Mutation in the LONGIFOLIA1 Gene Resulted in Suppressed Insensitivity of Arabidopsis thaliana proteolysis6 Mutant to Ethylene During Seed Germination
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. EMS Mutagenesis
2.3. Screening of Suppressors of prt6−1 Mutant
2.4. Population Construction and Bulk DNA Preparation
2.5. BSA-Seq Analysis
2.6. CRISPR-Cas9 Editing
2.7. Germination Assays with Ethylene Treatment
2.8. Silique Length Measurement
3. Results
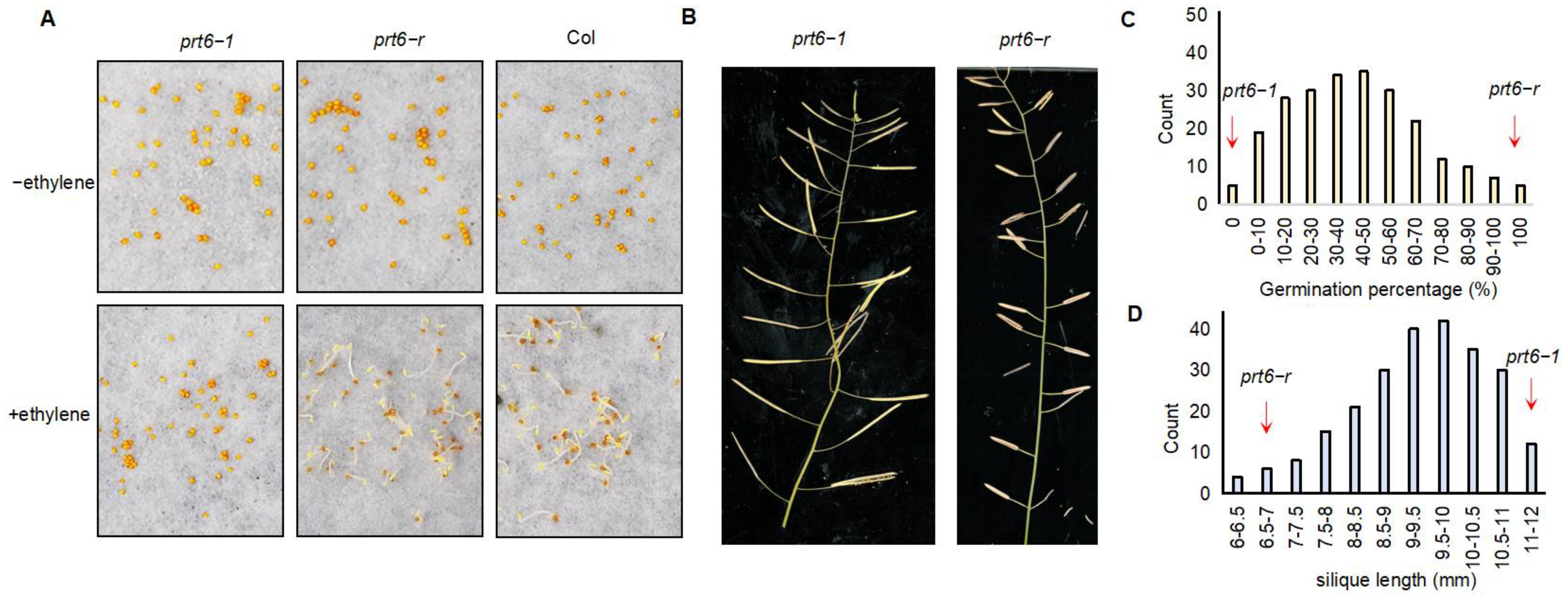
3.1. Phenotypic Characterization of prt6−r and Bulk Construction
3.2. Quality Control of Whole-Genome Sequencing Data
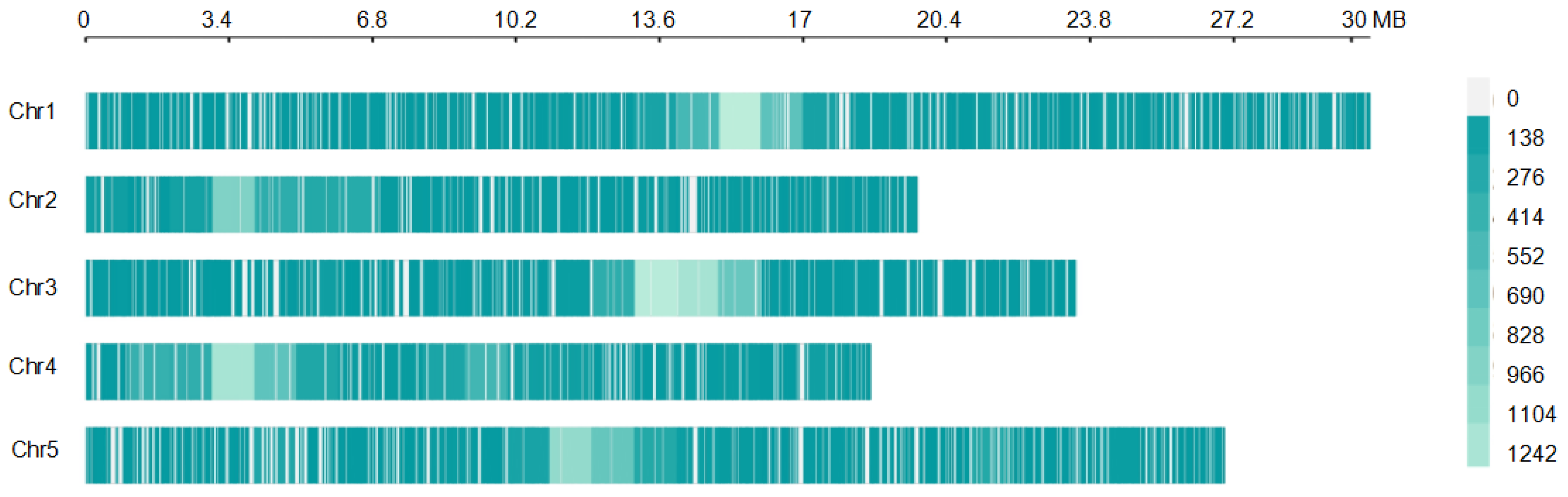
3.3. Genomic Variation Distribution and Annotation
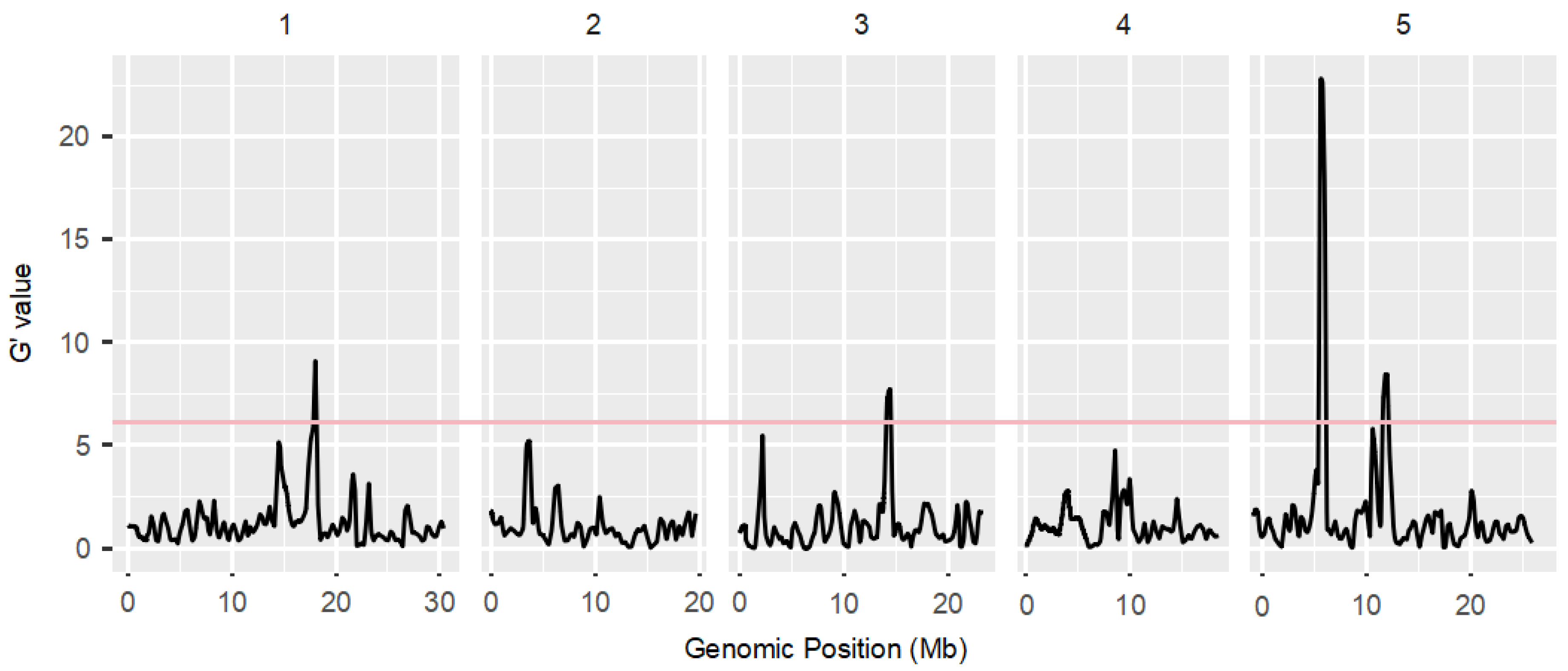
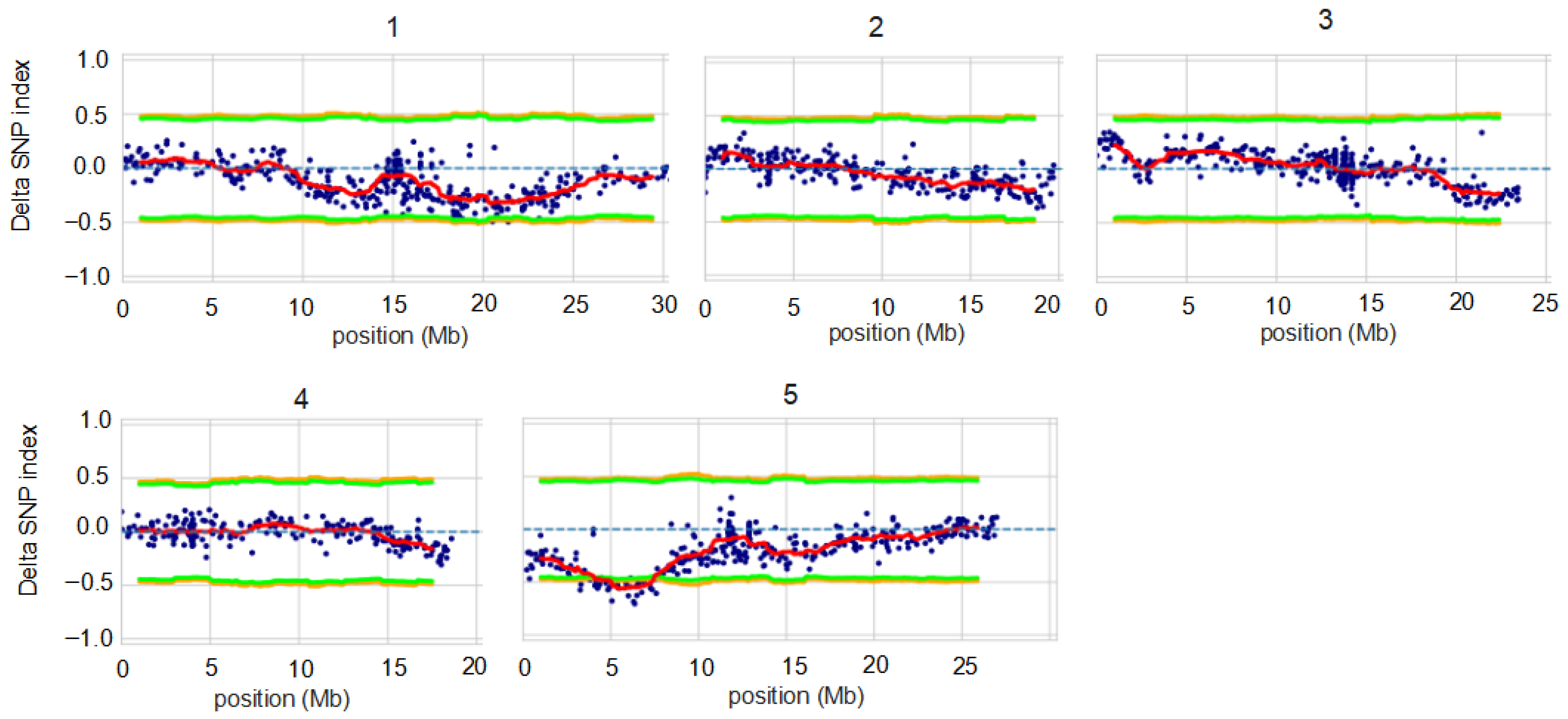
3.4. BSAseq Based on G’ Value Method and Delta SNP Method
3.5. Annotation of Candidate Variants and Candidate Gene Confirmation
4. Discussion
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Footitt, S. Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 2017, 68, 843–856. [Google Scholar] [CrossRef]
- Matilla, A.J.; Matilla-Vázquez, M.A. Involvement of ethylene in seed physiology. Plant Sci. 2008, 175, 87–97. [Google Scholar] [CrossRef]
- Arc, E.; Sechet, J.; Corbineau, F.; Rajjou, L.; Marion-Poll, A. ABA crosstalk with ethylene and nitric oxide in seed dormancy and germination. Front. Plant Sci. 2013, 4, 63. [Google Scholar] [CrossRef]
- Corbineau, F.; Xia, Q.; Bailly, C.; El-Maarouf-Bouteau, H. Ethylene, a key factor in the regulation of seed dormancy. Front. Plant Sci. 2013, 5, 539. [Google Scholar] [CrossRef] [PubMed]
- Corbineau, F. Ethylene, a signaling compound involved in seed germination and dormancy. Plants 2024, 13, 2674. [Google Scholar] [CrossRef] [PubMed]
