Abstract
Seed germination is the initial step in a plant’s life cycle; it is precisely regulated by many factors at the molecular and biological levels. Reversible protein phosphorylation, which is regulated by protein kinases and protein phosphatases, plays a key role in hormone signal transduction, energy metabolism, stress response, and plant growth and development, including seed germination. This review provides a comprehensive elucidation of the coordinated regulatory mechanisms mediated by kinases and phosphatases during seed germination, with particular emphasis on their dynamic interplay and reciprocal modulation within biological signaling networks. Through the systematic integration of current research findings, we mechanistically dissect the sophisticated phosphorylation–dephosphorylation circuitry that governs metabolic activation, hormonal signaling transduction, and cellular homeostasis in germinating seeds. Furthermore, we propose a novel conceptual framework that delineates the spatiotemporal cooperation between these opposing enzymatic activities in regulating dormancy release and developmental transitions. The current challenges in the field of seed germination research are critically examined, and potential future investigative trajectories are outlined, aiming to establish a robust theoretical framework for elucidating the molecular mechanisms underlying seed dormancy regulation, as well as translating these findings into innovative agricultural production practices.
1. The Biological Basis of Seed Germination
As the first step of a plant’s life cycle, seed germination is related to seedling morphology construction, their subsequent growth, and yield formation; it is the basis of agricultural production [1,2]. Seed germination is the most critical stage in a plant’s life cycle as it is the most vulnerable to environmental stresses and diseases [3]. Therefore, seed germination has important economic significance. Seed germination can be defined in both narrow and broad terms. In the narrow sense, it refers to the process whereby dry seeds with vitality break through the seed coat from the water absorption stage to the radicle emergence stage. In the broad sense, it refers to the process whereby dry seeds start from imbibition and eventually develop into complete seedlings with roots, stems, and leaves [4,5]. Seed germination is a complicated process. In this process, imbibed mature seeds must quickly change from the mature stage to the germination stage. Moreover, it is necessary to start multiple metabolic pathways, such as the degradation of stored substances, the synthesis and modification of proteins, and the synthesis and metabolism of hormones (Figure 1), in order to provide the necessary conditions for seed germination [3].
