Air–Liquid-Interface-Differentiated Human Nose Epithelium: The Benchmark Culture Model for SARS-CoV-2 Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Procurement of Human Material and Informed Consent
2.3. Primary Nasal Epithelium Culture and Differentiation
2.4. Polarization of Calu-3 Cells
2.5. SARS-CoV-2 Propagation and Infection
2.6. Immunofluorescence and Confocal Microscopy
2.7. Statistical Analyses
3. Results
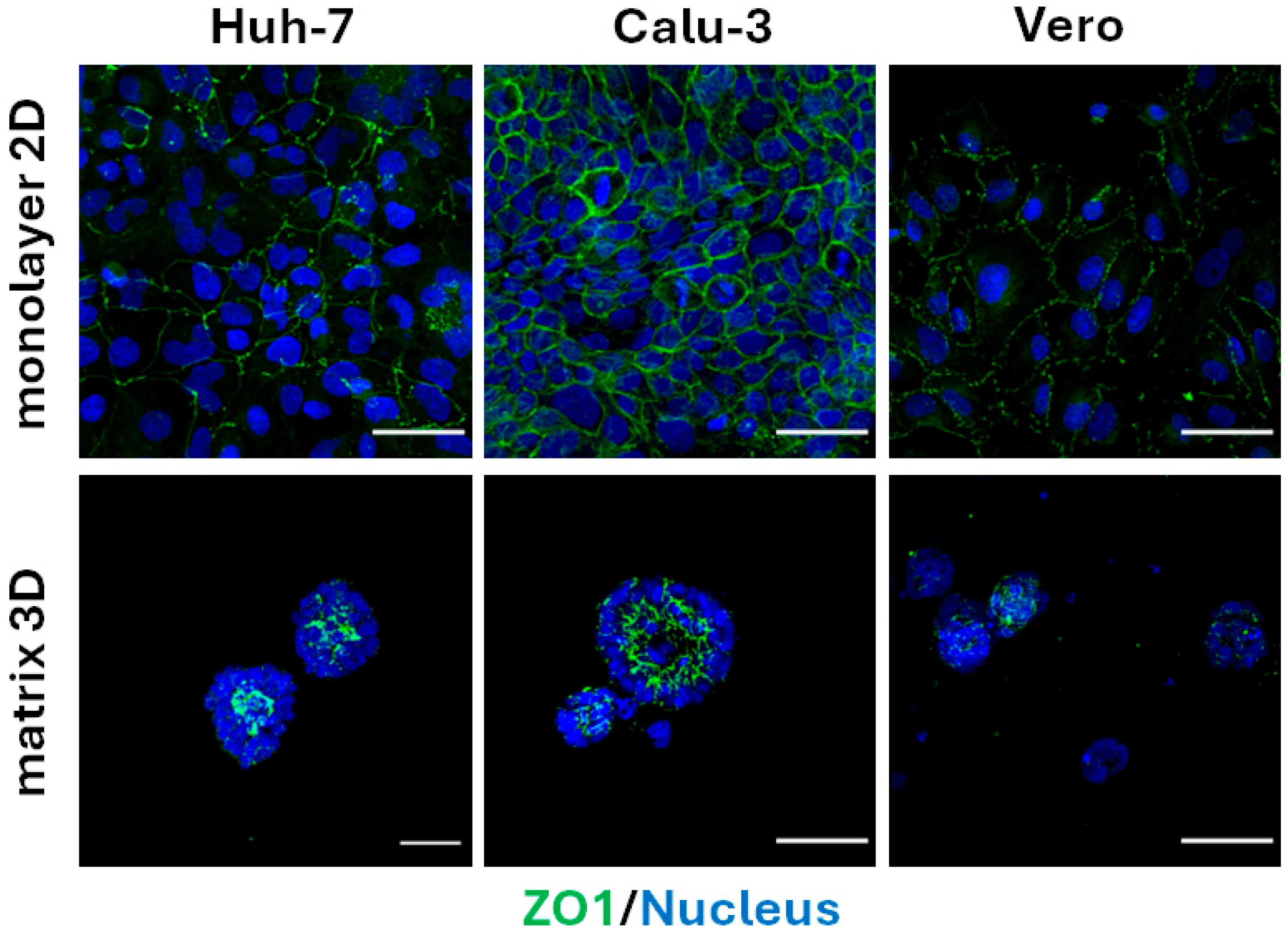
3.1. Calu-3 Cells Polarize Along the Apical–Basal Axis When Embedded in Matrix
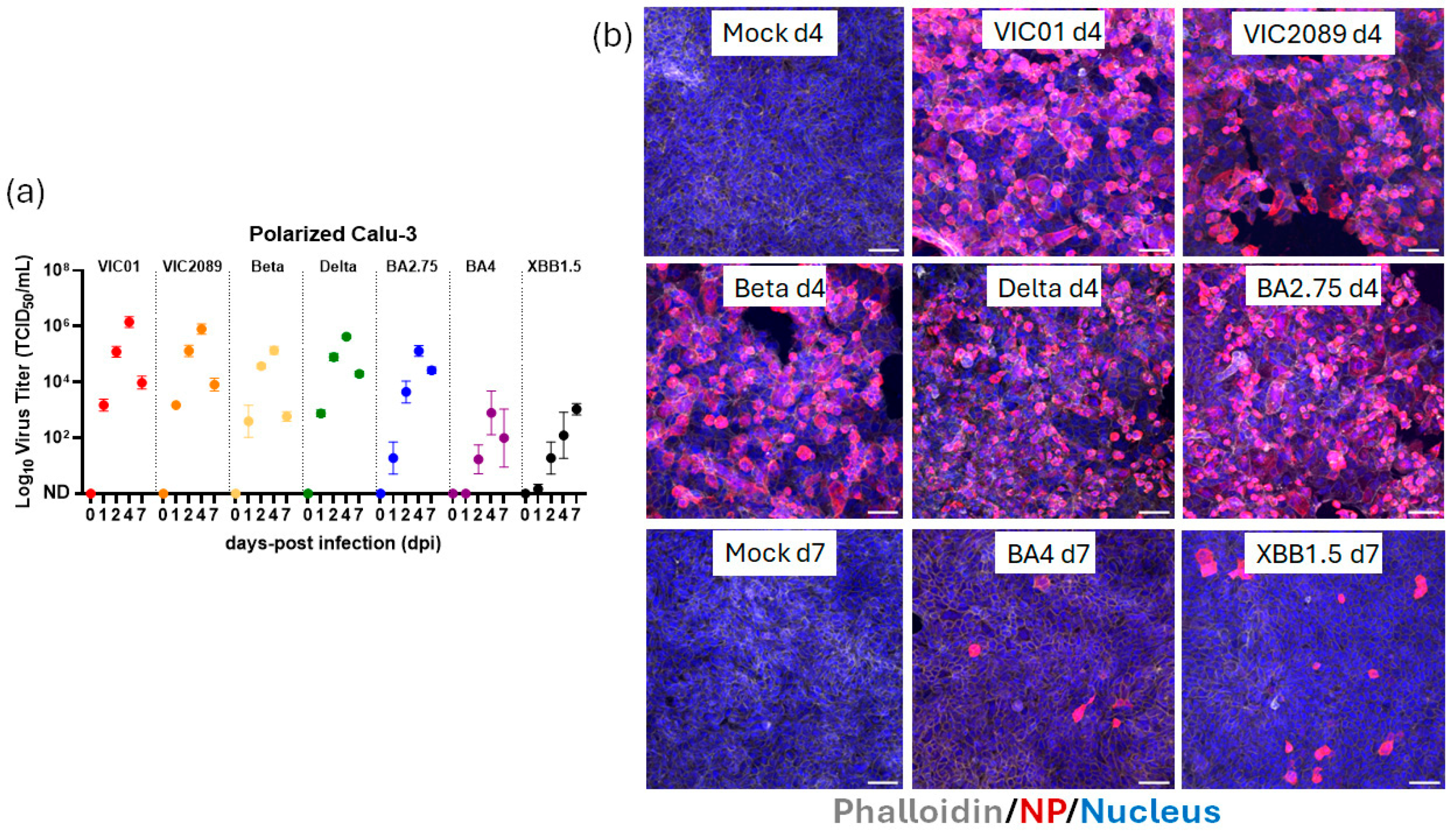
3.2. Calu-3 Cells Polarize on TranswellTM Inserts at ALI and Support Replication of Most, but Not All, SARS-CoV-2 Variants Tested
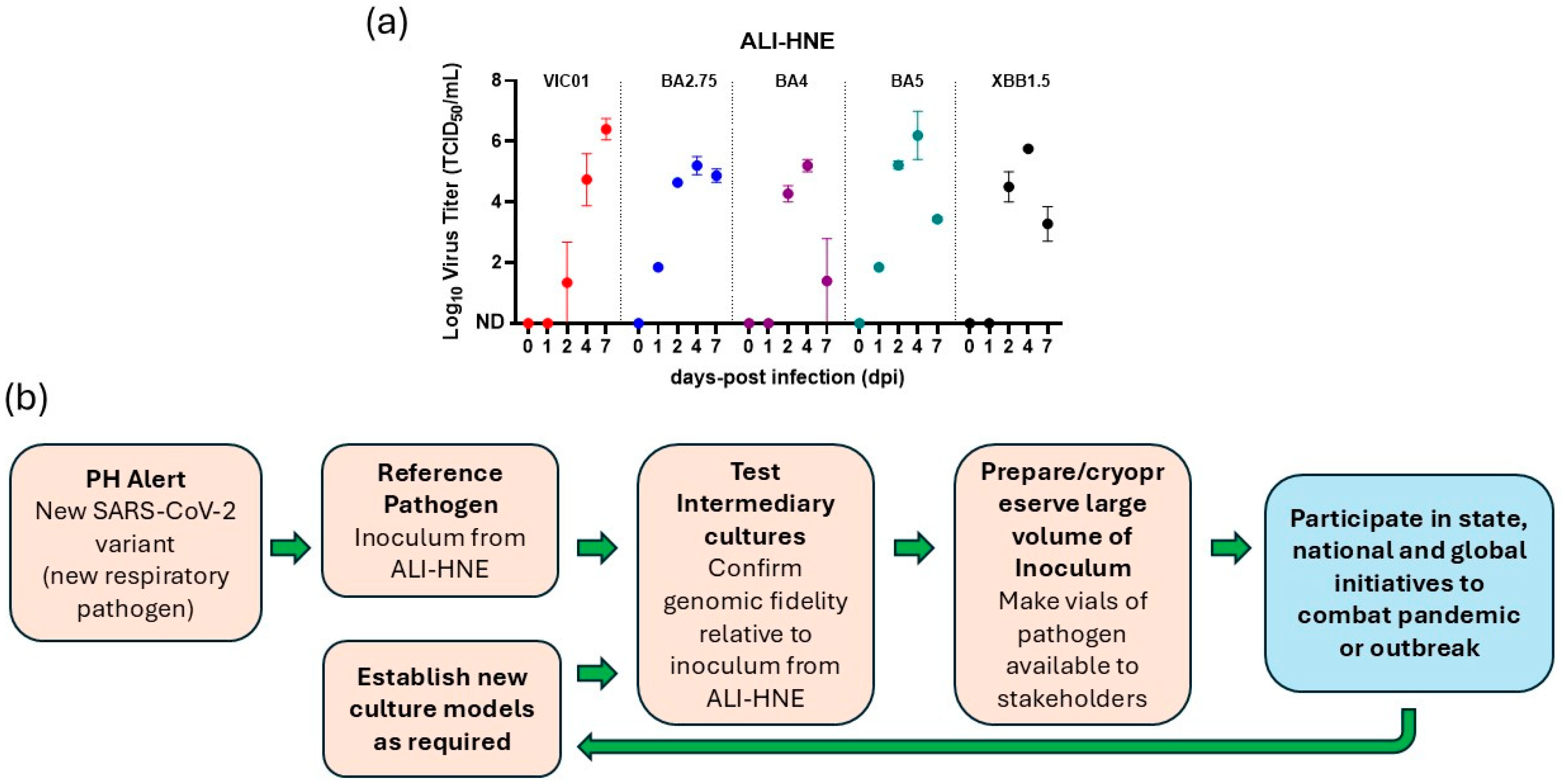
3.3. ALI-HNE Support Replication of All SARS-CoV-2 Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Quarleri, J.; Galvan, V.; Delpino, M.V. Omicron variant of the SARS-CoV-2: A quest to define the consequences of its high mutational load. Geroscience 2022, 44, 53–56. [Google Scholar] [CrossRef]
- Suzuki, R.; Yamasoba, D.; Kimura, I.; Wang, L.; Kishimoto, M.; Ito, J.; Morioka, Y.; Nao, N.; Nasser, H.; Uriu, K.; et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 2022, 603, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J.F.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M.S.; et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022, 602, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Suzuki, R.; Uriu, K.; Itakura, Y.; Zahradnik, J.; Kimura, K.T.; Deguchi, S.; Wang, L.; Lytras, S.; Tamura, T.; et al. Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant. Nat. Commun. 2023, 14, 2671. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddou, M.; Reuschl, A.K.; Polacco, B.J.; Thorne, L.G.; Ummadi, M.R.; Ye, C.; Rosales, R.; Pelin, A.; Batra, J.; Jang, G.M.; et al. SARS-CoV-2 variants evolve convergent strategies to remodel the host response. Cell 2023, 186, 4597–4614.e4526. [Google Scholar] [CrossRef]
- Khan, K.; Lustig, G.; Romer, C.; Reedoy, K.; Jule, Z.; Karim, F.; Ganga, Y.; Bernstein, M.; Baig, Z.; Jackson, L.; et al. Evolution and neutralization escape of the SARS-CoV-2 BA.2.86 subvariant. Nat. Commun. 2023, 14, 8078. [Google Scholar] [CrossRef]
- Tamura, T.; Ito, J.; Uriu, K.; Zahradnik, J.; Kida, I.; Anraku, Y.; Nasser, H.; Shofa, M.; Oda, Y.; Lytras, S.; et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat. Commun. 2023, 14, 2800. [Google Scholar] [CrossRef]
- Avdonin, P.P.; Rybakova, E.Y.; Trufanov, S.K.; Avdonin, P.V. SARS-CoV-2 Receptors and Their Involvement in Cell Infection. Biochem. Suppl. Ser. A Membr. Cell Biol. 2023, 17, 1–11. [Google Scholar] [CrossRef]
- Beumer, J.; Geurts, M.H.; Lamers, M.M.; Puschhof, J.; Zhang, J.; van der Vaart, J.; Mykytyn, A.Z.; Breugem, T.I.; Riesebosch, S.; Schipper, D.; et al. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat. Commun. 2021, 12, 5498. [Google Scholar] [CrossRef]
- Hou, Y.J.; Okuda, K.; Edwards, C.E.; Martinez, D.R.; Asakura, T.; Dinnon, K.H., 3rd; Kato, T.; Lee, R.E.; Yount, B.L.; Mascenik, T.M.; et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 2020, 182, 429–446.e14. [Google Scholar] [CrossRef]