- Kamaluddin, M.; Zwiazek, J.J. Ethylene enhances water transport in hypoxic Aspen. Plant Physiol. 2002, 128, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Arc, E.; Galland, M.; Godin, B.; Cueff, G.; Rajjou, L. Nitric oxide implication in the control of seed dormancy and germination. Front. Plant Sci. 2013, 4, 346. [Google Scholar] [CrossRef]
- Prusinski, J.; Khan, A.A. Relationship of ethylene production to stress alleviation in seeds of lettuce cultivars. J. Am. Soc. Hortic. Sci. 1990, 115, 294–298. [Google Scholar] [CrossRef]
- El-Maarouf-Bouteau, H.; Sajjad, Y.; Bazin, J.; Langlade, N.; Cristescu, S.M.; Balzergue, S.; Baudouin, E.; Bailly, C. Reactive oxygen species, abscisic acid and ethylene interact to regulate sunflower seed germination. Plant Cell Environ. 2015, 38, 364–374. [Google Scholar] [CrossRef]
- Wang, K.L.C.; Li, H.; Ecker, J.R. Ethylene Biosynthesis and Signaling Networks. Plant Cell 2002, 14, S131. [Google Scholar] [CrossRef] [PubMed]
- Merchante, C.; Alonso, J.M.; Stepanova, A.N. Ethylene signaling: Simple ligand, complex regulation. Curr. Opin. Plant Biol. 2013, 16, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, N.; Serizet, C.; Gosti, F.; Giraudat, J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 2000, 12, 1103–1115. [Google Scholar] [CrossRef]
- Siriwitayawan, G.; Geneve, R.L.; Downie, A. Seed germination of ethylene perception mutants of tomato and Arabidopsis. Seed Sci. Res. 2003, 13, 303–314. [Google Scholar] [CrossRef]
- Chiwocha, S.D.; Cutler, A.J.; Abrams, S.R.; Ambrose, S.J.; Yang, J.; Ross, A.R.; Kermode, A.R. The etr1−2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 2005, 42, 35–48. [Google Scholar] [CrossRef]
- Li, X.; Chen, T.; Li, Y.; Wang, Z.; Cao, H.; Chen, F.; Li, Y.; Soppe, W.J.J.; Li, W.; Liu, Y. ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell 2019, 31, 832–847. [Google Scholar] [CrossRef] [PubMed]
