scholar] [CrossRef] [PubMed]
- Ermakova, I.T.; Shushkova, T.V.; Sviridov, A.V.; Zelenkova, N.F.; Vinokurova, N.G.; Baskunov, B.P.; Leontievsky, A.A. Organophosphonates Utilization by Soil Strains of Ochrobactrum anthropi and Achromobacter Sp. Arch. Microbiol. 2017, 199, 665–675. [Google Scholar] [CrossRef]
- Hadi, F.; Mousavi, A.; Noghabi, K.A.; Tabar, H.G.; Salmanian, A.H. New Bacterial Strain of the Genus Ochrobactrum with Glyphosate-Degrading Activity. J. Environ. Sci. Health B 2013, 48, 208–213. [Google Scholar] [CrossRef]
- Mcauliffe, K.S.; Hallas, L.E.; Kulpa, C.F. Glyphosate Degradation by Agrobacterium radiobacter Isolated from Activated Sludge. J. Ind. Microbiol. 1990, 6, 219–221. [Google Scholar] [CrossRef]
- Parker, G.F.; Higgins, T.P.; Hawkes, T.; Robson, R.L. Rhizobium (Sinorhizobium) meliloti phn Genes: Characterization and Identification of Their Protein Products. J. Bacteriol. 1999, 181, 389–395. [Google Scholar] [CrossRef]
- Masotti, F.; Garavaglia, B.S.; Piazza, A.; Burdisso, P.; Altabe, S.; Gottig, N.; Ottado, J. Bacterial Isolates from Argentine Pampas and Their Ability to Degrade Glyphosate. Sci. Total Environ. 2021, 774, 145761. [Google Scholar] [CrossRef]
- Firdous, S.; Iqbal, S.; Anwar, S. Optimization and Modeling of Glyphosate Biodegradation by a Novel Comamonas odontotermitis P2 through Response Surface Methodology. Pedosphere Int. J. 2020, 30, 618–627. [Google Scholar] [CrossRef]
- Lerbs, W.; Stock, M.; Parthier, B. Physiological Aspects of Glyphosate Degradation in Alcaligenes Spec. Strain GL. Arch. Microbiol. 1990, 153, 146–150. [Google Scholar] [CrossRef]
- Manogaran, M.; Shukor, M.Y.; Yasid, N.A.; Khalil, K.A.; Ahmad, S.A. Optimisation of Culture Composition for Glyphosate Degradation by Burkholderia vietnamiensis Strain AQ5-12. 3Biotech 2018, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Tarlachkov, S.V.; Epiktetov, D.O.; Sviridov, A.V.; Shushkova, T.V.; Ermakova, I.T.; Leontievsky, A.A. Draft Genome Sequence of Glyphosate-Degrading Achromobacter insolitus Strain Kg 19 (VKM B-3295), Isolated from Agricultural Soil. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Jacob, G.S.; Garbow, J.R.; Hallas, L.E.; Kimack, N.M.; Kishore, G.M.; Schaefer, J. Metabolism of glyphosate in Pseudomonas sp. strain LBr. Appl. Environ. Microbiol. 1988, 54, 2953–2958. [Google Scholar] [CrossRef]
- Xu, B.; Sun, Q.J.; Lan, J.C.W.; Chen, W.M.; Hsueh, C.C.; Chen, B.Y. Exploring the Glyphosate-Degrading Characteristics of a Newly Isolated, Highly Adapted Indigenous Bacterial Strain, Providencia rettgeri GDB 1. J. Biosci. Bioeng. 2019, 128, 80–87. [Google Scholar] [CrossRef]
- Dick, R.E.; Quinn, J.P. Control of glyphosate uptake and metabolism in Pseudomonas sp. 4ASW. FEMS Microbiol. Lett. 1995, 134, 177–182. [Google Scholar] [CrossRef]
- Kryuchkova, Y.V.; Burygin, G.L.; Gogoleva, N.E.; Gogolev, Y.V.; Chernyshova, M.P.; Makarov, O.E.; Fedorov, E.E.; Turkovskaya, O.V. Isolation and Characterization of a Glyphosate-Degrading Rhizosphere Strain, Enterobacter cloacae K7. Microbiol. Res. 2014, 169, 99–105. [Google Scholar] [CrossRef]