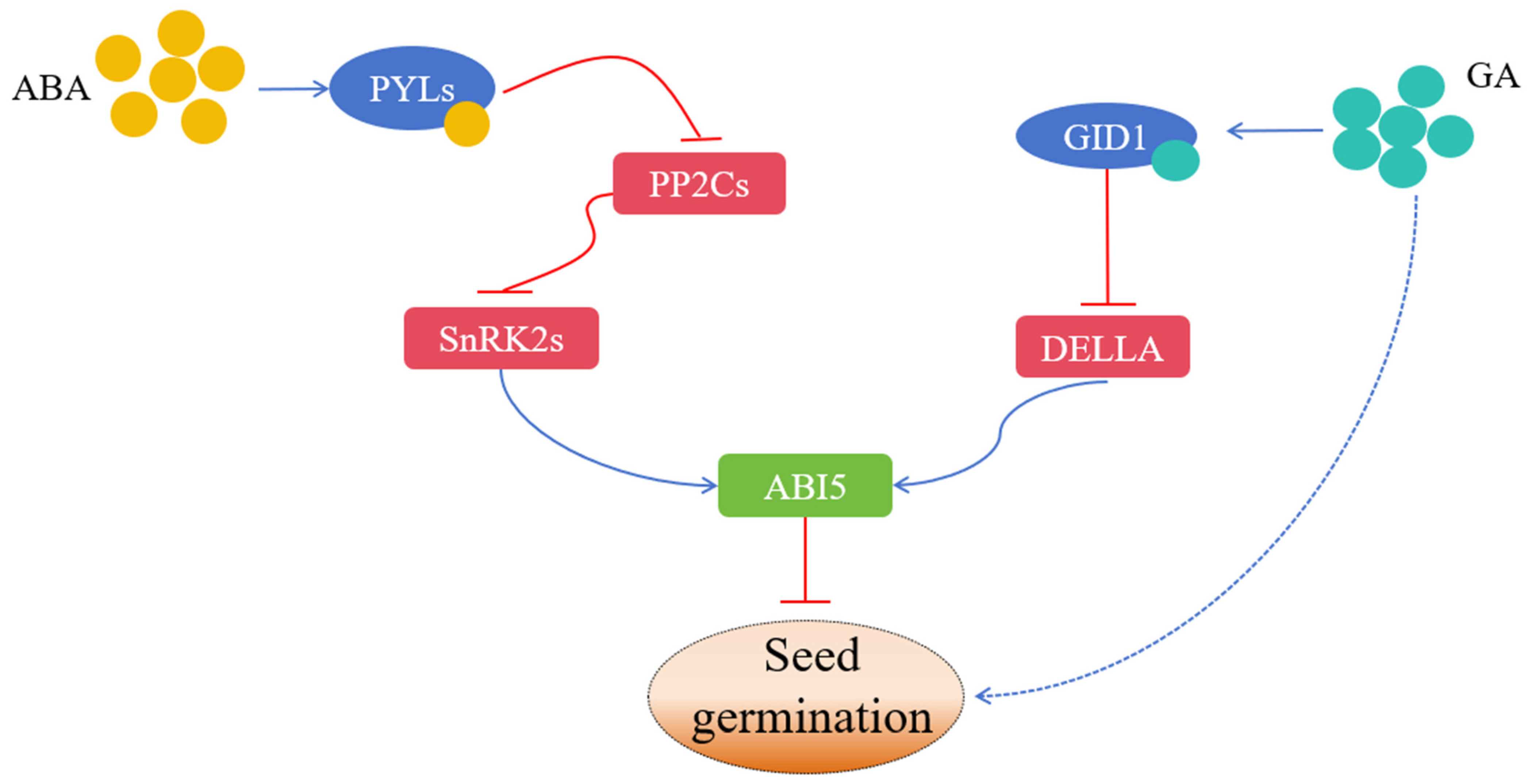
Figure 1.
The hormone regulatory network of seed germination. The blue and red lines indicate positive and negative effects, respectively.
- (1)
- Rapid water absorption (stage I) [6]: At this stage, the dry seeds quickly absorb water to soften or break the seed coat. The increase in air permeability and the relaxation of the cell membrane create conditions for the metabolism of stored substances and the recovery of key enzyme activities. However, the organic matter inside the seeds did not begin to change. In the process of seed maturation, dehydration will lead to the membrane system, protein, and DNA being damaged; this damage will be aggravated by the rapid absorption of water after imbibition [7]. The disturbance of the membrane structure will lead to the leakage of solutes and metabolites, accelerating water absorption [6], which promotes seed germination by reducing the concentration of inhibitors [8]. Soon after imbibition, oxygen will be absorbed and carbon dioxide will be released [4,7]. Mitochondria and respiration-related enzymes in cells provide ATP for cells through redox reactions. Glycolysis can also produce a small amount of ATP through substrate-level phosphorylation in the cytoplasm, providing a substrate for mitochondrial respiration. The pentose phosphate pathway mainly operates in the cytoplasm and its main function is to produce NADPH and ribose-5-phosphate, which provides a reducing power and a precursor for biosynthesis. When the mitochondrial activity is restricted, the glycolytic pathway will play a major role in providing ATP. When mitochondria are active, the pentose phosphate pathway plays a major role [9]. This shows that energy generation plays an important role in seed germination.
- (2)
- Stagnation of water absorption (stage II): In the second stage, the water content of the seeds is stable; the protoplasm state becomes a sol state, which readily reacts; and the seed coat gradually begins to break. With the repair of the mitochondrial structure and the synthesis of protein after a large number of new mRNA transcription and translation processes, various enzyme activities, respiration, and other metabolic activities are gradually enhanced [10].