- Jurdak, R.; Launay-Avon, A.; Paysant-Le Roux, C.; Bailly, C. Retrograde signalling from the mitochondria to the nucleus translates the positive effect of ethylene on dormancy breaking of Arabidopsis thaliana seeds. New Phytol. 2021, 229, 2192–2205. [Google Scholar] [CrossRef]
- Graciet, E.; Wellmer, F. The plant N-end rule pathway: Structure and functions. Trends Plant Sci. 2010, 15, 447–453. [Google Scholar] [CrossRef]
- Sriram, S.M.; Kim, B.Y.; Kwon, Y.T. The N-end rule pathway: Emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 2011, 12, 735–747. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Lee, S.C.; Isa, N.M.; Gramuglia, S.; Fukao, T.; Bassel, G.W.; Correia, C.S.; Corbineau, F.; Theodoulou, F.L.; Bailey-Serres, J.; et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 2011, 479, 415–418. [Google Scholar] [CrossRef]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Tedds, H.M.; Labandera, A.-M.; Bailey, M.; White, M.D.; Hartman, S.; Sprigg, C.; Mogg, S.L.; Osborne, R.; Dambire, C.; et al. Oxygen-dependent proteolysis regulates the stability of angiosperm polycomb repressive complex 2 subunit VERNALIZATION 2. Nat. Commun. 2018, 9, 5438. [Google Scholar] [CrossRef] [PubMed]
- Weits, D.A.; Kunkowska, A.B.; Kamps, N.C.W.; Portz, K.M.S.; Packbier, N.K.; Nemec Venza, Z.; Gaillochet, C.; Lohmann, J.U.; Pedersen, O.; van Dongen, J.T.; et al. An apical hypoxic niche sets the pace of shoot meristem activity. Nature 2019, 569, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Labandera, A.-M.; Tedds, H.M.; Bailey, M.; Sprigg, C.; Etherington, R.D.; Akintewe, O.; Kalleechurn, G.; Holdsworth, M.J.; Gibbs, D.J. The PRT6 N-degron pathway restricts VERNALIZATION 2 to endogenous hypoxic niches to modulate plant development. New Phytol. 2021, 229, 126–139. [Google Scholar] [CrossRef]