- Caly, L.; Druce, J.; Roberts, J.; Bond, K.; Tran, T.; Kostecki, R.; Yoga, Y.; Naughton, W.; Taiaroa, G.; Seemann, T.; et al. Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med. J. Aust. 2020, 212, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Rosenke, K.; Jarvis, M.A.; Feldmann, F.; Schwarz, B.; Okumura, A.; Lovaglio, J.; Saturday, G.; Hanley, P.W.; Meade-White, K.; Williamson, B.N.; et al. Hydroxychloroquine Proves Ineffective in Hamsters and Macaques Infected with SARS-CoV-2. bioRxiv 2020. [Google Scholar] [CrossRef]
- Boulware, D.R.; Pullen, M.F.; Bangdiwala, A.S.; Pastick, K.A.; Lofgren, S.M.; Okafor, E.C.; Skipper, C.P.; Nascene, A.A.; Nicol, M.R.; Abassi, M.; et al. A Randomized Trial of Hydroxychloroquine as Postexposure Prophylaxis for COVID-19. N. Engl. J. Med. 2020, 383, 517–525. [Google Scholar] [CrossRef]
- Tran, B.M.; Grimley, S.L.; McAuley, J.L.; Hachani, A.; Earnest, L.; Wong, S.L.; Caly, L.; Druce, J.; Purcell, D.F.J.; Jackson, D.C.; et al. Air-Liquid-Interface Differentiated Human Nose Epithelium: A Robust Primary Tissue Culture Model of SARS-CoV-2 Infection. Int. J. Mol. Sci. 2022, 23, 835. [Google Scholar] [CrossRef]
- Mykytyn, A.Z.; Breugem, T.I.; Geurts, M.H.; Beumer, J.; Schipper, D.; van Acker, R.; van den Doel, P.B.; van Royen, M.E.; Zhang, J.; Clevers, H.; et al. SARS-CoV-2 Omicron entry is type II transmembrane serine protease-mediated in human airway and intestinal organoid models. J. Virol. 2023, 97, e0085123. [Google Scholar] [CrossRef]