- (3)
- Hypocotyl water absorption (stage III): in the third stage, the seeds absorb a lot of water. Breathing decomposes macromolecular substances such as the starch, protein, and fat that are stored in seeds, which provides energy for the growth of young embryos and provides a material basis for the formation of new cells. With the growth of young embryos and the gradual weakening of the endosperm, the radicle finally protrudes [11]. The emergence of the hypocotyl and the elongation of the radicle mark the seed germination entering stage III [7,12].
When the seeds begin to absorb water, they will be accompanied by the generation of energy and the repair of DNA and protein. During seed development, various nutrients, including lipids, protein, and starch, accumulated. After imbibition, the cell structure and enzyme activity gradually recovered to stages I and II. Once enough water is absorbed, different kinds of metabolism will be activated. Protein omics analysis showed that a large number of enzymes that are needed for major metabolism accumulated in mature dry seeds, which remained stable or increased during seed germination [13,14,15,16].
2. The Regulatory Role of Kinases in Seed Germination
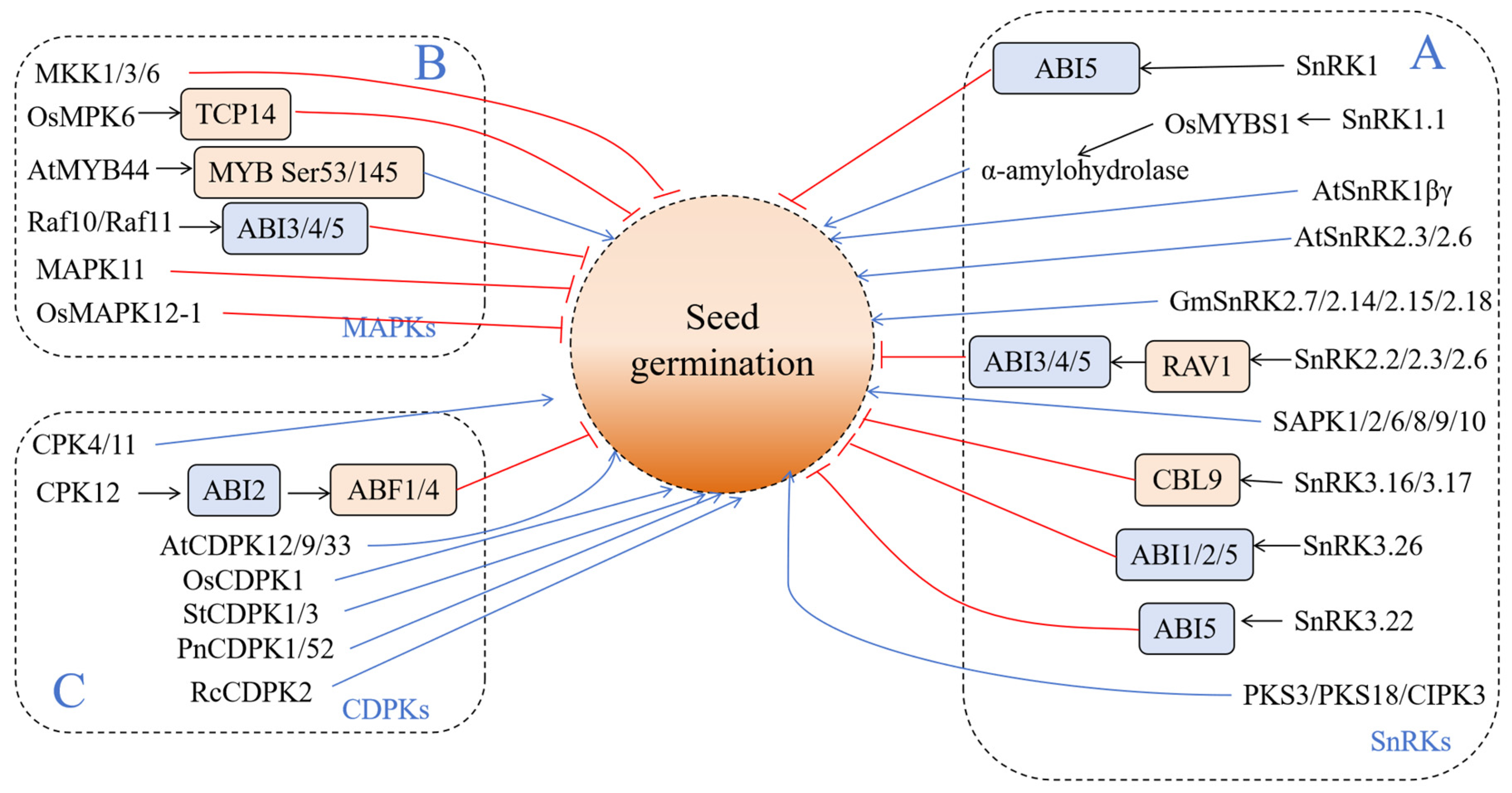
In the process of seed dormancy and germination regulation, protein kinases (PKs) can play a direct role, or they can indirectly regulate biological processes such as seed dormancy, germination, and stress response by affecting hormone signal transduction such as ABA/GA. At present, there are three types of kinases that have been studied in relation to seed dormancy and germination—sucrose non-fermentation 1-related protein kinases (SnRKs), mitogen-activated protein kinases (MAPKs), and calcium-dependent protein kinases (CDPKs) (Figure 2).
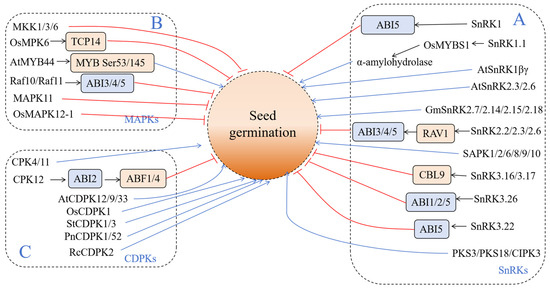
Figure 2.
Kinases involved in seed germination. (A) Sucrose non-fermentation 1-related protein kinases (SnRKs); (B) mitogen-activated protein kinases (MAPKs); (C) calcium-dependent protein kinases (CDPKs). The blue and red lines indicate positive and negative effects, respectively.
