Comparative Genomic Analysis of Phytopathogenic Xanthomonas Species Suggests High Level of Genome Plasticity Related to Virulence and Host Adaptation
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Annotation of Xanthomonas species
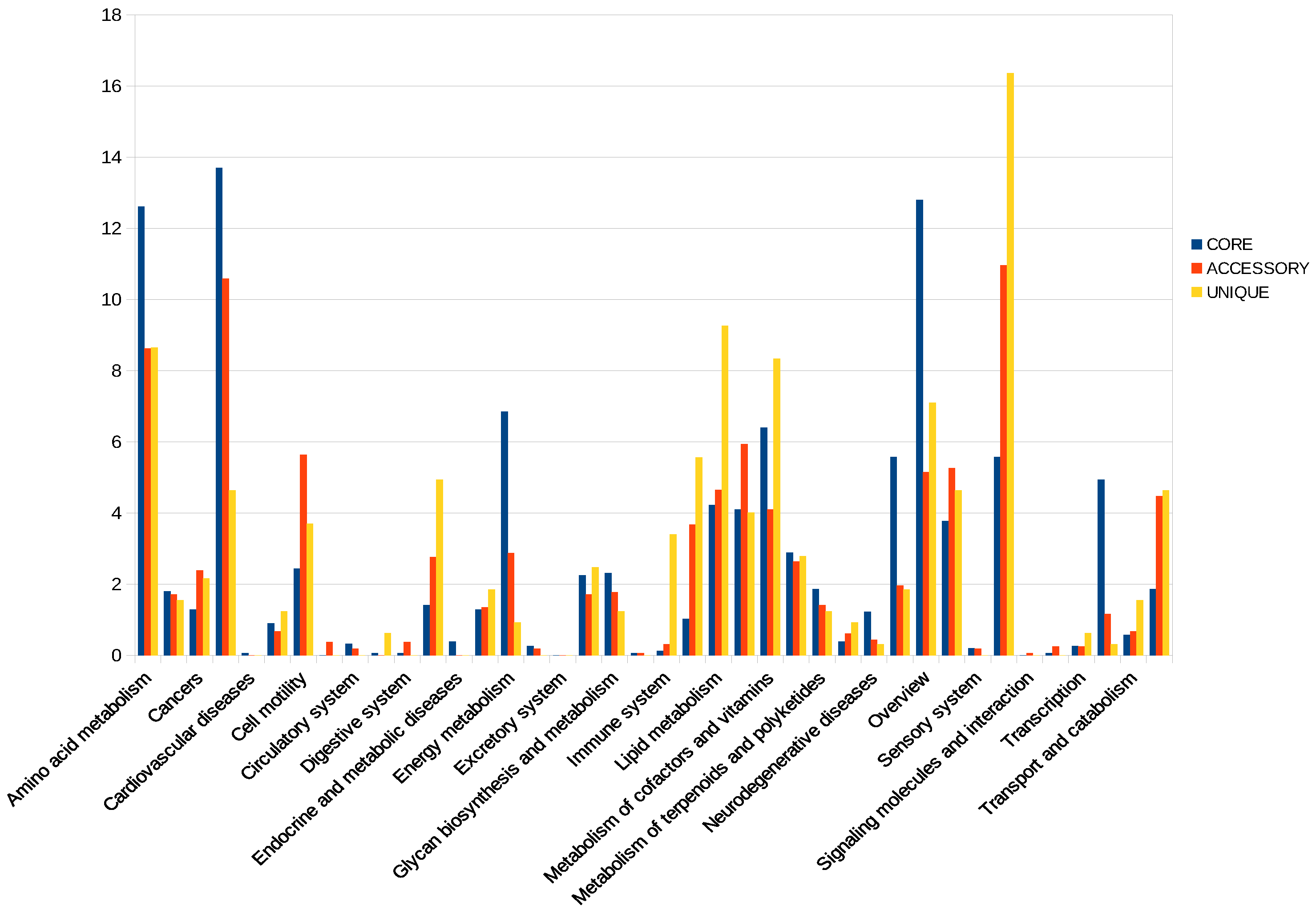
2.2. Genomic Characterization and Enrichment Analysis
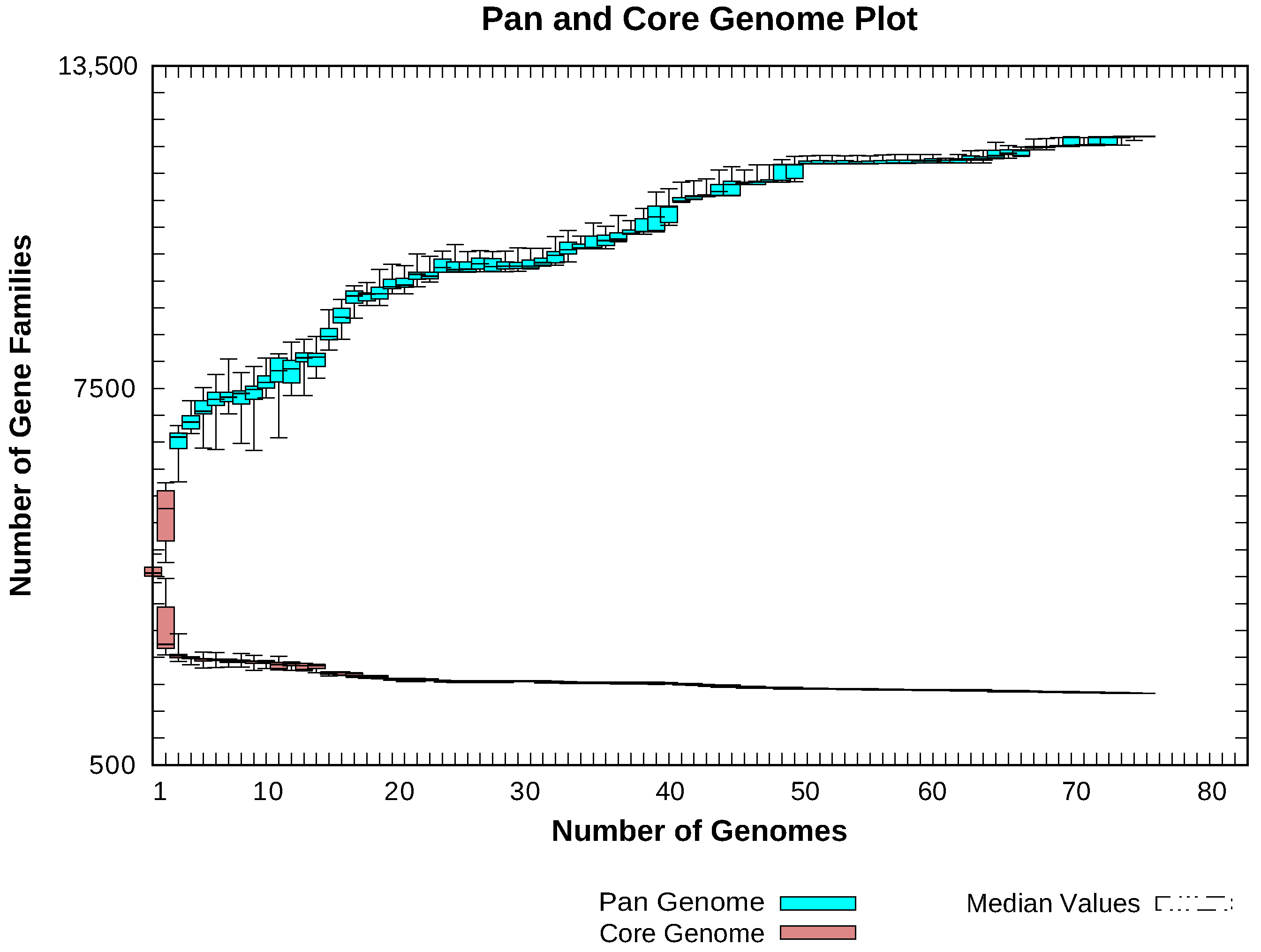
2.3. Pan-Genome Analyses
2.4. Prediction and Characterization of Pathogenicity Islands
2.5. Subcellular Localization of Proteins inside PAIs
2.6. Prediction of Prophages Regions in Xanthomonas genomes
3. Results
3.1. Functional Genomics and Comparative Genomics Insights of Xanthomonas species
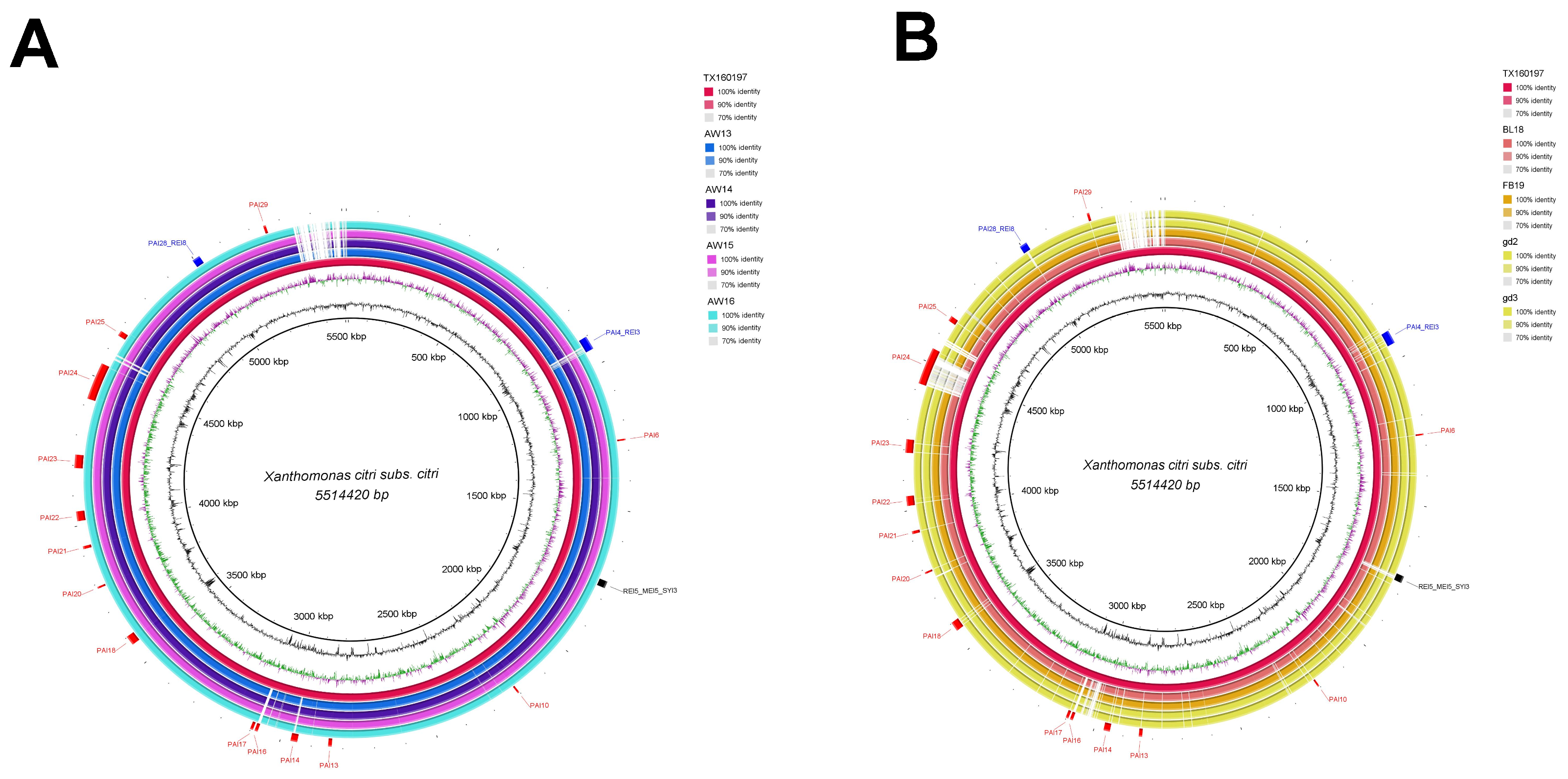
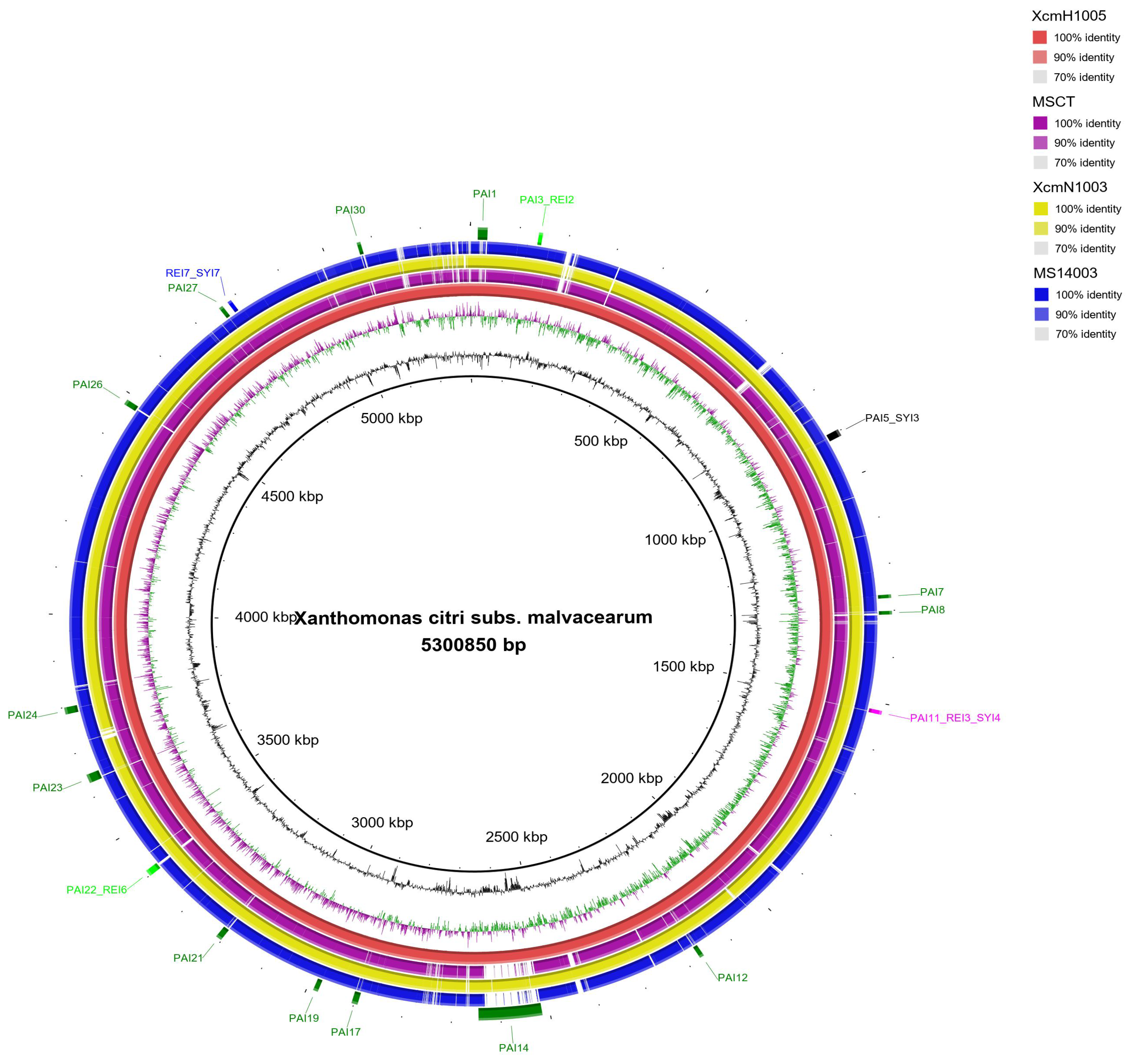
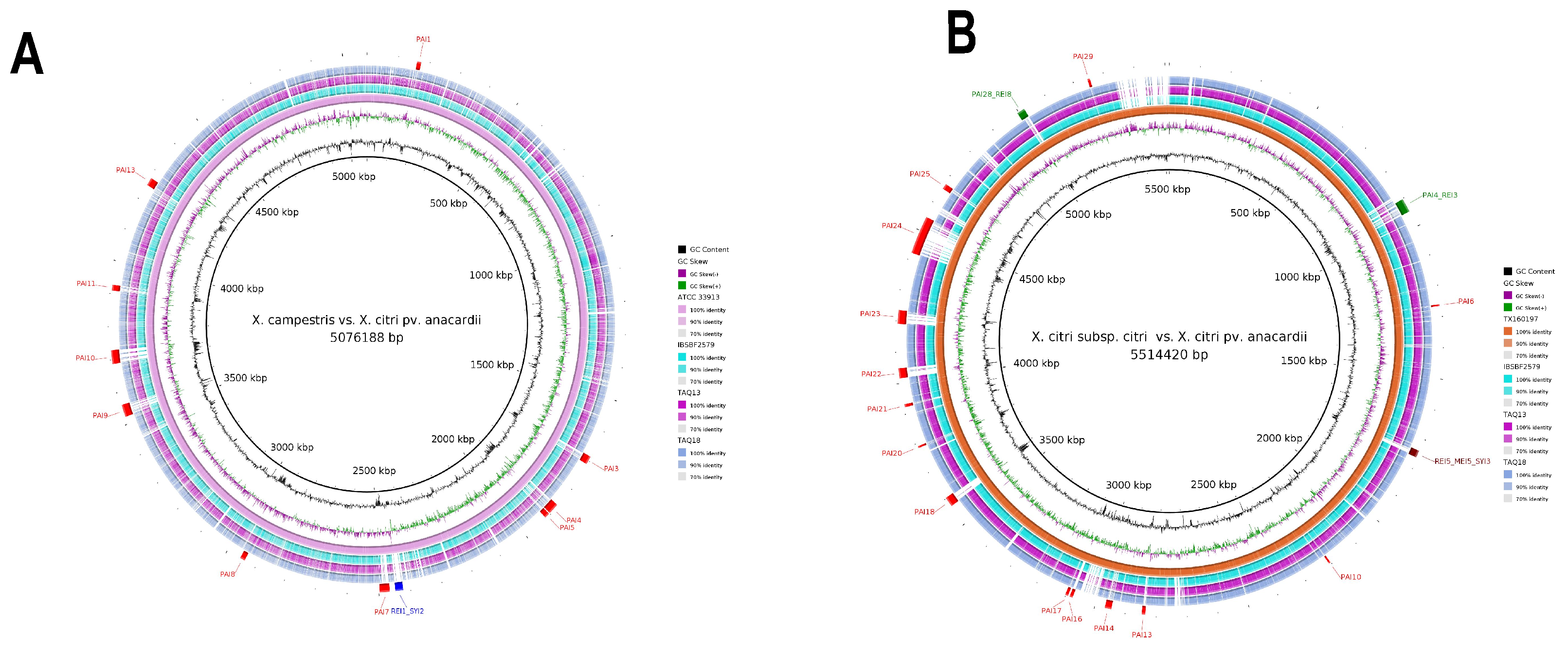
3.2. Genomic Islands Reveal a New Potential Regarding the Pathogenicity of the Genus
3.3. Cellular Localization and Ontology of Pathogenicity Islands
3.4. Prophage Regions, Transposable Elements and Their Relationship with Pathogenicity Islands
4. Discussion
4.1. Insights Provided by the Pan-Genome of Xanthomonas
4.2. Enrichment Analysis in Pathogenicity Islands
4.3. Prophage Regions and the Evolution of Bacterial Phytopathogens
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. General Information Regarding Genomes Used in This Work
| Species [18] | Strains | BioProject | Replicons | Assembly | Phytobacteriosis |
|---|---|---|---|---|---|
| Xanthomonas arboricola (Vauterin et al. 1995) | 17 | PRJNA280784 | CP011256.1 | GCA_000972745.1 | Black rot of crucifers |
| Xanthomonas campestris pv. campestris (Pammel) Dowson | 3811 | PRJNA428847 | NZ_CP025750.1 | GCA_002879955.1 | Black rot of crucifers |
| B100 | PRJNA29801 | NC_010688.1 | GCA_000070605.1 | ||
| ICMP 21080 | PRJNA283327 | NZ_CP012145.1 | GCA_001186415.1 | ||
| ICMP 4013 | PRJNA283326 | NZ_CP012146.1 | GCA_001186465.1 | ||
| 8004 | PRJNA15 | NC_007086.1 | GCA_000012105.1 | ||
| CN03 | PRJNA211759 | NZ_CP017308.1 | GCA_002776735.1 | ||
| CN18 | PRJNA343742 | NZ_CP017319.1 | GCA_002776835.1 | ||
| ATCC 33913 | PRJNA296 | NC_003902 | GCA_000007145.1 | ||
| CN12 | PRJEB4853 | NZ_CP017310.1 | GCA_002776775.1 | ||
| CN17 | PRJNA211780 | CP017307.1 | GCA_002776715.1 | ||
| Xanthomonas campestris pv. raphani (White) Dye | 756C | PRJNA63187 | NC_017271.1 | GCA_000221965.1 | Black rot of crucifers |
| Xanthomonas citri pv. anacardii (Ah-You et al. 2009) [62] | IBSBF2579 | PRJNA224116 | NZ_PESI01000001.1 | GCA_002837255.1 | Bacterial black spot |
| TAQ13 | PRJNA224116 | NZ_PESH01000000 | GCA_002898475.1 | ||
| TAQ18 | PRJNA224116 | NZ_PESG01000001.1 | GCA_002898415.1 | ||
| Xanthomonas citri pv. aurantifolii (Ah-You et al. 2009) | 1566 | PRJNA273983 | NZ_CP012002.1 | GCA_001610915.1 | Citrus canker |
| FDC 1559 | PRJNA273983 | NZ_CP011160.1 | GCA_001610795.1 | ||
| FDC 1561 | PRJNA273983 | NZ_CP011250.1 | GCA_002079965.1 | ||
| FDC 1609 | PRJNA273983 | NZ_CP011163.1 | GCA_001610815.1 | ||
| Xanthomonas citri pv. fuscans (Schaad et al. 2007) | ISO12C3 | PRJNA289080 | NZ_CP012055.1 | GCA_003999505.1 | Bacterial blight of bean |
| ISO18C1 | PRJNA289080 | NZ_CP012053.1 | GCA_003999485.1 | ||
| ISO18C5 | PRJNA289080 | NZ_CP012051.1 | GCA_004000475.1 | ||
| 4834-R | PRJNA176873 | NC_022539.1 | GCA_000969685.1 | ||
| Xanthomonas citri pv. glycines (Nakano, 2019) | 12-2 | PRJNA323439 | NZ_CP015972.1 | GCA_002163775.1 | Bacterial pustule of soybean |
| 8ra | PRJNA341902 | NZ_CP017188.1 | GCA_001854145.2 | ||
| Xanthomonas citri pv. phaseoli var. fuscans (ex Smith, 1987) Dye 1978 | CFBP4885 | PRJNA384182 | NZ_CP020992.1 | GCA_002759355.2 | Bacterial blight of bean |
| CFBP6165 | PRJNA384145 | NZ_CP020998.1 | GCA_002759215.3 | ||
| CFBP6166 | PRJNA384183 | NZ_CP021001.1 | GCA_002759235.2 | ||
| CFBP6167 | PRJNA384187 | NZ_CP021018.1 | GCA_002759415.2 | ||
| CFBP6975 | PRJNA384188 | NZ_CP021006.1 | GCA_002759255.3 | ||
| CFBP6988R | PRJNA384160 | NZ_CP020979.1 | GCA_002759275.2 | ||
| CFBP6989 | PRJNA384161 | NZ_CP020981.1 | GCA_002759295.2 | ||
| CFBP6990 | PRJNA384163 | NZ_CP020983.1 | GCA_002759315.2 | ||
| CFBP6991 | PRJNA384178 | NZ_CP021015.1 | GCA_002759395.2 | ||
| CFBP6992 | PRJNA384177 | NZ_CP020985.1 | GCA_002759335.2 | ||
| CFBP6994R | PRJNA384179 | NZ_CP020987.1 | GCA_002759175.2 | ||