- Tanneti, N.S.; Patel, A.K.; Tan, L.H.; Marques, A.D.; Perera, R.; Sherrill-Mix, S.; Kelly, B.J.; Renner, D.M.; Collman, R.G.; Rodino, K.; et al. Comparison of SARS-CoV-2 variants of concern in primary human nasal cultures demonstrates Delta as most cytopathic and Omicron as fastest replicating. mBio 2024, 15, e0312923. [Google Scholar] [CrossRef] [PubMed]
- Wickenhagen, A.; Flagg, M.; Port, J.R.; Yinda, C.K.; Goldin, K.; Gallogly, S.; Schulz, J.E.; Lutterman, T.; Williamson, B.N.; Kaiser, F.; et al. Evolution of Omicron lineage towards increased fitness in the upper respiratory tract in the absence of severe lung pathology. Nat. Commun. 2025, 16, 594. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.Q.; Finkbeiner, W.E.; Wine, J.J.; Mrsny, R.J.; Widdicombe, J.H. Calu-3: A human airway epithelial cell line that shows cAMP-dependent Cl- secretion. Am. J. Physiol. Lung Cell. Mol. Physiol. 1994, 266, L493–L501. [Google Scholar] [CrossRef]
- Mykytyn, A.Z.; Breugem, T.I.; Riesebosch, S.; Schipper, D.; van den Doel, P.B.; Rottier, R.J.; Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. eLife 2021, 10, e64508. [Google Scholar] [CrossRef]
- Lamers, M.M.; Mykytyn, A.Z.; Breugem, T.I.; Wang, Y.; Wu, D.C.; Riesebosch, S.; van den Doel, P.B.; Schipper, D.; Bestebroer, T.; Wu, N.C.; et al. Human airway cells prevent SARS-CoV-2 multibasic cleavage site cell culture adaptation. eLife 2021, 10, e66815. [Google Scholar] [CrossRef]