- Holman, T.J.; Jones, P.D.; Russell, L.; Medhurst, A.; Ubeda Tomas, S.; Talloji, P.; Marquez, J.; Schmuths, H.; Tung, S.A.; Taylor, I.; et al. The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 4549–4554. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Isa, N.M.; Movahedi, M.; Lozano-Juste, J.; Mendiondo, G.M.; Berckhan, S.; Marin-de la Rosa, N.; Conde, J.V.; Correia, C.S.; Pearce, S.P.; et al. Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 2014, 53, 369–379. [Google Scholar] [CrossRef]
- Wang, X.; Gomes, M.M.; Bailly, C.; Nambara, E.; Corbineau, F. Role of ethylene and proteolytic N-degron pathway in the regulation of Arabidopsis seed dormancy. J. Integr. Plant Biol. 2021, 63, 2110–2122. [Google Scholar] [CrossRef]
- Wang, X.; Yesbergenova-Cuny, Z.; Biniek, C.; Bailly, C.; El-Maarouf-Bouteau, H.; Corbineau, F. Revisiting the role of ethylene and N-end rule pathway on chilling-induced dormancy release in Arabidopsis seeds. Int. J. Mol. Sci. 2018, 19, 3577. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chory, J. Preparation of DNA from Arabidopsis. In Arabidopsis Protocols; Springer: Berlin/Heidelberg, Germany, 1998; pp. 55–60. [Google Scholar]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing Supgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Sugihara, Y.; Young, L.; Yaegashi, H.; Natsume, S.; Shea, D.J.; Takagi, H.; Booker, H.; Innan, H.; Terauchi, R.; Abe, A. High-performance pipeline for MutMap and QTL-seq. PeerJ 2022, 10, e13170. [Google Scholar] [CrossRef] [PubMed]
- Mansfeld, B.N.; Grumet, R. QTLseqr: An R Package for Bulk Segregant Analysis with Next-Generation Sequencing. Plant Genome 2018, 11, 180006. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Greene, E.A.; Codomo, C.A.; Taylor, N.E.; Henikoff, J.G.; Till, B.J.; Reynolds, S.H.; Enns, L.C.; Burtner, C.; Johnson, J.E.; Odden, A.R.; et al. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 2003, 164, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Magwene, P.M.; Willis, J.H.; Kelly, J.K. The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput. Biol. 2011, 7, e1002255. [Google Scholar] [CrossRef]