| CFBP6996R | PRJNA384180 | NZ_CP020989.1 | GCA_002759195.2 | ||
| CFBP7767 | PRJNA383319 | NZ_CP021012.1 | GCA_002759375.2 | ||
| Xanthomonas citri pv. vignicola (Burkholder, 1994) | CFBP7111 | PRJNA390891 | NZ_CP022263.1 | GCA_002218245.1 | Bacterial blight of cowpea |
| CFBP7112 | PRJNA390892 | NZ_CP022267.1 | GCA_002218265.1 | ||
| CFBP7113 | PRJNA390890 | NZ_CP022270.1 | GCA_002218285.1 | ||
| Xanthomonas citri subsp. citri (Gabriel et al. 1989) Schaad et al. 2007 | 03-1638-1-1 | PRJNA401937 | NZ_CP023285.1 | GCA_002952295.1 | Citrus canker |
| Xcc29-1 | PRJNA407058 | NZ_CP023661.1 | GCA_003665475.1 | ||
| Xcc49 | PRJNA407058 | NZ_CP023662.1 | GCA_003665455.1 | ||
| MSCT | PRJNA299817 | NZ_CP017020.1 | GCA_001719145.1 | ||
| 5208 | PRJNA255042 | NZ_CP009026.1 | GCA_000961415.1 | ||
| AW13 | PRJNA255042 | NZ_CP009029.1 | GCA_000961435.1 | ||
| AW14 | PRJNA255042 | NZ_CP009032.1 | GCA_000961455.1 | ||
| AW15 | PRJNA255042 | NZ_CP009035.1 | GCA_000961475.1 | ||
| AW16 | PRJNA255042 | NZ_CP009038.1 | GCA_000961495.1 | ||
| BL18 | PRJNA255042 | NZ_CP009023.1 | GCA_000961395.1 | ||
| FB19 | PRJNA255042 | NZ_CP009020.1 | GCA_000961375.1 | ||
| gd2 | PRJNA255042 | NZ_CP009017.1 | GCA_000961355.1 | ||
| gd3 | PRJNA255042 | NZ_CP009014.1 | GCA_000961335.1 | ||
| jx-6 | PRJNA286060 | NZ_CP011827.1 | GCA_001028285.3 | ||
| jx4 | PRJNA255042 | NZ_CP009011.1 | GCA_000961315.1 | ||
| jx5 | PRJNA255042 | NZ_CP009008.1 | GCA_000961295.1 | ||
| LH201 | PRJNA344031 | NZ_CP018858.1 | GCA_001922105.1 | ||
| LH276 | PRJNA344031 | NZ_CP018854.1 | GCA_001922065.1 | ||
| LJ207-7 | PRJNA344031 | NZ_CP018850.1 | GCA_001922085.1 | ||
| LL074-4 | PRJNA344031 | NZ_CP018847.1 | GCA_001922045.1 | ||
| mf20 | PRJNA255042 | NZ_CP009005.1 | GCA_000961275.1 | ||
| MN10 | PRJNA255042 | NZ_CP009002.1 | GCA_000961255.1 | ||
| MN11 | PRJNA255042 | NZ_CP008999.1 | GCA_000961235.1 | ||
| MN12 | PRJNA255042 | NZ_CP008996.1 | GCA_000961215.1 | ||
| NT17 | PRJNA255042 | NZ_CP008993.1 | GCA_000961195.1 | ||
| TX160042 | PRJNA381640 | NZ_CP020882.1 | GCA_002139975.1 | ||
| TX160149 | PRJNA381640 | NZ_CP020885.1 | GCA_002139955.1 | ||
| TX160197 | PRJNA381640 | NZ_CP020889.1 | GCA_002139995.1 | ||
| UI7 | PRJNA255042 | NZ_CP008987.1 | GCA_000961155.1 | ||
| 306 | PRJNA297 | NC_003919.1 | GCA_000007165.1 | ||
| UI6 | PRJNA255042 | NZ_CP008990.1 | GCA_000961175.1 | ||
| A306 | PRJNA193618 | NZ_CP006855.1 | GCA_000816885.1 | ||
| Aw12879 | PRJNA81931 | NC_020815. | GCA_000349225.1 | ||
| Xanthomonas citri subsp. malvacearum (ex Smith 1901) Schaad et al. 2007 | MS14003 | PRJNA396899 | NZ_CP023159.1 | GCA_002288585.1 | Bacterial blight of cotton |
| AR81009 | PRJNA396899 | NZ_CP023155.1 | GCA_002288565.1 | ||
| XcmH1005 | PRJNA298765 | NZ_CP013004.1 | GCA_002224525.1 | ||
| XcmN1003 | PRJNA298770 | NZ_CP013006.1 | GCA_002224545.1 | ||
| Xanthomonas vasicola pv. arecae (Rao & Mohan) Dye 1978 | NCPPB 2649 | PRJNA264725 | CP034653.1 | GCA_000770355.2 | Bacterial leaf streak of maize |
| Xanthomonas vasicola pv. musacearum (Yirgou & Bradbury) Dye 1978 | NCPPB 4379 | PRJNA73877 | NZ_CP034655.1 | GCA_000277895.2 | Xanthomonas wilt of banana |
Appendix B. Mobile Elements Found within Xanthomonas Genomes
| Species [18] | Strains | Number of PAIs |
|---|---|---|
| Xanthomonas arboricola | 17 | 3 |
| Xanthomonas campestris pv. campestris | 8004 | 10 |
| CN03 | 6 | |
| CN12 | 8 | |
| CN17 | 7 | |
| CN18 | 7 | |
| 3811 | 9 | |
| ATCC 33913 | 10 | |
| B100 | 7 | |
| ICMP 21080 | 8 | |
| ICMP 4013 | 7 | |
| Xanthomonas campestris pv. raphani | 756C | 4 |
| Xanthomonas citri pv. anacardii | IBSBF2579 | 8 |
| TAQ13 | 7 | |
| TAQ18 | 7 | |
| Xanthomonas citri pv. aurantifolii | 1566 | 7 |
| FDC1559 | 11 | |
| FDC1561 | 6 | |
| FDC1609 | 8 | |
| Xanthomonas citri pv. fuscans | ISO118C1 | 4 |
| ISO118C5 | 3 | |
| ISO12C3 | 3 | |
| 4834-R | 4 | |
| Xanthomonas citri pv. glycines | 12-2 | 7 |
| 8ra | 6 | |
| Xanthomonas citri pv. phaseoli var. fuscans | CFBP4885 | 7 |
| CFBP6165 | 4 | |
| CFBP6166 | 4 | |
| CFBP6167 | 3 | |
| CFBP6975 | 4 | |
| CFBP6988R | 3 | |
| CFBP6989 | 4 | |
| CFBP6990 | 4 | |
| CFBP6991 | 3 | |
| CFBP6992 | 3 | |
| CFBP6994R | 5 | |
| CFBP6996R | 4 | |
| CFBP7767 | 7 | |
| Xanthomonas citri pv. vignicola | CFBP7111 | 10 |
| CFBP7112 | 8 | |