- Ren, X.; Glende, J.; Al-Falah, M.; de Vries, V.; Schwegmann-Wessels, C.; Qu, X.; Tan, L.; Tschernig, T.; Deng, H.; Naim, H.Y.; et al. Analysis of ACE2 in polarized epithelial cells: Surface expression and function as receptor for severe acute respiratory syndrome-associated coronavirus. J. Gen. Virol. 2006, 87, 1691–1695. [Google Scholar] [CrossRef] [PubMed]
- Baczenas, J.J.; Andersen, H.; Rashid, S.; Yarmosh, D.; Puthuveetil, N.; Parker, M.; Bradford, R.; Florence, C.; Stemple, K.J.; Lewis, M.G.; et al. Propagation of SARS-CoV-2 in Calu-3 Cells to Eliminate Mutations in the Furin Cleavage Site of Spike. Viruses 2021, 13, 2434. [Google Scholar] [CrossRef]
- Lee, G.Y.; Kenny, P.A.; Lee, E.H.; Bissell, M.J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 2007, 4, 359–365. [Google Scholar] [CrossRef]
- Collett, S.; Torresi, J.; Silveira, L.E.; Truong, V.K.; Christiansen, D.; Tran, B.M.; Vincan, E.; Ramsland, P.A.; Elbourne, A. Investigating virus-host cell interactions: Comparative binding forces between hepatitis C virus-like particles and host cell receptors in 2D and 3D cell culture models. J. Colloid Interface Sci. 2021, 592, 371–384. [Google Scholar] [CrossRef]
- Harbach, S.L.; Tran, B.M.; Kastrappis, G.; Tran, H.; Grimley, S.L.; McAuley, J.L.; Hachani, A.; Vincan, E. Polarized Calu-3 Cells Serve as an Intermediary Model for SARS-CoV-2 Infection. In Methods in Molecular Biology; Springer: New York, NY, USA, 2025. [Google Scholar] [CrossRef]
- Lee, J.Y.H.; Best, N.; McAuley, J.; Porter, J.L.; Seemann, T.; Schultz, M.B.; Sait, M.; Orlando, N.; Mercoulia, K.; Ballard, S.A.; et al. Validation of a single-step, single-tube reverse transcription loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. J. Med. Microbiol. 