- Benslama, O.; Boulahrouf, A. High-Quality Draft Genome Sequence of Enterobacter Sp. Bisph2, a Glyphosate-Degrading Bacterium Isolated from a Sandy Soil of Biskra, Algeria. Genom. Data 2016, 8, 61–66. [Google Scholar] [CrossRef]
- Peñaloza-Vazquez, A.; Mena, G.L.; Herrera-Estrella, L.; Bailey, A.M. Cloning and sequencing of the genes. involved in glyphosate utilization by Pseudomonas pseudomallei. Appl. Environ. Microbiol. 1995, 61, 538–543. [Google Scholar] [CrossRef]
- Selvapandiyan, A.; Bhatnagar, R.K. Isolation of a Glyphosate-Metabolising Pseudomonas: Detection, Partial Purification and Localisation of Carbon-Phosphorus Lyase. Appl. Microbiol. Biotechnol. 1994, 40, 876–882. [Google Scholar] [CrossRef]
- Kishore, G.M.; Jacob, G.S. Degradation of Glyphosate by Pseudomonas Sp. PG2982 via a Sarcosine Intermediate. J. Biol. Chem. 1987, 262, 12164–12168. [Google Scholar] [CrossRef]
- Talbot, H.W.; Johnson, L.M.; Munnecke, D.M. Glyphosate Utilization by Pseudomonas Sp. and Alcaligenes Sp. Isolated from Environmental Sources. Curr. Microbiol. 1984, 10, 255–259. [Google Scholar] [CrossRef]
- Bethesda (MD): National Library of Medicine (US)-National Center for Biotechnology Information National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 14 November 2023).
- Nicolet, Y. Structure–Function Relationships of Radical SAM Enzymes. Nat. Catal. 2020, 3, 337–350. [Google Scholar] [CrossRef]
- Kryshtafovych, A.; Schwede, T.; Topf, M.; Fidelis, K.; Moult, J. Critical Assessment of Methods of Protein Structure Prediction (CASP)—Round XIV. Proteins Struct. Funct. Bioinform. 2021, 89, 1607–1617. [Google Scholar] [CrossRef] [PubMed]
- Husain, Q.; Fahad Ullah, M. Oxidoreductases: Overview and Practical Applications. In Biocatalysis: Enzymatic Basics and Applications; Springer International Publishing: Cham, Switzerland, 2019; pp. 39–55. ISBN 9783030250232. [Google Scholar]
| PhnJ | ||||
|---|---|---|---|---|
| Secondary Structure | MUSCLE Alignment Residues Identified in Alignments from This Study | Conserved Residues as Given by Seweryn et al. [37] | Residues Predicted In Silico to Participate in Interaction with AMPA and GLP [43]. HB: H Bond HI: Hydrophobic Interaction | |
| Residues Surrounding the Fe4S4 Cluster | ||||
| loop 1 | A207, G208, R209, E210 | - | AMPA: G208 (HB), R209 (HB) | |
| loop 2 | P36 *, P40, P43 Near the cluster: G47, G49 | - | AMPA: G47 (HB), G49 (HB) | |
| loop 3 | Q124, V125, P126 | - | AMPA: V125 (HB) | |
| helix 1 | T50, G51, G52 More distant: Q54 | - | AMPA: T50 (HB), G51 (HB) | |
| Residues surrounding Zn | ||||
| loop 4 | G245 | C241, C244 | Y250 (HI); more distant to Zn, S267 (HB) | |
| β sheet loop | C266 | |||
| helix6 | C272 | |||
| Residues surrounding universally conserved residue G32 | ||||
| loop 5 | P31, Q34 | G32 | ||
| CID—Central insertion domain—contacts both of the central PhnI molecules | ||||
| loop 3 | L131, E135 | 129–169 | ||
| helix 2 | H145 | |||
| helix 3 | Y150, L157 | |||
| CMD—C-terminal mini domain—stabilized by Zn ion and conserved C residues | ||||
| β hairpin | E253 (loop), G260 (β sheet) | 236–281 | ||
| His108 coordinate Zn ion with 2 His from PhnI | ||||
| loop 6 | R107, R109 More distant: P111, F112, L115 | H108 | GLP: H108 (HB), R109 (HI) | |