| CFBP7113 | 7 | |
| Xanthomonas citri subsp. citri (Gabriel et al. 1989) Schaad et al. 2007 | 306 | 8 |
| 03-1638-1-1 | 9 | |
| 5208 | 8 | |
| A306 | 9 | |
| Aw12879 | 12 | |
| AW13 | 13 | |
| AW14 | 14 | |
| AW15 | 11 | |
| AW16 | 13 | |
| BL18 | 9 | |
| FB19 | 9 | |
| gd2 | 7 | |
| gd3 | 7 | |
| jx-6 | 9 | |
| jx4 | 8 | |
| jx5 | 9 | |
| LH201 | 9 | |
| LH276 | 9 | |
| LJ207-7 | 8 | |
| LL074-4 | 8 | |
| mf20 | 9 | |
| MN10 | 9 | |
| MN11 | 9 | |
| MN12 | 9 | |
| NT17 | 8 | |
| TX160042 | 13 | |
| TX160149 | 10 | |
| TX160197 | 14 | |
| UI6 | 7 | |
| UI7 | 7 | |
| Xcc29-1 | 9 | |
| Xcc49 | 8 | |
| Xanthomonas citri subsp. malvacearum (ex Smith 1901) Schaad et al. 2007 | AR81009 | 15 |
| MS14003 | 4 | |
| MSCT | 5 | |
| XcmH1005 | 13 | |
| XcmN1003 | 10 | |
| Xanthomonas vasicola pv. arecae | NCPPB2649 | 8 |
| Xanthomonas vasicola pv. musacearum | NCPPB 4379 | 6 |
| Strains | Intact Region | Questionable Region | Incomplete Region | Total Prophage Regions |
|---|---|---|---|---|
| Xanthomonas arboricola 17 | 4 | 0 | 0 | 4 |
| Xanthomonas campestrispv. campestris CN17 | 1 | 1 | 0 | 2 |
| Xanthomonas campestris pv. campestris 3811 | 0 | 1 | 0 | 1 |
| Xanthomonas campestris pv. campestris 8004 | 2 | 0 | 0 | 2 |
| Xanthomonas campestris pv. campestris ATCC 33913 | 2 | 1 | 0 | 3 |
| Xanthomonas campestris pv. campestris B100 | 1 | 0 | 0 | 1 |
| Xanthomonas campestris pv. campestris CN03 | 1 | 2 | 0 | 3 |
| Xanthomonas campestris pv. campestris CN12 | 0 | 3 | 0 | 3 |
| Xanthomonas campestris pv. campestris CN18 | 0 | 0 | 0 | 0 |
| Xanthomonas campestris pv. campestris ICMP 21080 | 2 | 1 | 0 | 3 |
| Xanthomonas campestris pv. campestris ICMP 4013 | 0 | 1 | 0 | 1 |
| Xanthomonas campestris pv. raphani 756C | 0 | 1 | 0 | 1 |
| Xanthomonas citri pv. anacardii IBSBF2579 | 1 | 0 | 1 | 2 |
| Xanthomonas citri pv. anacardii TAQ13 | 1 | 0 | 1 | 2 |
| Xanthomonas citri pv. anacardii TAQ18 | 1 | 0 | 1 | 2 |
| Xanthomonas citri pv. aurantifolii 1566 | 2 | 3 | 3 | 8 |
| Xanthomonas citri pv. aurantifolii FDC1559 | 2 | 2 | 3 | 7 |
| Xanthomonas citri pv. aurantifolii FDC1561 | 1 | 1 | 0 | 2 |
| Xanthomonas citri pv. aurantifolii FDC1609 | 3 | 2 | 7 | 12 |
| Xanthomonas citri pv. fuscans 4834-R | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. fuscans ISO118C1 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. fuscans ISO118C5 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. fuscans ISO12C3 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. glycines 12-2 | 3 | 1 | 1 | 5 |
| Xanthomonas citri pv. glycines 8ra | 4 | 1 | 0 | 5 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP4885 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6165 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6166 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6167 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6975 | 2 | 0 | 0 | 2 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6988R | 2 | 0 | 1 | 3 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6989 | 2 | 0 | 1 | 3 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6990 | 2 | 0 | 1 | 3 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6991 | 2 | 0 | 1 | 3 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6992 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6994R | 2 | 0 | 0 | 2 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP6996R | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. phaseoli var. fuscans CFBP7767 | 1 | 0 | 0 | 1 |
| Xanthomonas citri pv. vignicola CFBP7111 | 4 | 0 | 4 | 8 |
| Xanthomonas citri pv. vignicola CFBP7112 | 1 | 1 | 0 | 2 |
| Xanthomonas citri pv. vignicola CFBP7113 | 2 | 1 | 3 | 6 |
| Xanthomonas citri subsp. citri 03-1638-1-1 | 1 | 2 | 1 | 4 |
| Xanthomonas citri subsp. citri 5208 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri A306 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri Aw12879 | 3 | 1 | 2 | 6 |
| Xanthomonas citri subsp. citri AW13 | 3 | 1 | 2 | 6 |
| Xanthomonas citri subsp. citri AW14 | 3 | 1 | 2 | 6 |
| Xanthomonas citri subsp. citri AW15 | 3 | 1 | 2 | 6 |
| Xanthomonas citri subsp. citri AW16 | 3 | 1 | 2 | 6 |
| Xanthomonas citri subsp. citri BL18 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri FB19 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri gd2 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri gd3 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri jx-6 | 1 | 1 | 1 | 3 |