- Lee, Y.K.; Kim, G.T.; Kim, I.J.; Park, J.; Kwak, S.S.; Choi, G.; Chung, W.I. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 2006, 133, 4305–4314. [Google Scholar] [CrossRef]
- Zou, C.; Wang, P.; Xu, Y. Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol. J. 2016, 14, 1941–1955. [Google Scholar] [CrossRef]
- Takagi, H.; Abe, A.; Yoshida, K.; Kosugi, S.; Natsume, S.; Mitsuoka, C.; Uemura, A.; Utsushi, H.; Tamiru, M.; Takuno, S.; et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013, 74, 174–183. [Google Scholar] [CrossRef] [PubMed]

| Clean Base/G | Q30/% | GC Content/% | Mapped Reads | Mapped Rate/% | Coverage/% | Average Depth | |
|---|---|---|---|---|---|---|---|
| bulk1 | 23.84 | 87.87 | 37 | 154,147,804 | 99.16 | 99.69 | 137 |
| bulk2 | 19.75 | 86.45 | 36 | 125,074,857 | 99.42 | 99.68 | 113 |
| prt6 | 24.41 | 87.23 | 36 | 151,180,273 | 99.75 | 99.68 | 152 |
| prt6−r | 30.71 | 92.46 | 41 | 124,972,640 | 63.44 | 99.69 | 126 |
| Pos | Ref | Alt | Annotation | Annotation | SNP Index bulk1 | SNP Index bulk2 | deltaSNP | Short_Description |
|---|---|---|---|---|---|---|---|---|
| 4,251,301 | G | A | nonsynonymous | AT5G13280 | 0.33 | 0.88 | −0.55 | Aspartate kinase 1 |
| 4,469,182 | G | A | Stopgain | AT5G13840 | 0.43 | 0.87 | −0.43 | FIZZY-related 3 |
| 4,922,519 | G | A | nonsynonymous | AT5G15160 | 0.36 | 0.93 | −0.57 | BANQUO 2 |
| 5,070,109 | G | A | Stopgain | AT5G15580 | 0.34 | 0.88 | −0.53 | Longifolia1 (LNG1) |
| 5,361,835 | G | A | nonsynonymous | AT5G16390 | 0.42 | 0.94 | −0.52 | Chloroplastic acetylcoenzyme A carboxylase 1 |