2020, 69, 1169–1178. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- To, C.Z.; Bhunia, A.K. Three Dimensional Vero Cell-Platform for Rapid and Sensitive Screening of Shiga-Toxin Producing Escherichia coli. Front. Microbiol. 2019, 10, 949. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; Paul van Schayck, J.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef]
- Gu, H.; Chen, Q.; Yang, G.; He, L.; Fan, H.; Deng, Y.Q.; Wang, Y.; Teng, Y.; Zhao, Z.; Cui, Y.; et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020, 369, 1603–1607. [Google Scholar] [CrossRef]
- Pymm, P.; Redmond, S.J.; Dolezal, O.; Mordant, F.; Lopez, E.; Cooney, J.P.; Davidson, K.C.; Haycroft, E.R.; Tan, C.W.; Seneviratna, R.; et al. Biparatopic nanobodies targeting the receptor binding domain efficiently neutralize SARS-CoV-2. iScience 2022, 25, 105259. [Google Scholar] [CrossRef]
- Bader, S.M.; Cooney, J.P.; Sheerin, D.; Taiaroa, G.; Harty, L.; Davidson, K.C.; Mackiewicz, L.; Dayton, M.; Wilcox, S.; Whitehead, L.; et al. SARS-CoV-2 mouse adaptation selects virulence mutations that cause TNF-driven age-dependent severe disease with human correlates. Proc. Natl. Acad. Sci. USA 2023, 120, e2301689120. [Google Scholar] [CrossRef]
- Meijers, M.; Ruchnewitz, D.; Eberhardt, J.; Luksza, M.; Lassig, M. Population immunity predicts evolutionary trajectories of SARS-CoV-2. Cell 2023, 186, 5151–5164.e13. [Google Scholar] [CrossRef] [PubMed]
- WHO. Tracking SARS-CoV-2. Available online: https://www.who.int/activities/tracking-SARS-CoV-2-variants (accessed on 5 June 2025).