| Xanthomonas citri subsp. citri jx4 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri jx5 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri LH201 | 1 | 1 | 1 | 3 |
| Xanthomonas citri subsp. citri LH276 | 1 | 2 | 1 | 4 |
| Xanthomonas citri subsp. citri LJ207-7 | 1 | 2 | 1 | 4 |
| Xanthomonas citri subsp. citri LL074-4 | 1 | 2 | 1 | 4 |
| Xanthomonas citri subsp. citri mf20 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri MN10 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri MN11 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri MN12 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri NT17 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri str. 306 | 0 | 0 | 0 | 0 |
| Xanthomonas citri subsp. citri TX160042 | 3 | 1 | 0 | 4 |
| Xanthomonas citri subsp. citri TX160149 | 2 | 1 | 3 | 6 |
| Xanthomonas citri subsp. citri TX160197 | 3 | 1 | 0 | 4 |
| Xanthomonas citri subsp. citri UI6 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri UI7 | 1 | 1 | 2 | 4 |
| Xanthomonas citri subsp. citri Xcc29-1 | 1 | 1 | 1 | 3 |
| Xanthomonas citri subsp. citri Xcc49 | 1 | 1 | 1 | 3 |
| Xanthomonas citri subsp. malvacearum AR81009 | 0 | 0 | 0 | 0 |
| Xanthomonas citri subsp. malvacearum MS14003 | 2 | 0 | 1 | 3 |
| Xanthomonas citri subsp. malvacearum MSCT | 1 | 0 | 0 | 1 |
| Xanthomonas citri subsp. malvacearum XcmH1005 | 0 | 2 | 0 | 2 |
| Xanthomonas citri subsp. malvacearum XcmN1003 | 3 | 1 | 1 | 5 |
| Xanthomonas vasicola pv. arecae NCPPB2649 | 1 | 2 | 0 | 3 |
| Xanthomonas vasicola pv. musacearum NCPPB 4379 | 2 | 0 | 1 | 3 |
References
- Chagas, M.C.M.; Parra, J.R.P.; Namekata, T.; Hartung, J.S.; Yamamoto, P.T. Phyllocnistis citrella Stainton (Lepidoptera: Gracillariidae) and its relationship with the citrus canker bacterium Xanthomonas axonopodis pv citri in Brazil. Neotrop. Entomol. 2001, 30, 55–59. [Google Scholar] [CrossRef]
- Nascimento, A.R.P.; Mariano, R.d.L.R. Cancro bacteriano da videira: Etiologia, epidemiologia e medidas de controle. Cienc. Rural 2004, 34, 301–307. [Google Scholar] [CrossRef]
- Iglesias-Bernabé, L.; Madloo, P.; Rodríguez, V.M.; Francisco, M.; Soengas, P. Dissecting quantitative resistance to Xanthomonas campestris pv. campestris in leaves of Brassica oleracea by QTL analysis. Sci. Rep. 2019, 9, 2015. [Google Scholar] [CrossRef] [PubMed]
- Batista, J.N.G.; Ferreira, M.A.d.S.V.; Quezado-Duval, A.M. Molecular and phenotypic characterization of Xanthomonas campestris pv. campestris causing black rot in Brassica crops in Brazil. Trop. Plant Pathol. 2021, 46, 684–701. [Google Scholar] [CrossRef]
- Nuñez, A.M.P.; Rodríguez, G.A.A.; Monteiro, F.P.; Faria, A.F.; Silva, J.C.P.; Monteiro, A.C.A.; Carvalho, C.V.; Gomes, L.A.A.; Souza, R.M.; de Souza, J.T.; et al. Bio-based products control black rot (Xanthomonas campestris pv. campestris) and increase the nutraceutical and antioxidant components in kale. Sci. Rep. 2018, 8, 10199. [Google Scholar] [CrossRef] [PubMed]
- Behlau, F. An overview of citrus canker in Brazil. Trop. Plant Pathol. 2021, 46, 1–12. [Google Scholar] [CrossRef]
- Ference, C.M.; Gochez, A.M.; Behlau, F.; Wang, N.; Graham, J.H.; Jones, J.B. Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 2018, 19, 1302–1318. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Patil, P.; Van Sluys, M.A.; White, F.F.; Ryan, R.P.; Dow, J.M.; Rabinowicz, P.; Salzberg, S.L.; Leach, J.E.; Sonti, R.; et al. Acquisition and Evolution of Plant Pathogenesis–Associated Gene Clusters and Candidate Determinants of Tissue-Specificity in Xanthomonas. PLoS ONE 2008, 3, e3828. [Google Scholar] [CrossRef] [PubMed]
- Hersemann, L.; Wibberg, D.; Blom, J.; Goesmann, A.; Widmer, F.; Vorhölter, F.J.; Kölliker, R. Comparative genomics of host adaptive traits in Xanthomonas translucens pv. graminis. BMC Genom. 2017, 18, 35. [Google Scholar] [CrossRef] [PubMed]