| 6,113,584 | G | A | nonsynonymous | AT5G18440 | 0.36 | 0.91 | −0.55 | NULL |
| 6,133,091 | G | A | nonsynonymous | AT5G18480 | 0.41 | 0.89 | −0.48 | Plant glycogenin-like starch initiation protein 6 |
| 6,362,987 | G | A | nonsynonymous | AT5G19040 | 0.28 | 0.97 | −0.69 | Isopentenyltransferase 5 |
| 6,467,895 | G | A | nonsynonymous | AT5G19230 | 0.45 | 0.97 | −0.52 | Glycoprotein membrane precursor |
| 6,854,241 | G | A | nonsynonymous | AT5G20300 | 0.41 | 0.92 | −0.51 | Avirulence induced gene (AIG1) family protein |
| 7,354,380 | G | A | nonsynonymous | AT5G22190 | 0.35 | 0.94 | −0.58 | NULL |
| 7,610,318 | A | T | nonsynonymous | AT5G22794 | 0.99 | 0.99 | 0.00 | NULL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Luo, Y.; Cao, Y.; Gong, Y.; Corbineau, F.; Xiang, Y. Mutation in the LONGIFOLIA1 Gene Resulted in Suppressed Insensitivity of Arabidopsis thaliana proteolysis6 Mutant to Ethylene During Seed Germination. Seeds 2025, 4, 48. https://doi.org/10.3390/seeds4040048
Wang X, Luo Y, Cao Y, Gong Y, Corbineau F, Xiang Y. Mutation in the LONGIFOLIA1 Gene Resulted in Suppressed Insensitivity of Arabidopsis thaliana proteolysis6 Mutant to Ethylene During Seed Germination. Seeds. 2025; 4(4):48. https://doi.org/10.3390/seeds4040048
Chicago/Turabian StyleWang, Xu, Ying Luo, Yuan Cao, Yujin Gong, Francoise Corbineau, and Yong Xiang. 2025. "Mutation in the LONGIFOLIA1 Gene Resulted in Suppressed Insensitivity of Arabidopsis thaliana proteolysis6 Mutant to Ethylene During Seed Germination" Seeds 4, no. 4: 48. https://doi.org/10.3390/seeds4040048
APA StyleWang, X., Luo, Y., Cao, Y., Gong, Y., Corbineau, F., & Xiang, Y. (2025). Mutation in the LONGIFOLIA1 Gene Resulted in Suppressed Insensitivity of Arabidopsis thaliana proteolysis6 Mutant to Ethylene During Seed Germination. Seeds, 4(4), 48. https://doi.org/10.3390/seeds4040048