| loop 7 | P187 | - | ||
| helix 5 | P189, D192 | - | GLP: P189 (HI) | |
| loop 8 | D70, Q71, G72 | - | GLP: Q71 (HB) | |
| Two of the residues (PhnI His328 and His333) coordinate the zinc ion directly PhnJ His108, PhnI His328, and PhnI His333 are located in a cavity between PhnI and PhnJ | ||||
| β strand 1 | K67, V68, I69, D70 | |||
| loop | Q71, G72 | |||
| β strand 2 | I104, Q105, R107 | |||
| loop | R109 | (H108) | ||
| loop (Bhatt) | P174, F232 | Y171 (HI) | ||
| PhnI | ||||
| Secondary structure | MUSCLE alignment Residues not reported by [37], identified in alignments from this study | Conserved residues [37] | ||
| Residues surrounding the Fe4S4 cluster | ||||
| loop 1 | A55 | G278 | - | |
| loop 2 | ||||
| NTD—N-terminal domain | ||||
| β strand 1 | Y2, A4 | 1–88 | ||
| helix 2 | G7, G8, A11 | |||
| helix 3 | E51 | |||
| loop 1 | A55 | |||
| helix 4 | A60, A63, Q66 | |||
| loop 4 | G69 | |||
| helix 5 | E73, A74, R82, T84 | |||
| β strand 2 | K87 | |||
| His108 coordinate Zn ion with 1 His from PhnJ | ||||
| loop 5 | P332 | - | ||
| helix 6 | Y334 | H333 | ||
| helix 7 | F325, K330 Further away: D318, G324 | H328 | ||
| C-terminal | ||||
| helix 8 | R189, R198, Y209, R213 | 188–318 | ||
| loop 6 | G214, G216, H219 | |||
| β strand 3 | G223, E224, R226, G228 | |||
| β strand 4 | G246, E252 | |||
| β strand 5 | G272, G274 | |||
| loop 7 | G278 | |||
| helix 9 | E281, K283, M287 | |||
| helix 10 | F312, H316 | |||
| helix 11 | D318 | |||
| PhnG | ||||
| Secondary structure | MUSCLE alignment Residues not reported by [37], identified in alignments from this study | Conserved residues [37] | ||
| β-hairpin and C-terminal helix form a molecular clamp that connects to a groove in PhnI | ||||
| β strand 1 | G63 | - | - | |
| β strand 2 | G80 | - | - | |
| helix | A96, D99, A100 | - | - | |
| loop | V139, F141 | - | - | |
| Bhatt et al., 2021 | BLASTp_nr_NOGROUP | BLASTp_ref_NOGROUP | BLASTp_nr_GROUP | BLASTp_ref_GROUP |
|---|---|---|---|---|
| G14 | + | + | ||
| I15 | ||||
| V16 | ||||
| A43 | + | |||
| S44 | ||||
| N47 | + | + | ||
| R358 | + | + | ||
| G384 | ||||
| G387 | + | + | ||
| M388 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannakara, M.; Koumandou, V.L. New Insights on the Glyphosate-Degrading Enzymes C-P Lyase and Glyphosate Oxidoreductase Based on Bioinformatics. Bacteria 2024, 3, 314-329. https://doi.org/10.3390/bacteria3040021
Giannakara M, Koumandou VL. New Insights on the Glyphosate-Degrading Enzymes C-P Lyase and Glyphosate Oxidoreductase Based on Bioinformatics. Bacteria. 2024; 3(4):314-329. https://doi.org/10.3390/bacteria3040021
Chicago/Turabian StyleGiannakara, Marina, and Vassiliki Lila Koumandou. 2024. "New Insights on the Glyphosate-Degrading Enzymes C-P Lyase and Glyphosate Oxidoreductase Based on Bioinformatics" Bacteria 3, no. 4: 314-329. https://doi.org/10.3390/bacteria3040021
APA StyleGiannakara, M., & Koumandou, V. L. (2024). New Insights on the Glyphosate-Degrading Enzymes C-P Lyase and Glyphosate Oxidoreductase Based on Bioinformatics. Bacteria, 3(4), 314-329. https://doi.org/10.3390/bacteria3040021