- Jacques, M.A.; Arlat, M.; Boulanger, A.; Boureau, T.; Carrère, S.; Cesbron, S.; Chen, N.W.; Cociancich, S.; Darrasse, A.; Denancé, N.; et al. Using Ecology, Physiology, and Genomics to Understand Host Specificity in Xanthomonas. Annu. Rev. Phytopathol. 2016, 54, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Assis, R.A.B.; Varani, A.M.; Sagawa, C.H.D.; Patané, J.S.L.; Setubal, J.C.; Uceda-Campos, G.; da Silva, A.M.; Zaini, P.A.; Almeida, N.F.; Moreira, L.M.; et al. A comparative genomic analysis of Xanthomonas arboricola pv. juglandis strains reveal hallmarks of mobile genetic elements in the adaptation and accelerated evolution of virulence. Genomics 2021, 113, 2513–2525. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.; Wu, T.L.; Zheng, P.X.; Ou, J.Y.; Ni, H.F.; Lin, Y.C. Comparative Genomic Analysis Uncovered Evolution of Pathogenicity Factors, Horizontal Gene Transfer Events, and Heavy Metal Resistance Traits in Citrus Canker Bacterium Xanthomonas citri subsp. citri. Front. Microbiol. 2021, 12, 731711. [Google Scholar] [CrossRef] [PubMed]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis, E.W.; Lim, C.K.; Shaffer, B.T.; Elbourne, L.D.H.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K.; et al. Comparative Genomics of Plant-Associated Pseudomonas spp.: Insights into Diversity and Inheritance of Traits Involved in Multitrophic Interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef]
- Hajri, A.; Brin, C.; Hunault, G.; Lardeux, F.; Lemaire, C.; Manceau, C.; Boureau, T.; Poussier, S. A «repertoire for repertoire» hypothesis: Repertoires of type three effectors are candidate determinants of host specificity in Xanthomonas. PLoS ONE 2009, 4, e6632. [Google Scholar] [CrossRef]
- Kay, S.; Bonas, U. How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 2009, 12, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Timilsina, S.; Pereira-Martin, J.A.; Minsavage, G.V.; Iruegas-Bocardo, F.; Abrahamian, P.; Potnis, N.; Kolaczkowski, B.; Vallad, G.E.; Goss, E.M.; Jones, J.B. Multiple Recombination Events Drive the Current Genetic Structure of Xanthomonas perforans in Florida. Front. Microbiol. 2019, 10, 448. [Google Scholar] [CrossRef] [PubMed]
- da Gama, M.A.S.; Mariano, R.d.L.R.; da Silva Júnior, W.J.; de Farias, A.R.G.; Barbosa, M.A.G.; Ferreira, M.Á.d.S.V.; Costa Júnior, C.R.L.; Santos, L.A.; de Souza, E.B. Taxonomic Repositioning of Xanthomonas campestris pv. viticola (Nayudu 1972) Dye 1978 as Xanthomonas citri pv. viticola (Nayudu 1972) Dye 1978 comb. nov. and Emendation of the Description of Xanthomonas citri pv. anacardii to Include Pigmented Isolates Pathogenic to Cashew Plant. Phytopathology 2018, 108, 1143–1153. [Google Scholar] [CrossRef] [PubMed]
- Constantin, E.C.; Cleenwerck, I.; Maes, M.; Baeyen, S.; Van Malderghem, C.; De Vos, P.; Cottyn, B. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 2016, 65, 792–806. [Google Scholar] [CrossRef]
- Cesbron, S.; Briand, M.; Essakhi, S.; Gironde, S.; Boureau, T.; Manceau, C.; Fischer-Le Saux, M.; Jacques, M.A. Comparative Genomics of Pathogenic and Nonpathogenic Strains of Xanthomonas arboricola Unveil Molecular and Evolutionary Events Linked to Pathoadaptation. Front. Plant Sci. 2015, 6, 1126. [Google Scholar] [CrossRef] [PubMed]
- Studholme, D.J.; Wicker, E.; Abrare, S.M.; Aspin, A.; Bogdanove, A.; Broders, K.; Dubrow, Z.; Grant, M.; Jones, J.B.; Karamura, G.; et al. Transfer of Xanthomonas campestris pv. arecae and X. campestris pv. musacearum to X. vasicola (Vauterin) as X. vasicola pv. arecae comb. nov. and X. vasicola pv. musacearum comb. nov. and Description of X. vasicola pv. vasculorum pv. nov. Phytopathology 2020, 110, 1153–1160. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Berriman, M.; Tivey, A.; Patel, C.; Böhme, U.; Barrell, B.G.; Parkhill, J.; Rajandream, M.A. Artemis and ACT: Viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 2008, 24, 2672–2676. [Google Scholar] [CrossRef] [PubMed]
- Araujo, F.A.; Barh, D.; Silva, A.; Guimarães, L.; Ramos, R.T.J. GO FEAT: A rapid web-based functional annotation tool for genomic and transcriptomic data. Sci. Rep. 2018, 8, 1794. [Google Scholar] [CrossRef]
- Kanehisa, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277-80. [Google Scholar] [CrossRef]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA—An ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree—Version 1.4. 3, a Graphical Viewer of Phylogenetic Trees. 2017. Computer program distributed by the author. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 10 September 2022).