- Planas, D.; Staropoli, I.; Michel, V.; Lemoine, F.; Donati, F.; Prot, M.; Porrot, F.; Guivel-Benhassine, F.; Jeyarajah, B.; Brisebarre, A.; et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat. Commun. 2024, 15, 2254. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.J.; Grimley, S.L.; Tran, B.M.; Deliyannis, G.; Tumpach, C.; Nguyen, A.N.T.; Steinig, E.; Zhang, J.; Schroder, J.; Caly, L.; et al. Uncovering strain- and age-dependent innate immune responses to SARS-CoV-2 infection in air-liquid-interface cultured nasal epithelia. iScience 2024, 27, 110009. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.Y.; Wang, P.; Mok, B.W.; Zhang, A.J.; Chu, H.; Lee, A.C.; Deng, S.; Chen, P.; Chan, K.H.; Song, W.; et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg. Microbes Infect. 2020, 9, 837–842. [Google Scholar] [CrossRef]
- WHO. Research Response to Pathogen X During a Pandemic. Available online: https://www.who.int/news-room/events/detail/2024/01/19/default-calendar/Research-response-to-pathogen-X-during-a-pandemic (accessed on 11 June 2025).
- Wadman, M. FDA no longer has to require animal testing for new drugs. Science 2023, 379, 127–128. [Google Scholar] [CrossRef]
- Carrera Montoya, J.; Collett, S.; Fernandez Ruiz, D.; Earnest, L.; Edeling, M.A.; Yap, A.H.Y.; Wong, C.Y.; Cooney, J.P.; Davidson, K.C.; Roberts, J.; et al. Human Nasal Epithelium Organoids for Assessing Neutralizing Antibodies to a Protective SARS-CoV-2 Virus-like Particle Vaccine. Organoids 2024, 3, 18–31. [Google Scholar] [CrossRef]
- Wan, Z.X.; Li, C.; Zhou, Y.; Chiu, M.C.; Huang, J.J.; Zhang, S.X.; Zhu, X.X.; Lan, Q.S.; Deng, Y.L.; Xue, W.; et al. Organoid-based neutralization assays reveal a distinctive profile of SARS-CoV-2 antibodies and recapitulate the real-world efficacy. Proc. Natl. Acad. Sci. USA 2025, 122, e2509616122. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harbach, S.L.; Tran, B.M.; Hachani, A.; Grimley, S.L.; Purcell, D.F.J.; Deliyannis, G.; Torresi, J.; McAuley, J.L.; Vincan, E. Air–Liquid-Interface-Differentiated Human Nose Epithelium: The Benchmark Culture Model for SARS-CoV-2 Infection. Organoids 2025, 4, 21. https://doi.org/10.3390/organoids4030021
Harbach SL, Tran BM, Hachani A, Grimley SL, Purcell DFJ, Deliyannis G, Torresi J, McAuley JL, Vincan E. Air–Liquid-Interface-Differentiated Human Nose Epithelium: The Benchmark Culture Model for SARS-CoV-2 Infection. Organoids. 2025; 4(3):21. https://doi.org/10.3390/organoids4030021
Chicago/Turabian StyleHarbach, Sarah L., Bang M. Tran, Abderrahman Hachani, Samantha Leigh Grimley, Damian F. J. Purcell, Georgia Deliyannis, Joseph Torresi, Julie L. McAuley, and Elizabeth Vincan. 2025. "Air–Liquid-Interface-Differentiated Human Nose Epithelium: The Benchmark Culture Model for SARS-CoV-2 Infection" Organoids 4, no. 3: 21. https://doi.org/10.3390/organoids4030021
APA StyleHarbach, S. L., Tran, B. M., Hachani, A., Grimley, S. L., Purcell, D. F. J., Deliyannis, G., Torresi, J., McAuley, J. L., & Vincan, E. (2025). Air–Liquid-Interface-Differentiated Human Nose Epithelium: The Benchmark Culture Model for SARS-CoV-2 Infection. Organoids, 4(3), 21. https://doi.org/10.3390/organoids4030021