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.C.; Geyik, H.; Ramos, R.T.J.; de Sá, P.H.C.G.; Barbosa, E.G.V.; Baumbach, J.; Figueiredo, H.C.P.; Miyoshi, A.; Tauch, A.; Silva, A.; et al. GIPSy: Genomic island prediction software. J. Biotechnol. 2016, 232, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Brinkman, F.S.L. Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 2018, 34, 2161–2167. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Singh, A.; Bansal, K.; Kumar, S.; Patil, P.B. Deep Population Genomics Reveals Systematic and Parallel Evolution at a Lipopolysaccharide Biosynthetic Locus in Xanthomonas Pathogens That Infect Rice and Sugarcane. Appl. Environ. Microbiol. 2022, 88, e00550-22. [Google Scholar] [CrossRef]
- Zarei, S.; Taghavi, S.M.; Rahimi, T.; Mafakheri, H.; Potnis, N.; Koebnik, R.; Fischer-Le Saux, M.; Pothier, J.F.; Palacio Bielsa, A.; Cubero, J.; et al. Taxonomic Refinement of Xanthomonas arboricola. Phytopathology 2022, 112, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Rana, R.; Madhavan, V.N.; Saroha, T.; Bansal, K.; Kaur, A.; Sonti, R.V.; Patel, H.K.; Patil, P.B. Xanthomonas indica sp. nov., a Novel Member of Non-Pathogenic Xanthomonas Community from Healthy Rice Seeds. Curr. Microbiol. 2022, 79, 304. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Song, E.S.; Heu, S.; Baek, J.; Lee, Y.K.; Lee, S.; Lee, S.W.; Park, D.S.; Lee, T.H.; Kim, J.G.; et al. Prediction of Host-Specific Genes by Pan-Genome Analyses of the Korean Ralstonia solanacearum Species Complex. Front. Microbiol. 2019, 10, 506. [Google Scholar] [CrossRef]
- Kang, I.J.; Kim, K.S.; Beattie, G.A.; Yang, J.W.; Sohn, K.H.; Heu, S.; Hwang, I. Pan-Genome Analysis of Effectors in Korean Strains of the Soybean Pathogen Xanthomonas citri pv. glycines. Microorganisms 2021, 9, 2065. [Google Scholar] [CrossRef]
- Patil, P.P.; Midha, S.; Kumar, S.; Patil, P.B. Genome Sequence of Type Strains of Genus Stenotrophomonas. Front. Microbiol. 2016, 7, 309. [Google Scholar] [CrossRef]
- Thieme, F.; Koebnik, R.; Bekel, T.; Berger, C.; Boch, J.; Büttner, D.; Caldana, C.; Gaigalat, L.; Goesmann, A.; Kay, S.; et al. Insights into Genome Plasticity and Pathogenicity of the Plant Pathogenic Bacterium Xanthomonas campestris pv. vesicatoria Revealed by the Complete Genome Sequence. J. Bacteriol. 2005, 187, 7254–7266. [Google Scholar] [CrossRef]
- Souza, D.P.; Oka, G.U.; Alvarez-Martinez, C.E.; Bisson-Filho, A.W.; Dunger, G.; Hobeika, L.; Cavalcante, N.S.; Alegria, M.C.; Barbosa, L.R.; Salinas, R.K.; et al. Bacterial killing via a type IV secretion system. Nat. Commun. 2015, 6, 6453. [Google Scholar] [CrossRef] [PubMed]
- Bansal, K.; Midha, S.; Kumar, S.; Patil, P.B. Ecological and Evolutionary Insights into Xanthomonas citri Pathovar Diversity. Appl. Environ. Microbiol. 2017, 83, e02993-16. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Grossman, A.D. Integrative and Conjugative Elements (ICEs): What They Do and How They Work. Annu. Rev. Genet. 2015, 49, 577–601. [Google Scholar] [CrossRef] [PubMed]
- Hallstrom, K.N.; McCormick, B.A. Pathogenicity Islands. In Molecular Medical Microbiology; Elsevier: Amsterdam, The Netherlands, 2015; pp. 303–314. [Google Scholar] [CrossRef]
- Sgro, G.G.; Oka, G.U.; Souza, D.P.; Cenens, W.; Bayer-Santos, E.; Matsuyama, B.Y.; Bueno, N.F.; dos Santos, T.R.; Alvarez-Martinez, C.E.; Salinas, R.K.; et al. Bacteria-Killing Type IV Secretion Systems. Front. Microbiol. 2019, 10, 1078. [Google Scholar] [CrossRef] [PubMed]
- Katzen, F.; Ferreiro, D.U.; Oddo, C.G.; Ielmini, M.V.; Becker, A.; Pühler, A.; Ielpi, L. Xanthomonas campestris pv. campestris gum Mutants: Effects on Xanthan Biosynthesis and Plant Virulence. J. Bacteriol. 1998, 180, 1607–1617. [Google Scholar] [CrossRef] [PubMed]
- Capage, M.; Doherty, D.H.; Betlach, M.; Vanderslice, R.W. Recombinant-DNA mediated production of xanthan gum. Biotechnology Advances 1997, 15, 547. [Google Scholar]
- Büttner, D.; Noël, L.; Thieme, F.; Bonas, U. Genomic approaches in Xanthomonas campestris pv. vesicatoria allow fishing for virulence genes. J. Biotechnol. 2003, 106, 203–214. [Google Scholar] [CrossRef]
- Han, S.W.; Hwang, B.K. Molecular functions of Xanthomonas type III effector AvrBsT and its plant interactors in cell death and defense signaling. Planta 2017, 245, 237–253. [Google Scholar] [CrossRef]
- Dong, Q.; Ebright, R.H. DNA binding specificity and sequence of Xanthomonas campestris catabolite gene activator protein-like protein. J. Bacteriol. 1992, 174, 5457–5461. [Google Scholar] [CrossRef][Green Version]
- de Crecy-Lagard, V.; Glaser, P.; Lejeune, P.; Sismeiro, O.; Barber, C.E.; Daniels, M.J.; Danchin, A. A Xanthomonas campestris pv. campestris protein similar to catabolite activation factor is involved in regulation of phytopathogenicity. J. Bacteriol. 1990, 172, 5877–5883. [Google Scholar] [CrossRef]
- Payne, S.M. Iron acquisition in microbial pathogenesis. Trends Microbiol. 1993, 1, 66–69. [Google Scholar] [CrossRef]
- Jordan, M.R.; Wang, J.; Capdevila, D.A.; Giedroc, D.P. Multi-metal nutrient restriction and crosstalk in metallostasis systems in microbial pathogens. Curr. Opin. Microbiol. 2020, 55, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Jiang, D.; Wang, Y.; Yao, X.; Luo, Y.; Yang, Z. Comparison of De Novo Assembly Strategies for Bacterial Genomes. Int. J. Mol. Sci. 2021, 22, 7668. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sharma, D.; Verma, S.K. In silico identification of copper-binding proteins of Xanthomonas translucens pv. undulosa for their probable role in plant-pathogen interactions. PMPP Physiol. Mol. Plant Pathol. 2019, 106, 187–195. [Google Scholar] [CrossRef]
- Lima, W.C.; Paquola, A.C.; Varani, A.M.; Van Sluys, M.A.; Menck, C.F. Laterally transferred genomic islands in Xanthomonadales related to pathogenicity and primary metabolism: LGT islands in Xanthomonas. FEMS Microbiol. Lett. 2008, 281, 87–97. [Google Scholar] [CrossRef]
- Cabrejos, D.A.L.; Alexandrino, A.V.; Pereira, C.M.; Mendonça, D.C.; Pereira, H.D.; Novo-Mansur, M.T.M.; Garratt, R.C.; Goto, L.S. Structural characterization of a pathogenicity-related superoxide dismutase codified by a probably essential gene in Xanthomonas citri subsp. citri. PLoS ONE 2019, 14, e0209988. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Daudi, A.; Butt, V.S.; Paul Bolwell, G. Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 2012, 236, 765–779. [Google Scholar] [CrossRef]
- Canchaya, C.; Fournous, G.; Brüssow, H. The impact of prophages on bacterial chromosomes: Prophage-chromosome interaction. Mol. Microbiol. 2004, 53, 9–18. [Google Scholar] [CrossRef]
- da Silva, A.C.R.; Ferro, J.A.; Reinach, F.C.; Farah, C.S.; Furlan, L.R.; Quaggio, R.B.; Monteiro-Vitorello, C.B.; Van Sluys, M.A.; Almeida, N.F.; Alves, L.M.C.; et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002, 417, 459–463. [Google Scholar] [CrossRef]
- Silva Junior, W.J.; Farias, A.R.G.; Lima, N.B.; Benko-Iseppon, A.M.; Aburjaile, F.; Balbino, V.Q.; Falcão, R.M.; Leitão Paiva Júnior, S.d.S.; Sousa-Paula, L.C.; Mariano, R.L.R.; et al. Complete genome sequence of Xanthomonas citri pv. anacardii strain IBSBF2579 from Brazil. Genome Announc. 2018, 6, e01574-17. [Google Scholar] [CrossRef]

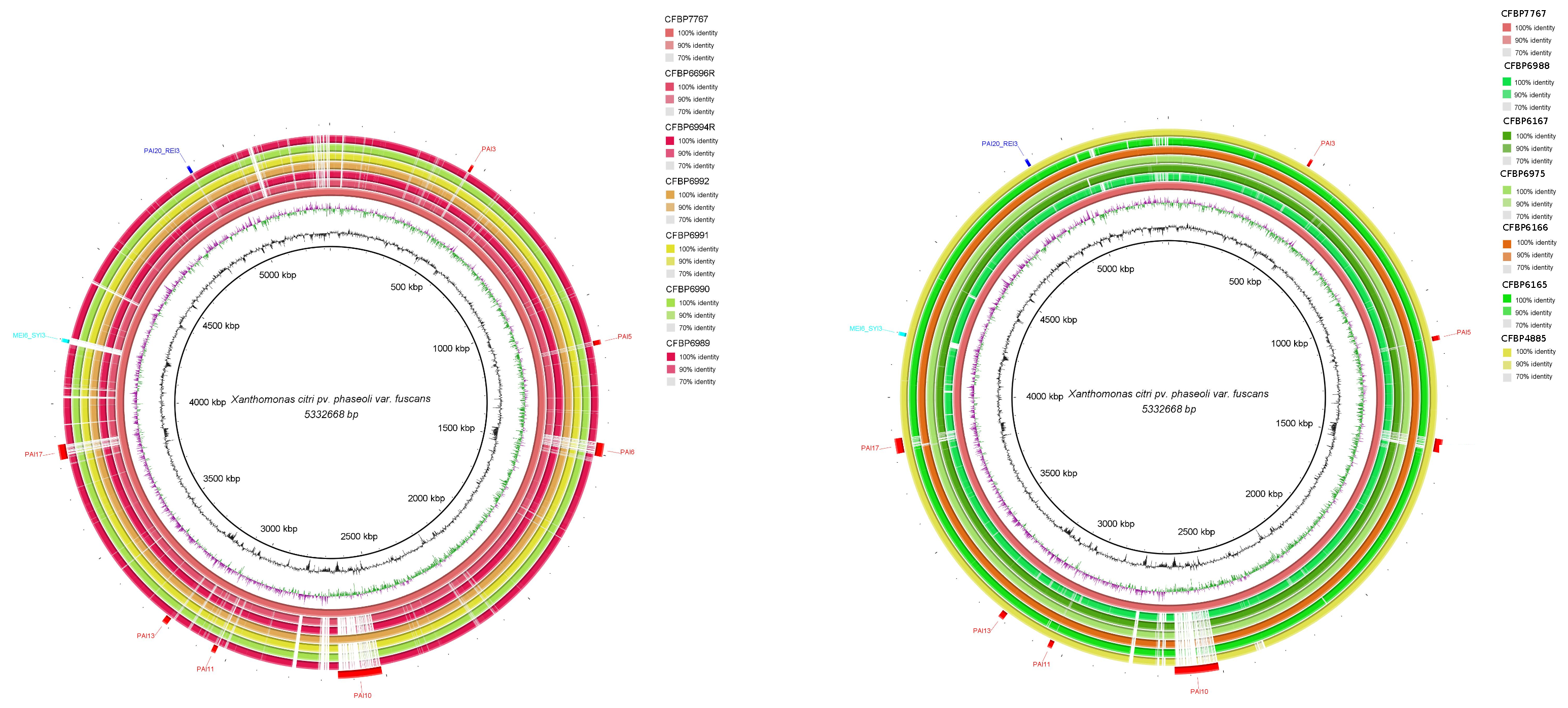
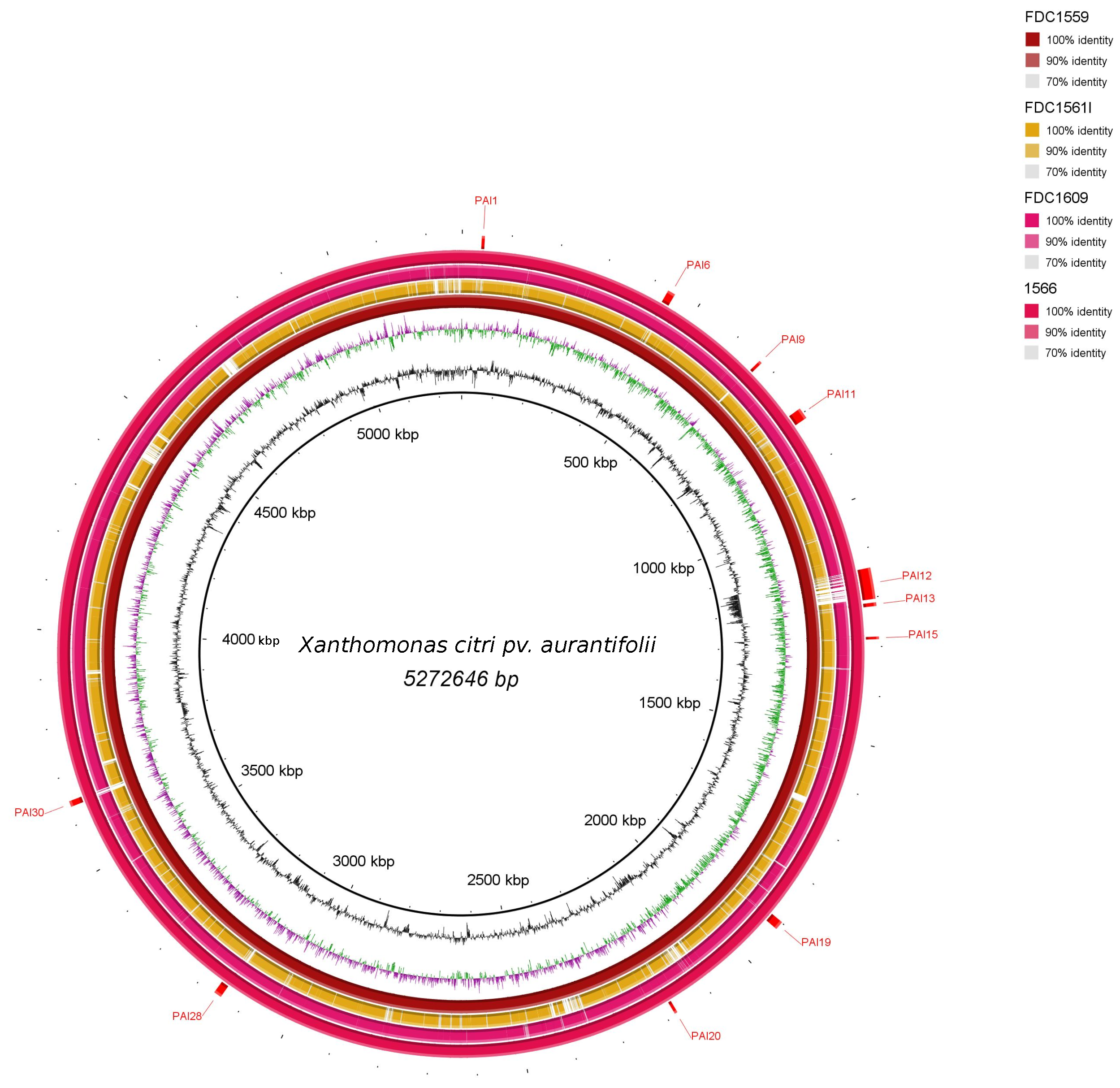
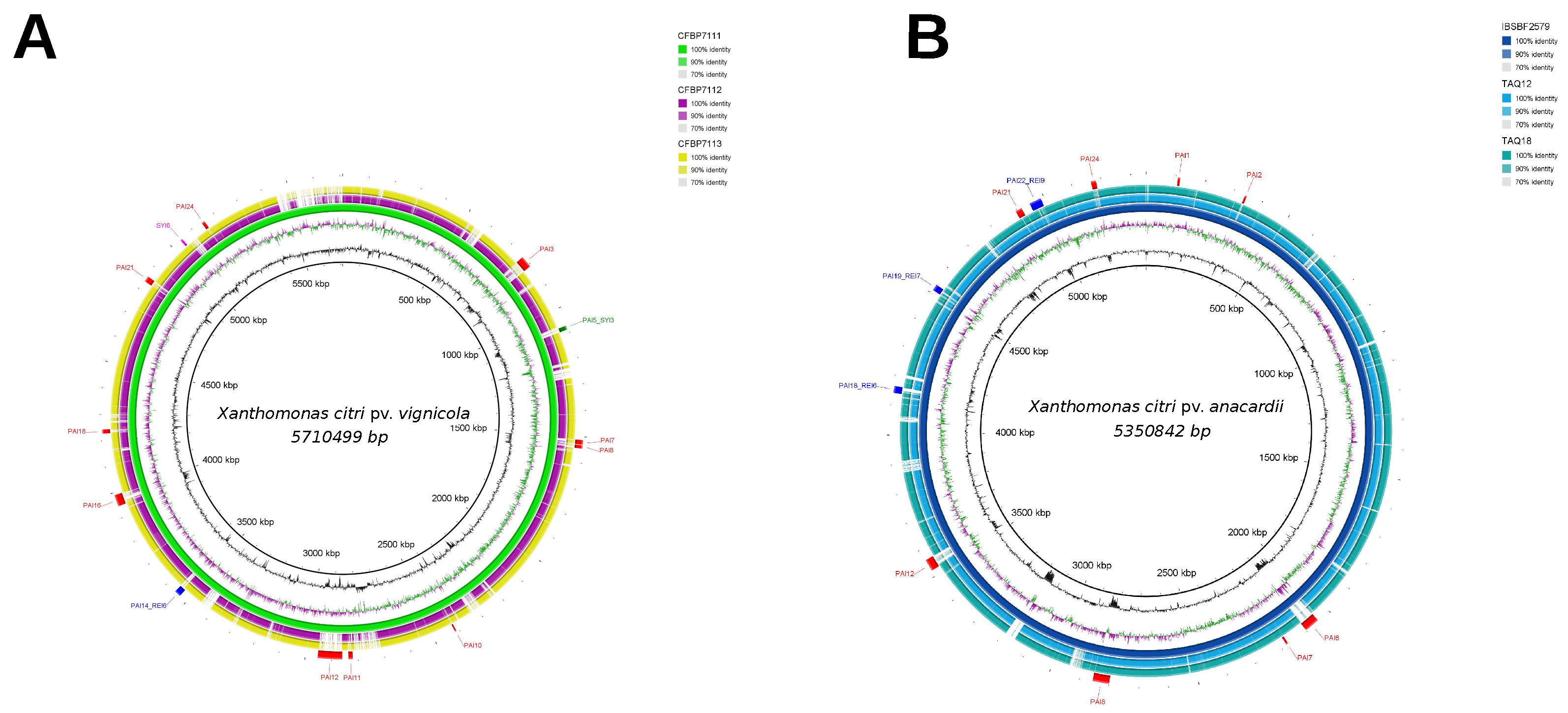
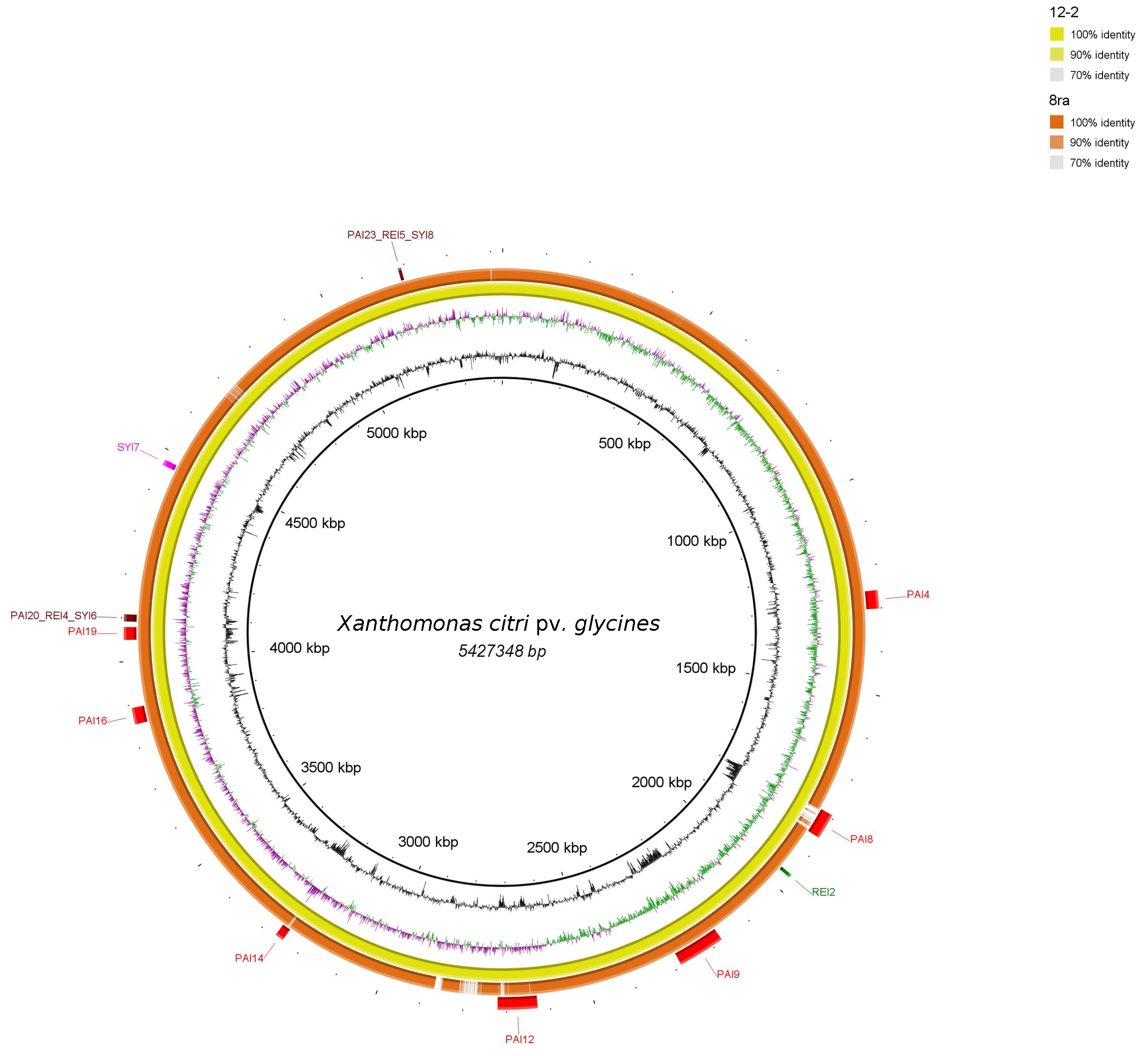

| Core | Acessory | Unique | |
|---|---|---|---|
| Xanthomonas | 2585 | 2750 | 11,562 |
| X. campestris | 3413 | 1813 | 1173 |
| X. citri | 2936 | 2561 | 9055 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ariute, J.C.; Rodrigues, D.L.N.; de Castro Soares, S.; Azevedo, V.; Benko-Iseppon, A.M.; Aburjaile, F.F. Comparative Genomic Analysis of Phytopathogenic Xanthomonas Species Suggests High Level of Genome Plasticity Related to Virulence and Host Adaptation. Bacteria 2022, 1, 218-241. https://doi.org/10.3390/bacteria1040017
Ariute JC, Rodrigues DLN, de Castro Soares S, Azevedo V, Benko-Iseppon AM, Aburjaile FF. Comparative Genomic Analysis of Phytopathogenic Xanthomonas Species Suggests High Level of Genome Plasticity Related to Virulence and Host Adaptation. Bacteria. 2022; 1(4):218-241. https://doi.org/10.3390/bacteria1040017
Chicago/Turabian StyleAriute, Juan Carlos, Diego Lucas Neres Rodrigues, Siomar de Castro Soares, Vasco Azevedo, Ana Maria Benko-Iseppon, and Flávia Figueira Aburjaile. 2022. "Comparative Genomic Analysis of Phytopathogenic Xanthomonas Species Suggests High Level of Genome Plasticity Related to Virulence and Host Adaptation" Bacteria 1, no. 4: 218-241. https://doi.org/10.3390/bacteria1040017
APA StyleAriute, J. C., Rodrigues, D. L. N., de Castro Soares, S., Azevedo, V., Benko-Iseppon, A. M., & Aburjaile, F. F. (2022). Comparative Genomic Analysis of Phytopathogenic Xanthomonas Species Suggests High Level of Genome Plasticity Related to Virulence and Host Adaptation. Bacteria, 1(4), 218-241. https://doi.org/10.3390/bacteria1040017