2.1. Sucrose Non-Fermentation 1-Related Protein Kinases (SnRKs)
Sucrose non-Fermentation 1-related protein kinases (SnRKs) are a type of widely prevalent Ser/Thr protein kinase in plants; they participate in various stress signal pathways and respond to complex environmental changes. The SnRK protein family in plants is divided into three subfamilies—SnRK1, SnRK2, and SnRK3 (Figure 2).
In plants, the SnRK1 protein kinase complex exists in the form of a heterotrimer, which includes three subunits—α, β, and λ [17]. The SnRK1 protein kinase is one of the hubs of life activities in plants and participates in various activities such as metabolism, development, and stress response [18]. First of all, SnRK1 is a receptor of sugar metabolism, which regulates various life processes such as primary metabolism and secondary metabolism in plants [19]. In Arabidopsis thaliana, SnRK1 is reported to be involved in responding to sugar metabolism and ABA signals, thus regulating plant growth and stress response [20,21,22,23]. Radchuk et al. [24] found that SnRK1 regulates the growth and development of Pisum sativum L seeds by coordinating sugar metabolism with plant hormone signals such as auxin, cytokinin, and abscisic acid. Lu et al. [25] found that SnRK1A (SnRK1.1) plays a key role in regulating the pathway of rice seed germination and seedling growth through sugar signals. It can also affect the expression of the downstream α-amylohydrolase gene by phosphorylating transcription factor OSMYBS1. Hu et al. [26] found that the SnRK1 kinase promoted rice seed germination by inhibiting ABA and activating the ethylene signaling pathway. Song et al. [27] also found that in tomato seeds, tomato SnRK1 downregulated the transcription expression of ABI5 by combining with the positive regulator of the ABA signal, thus affecting ABA signal transduction and seed germination regulation. AtSnRK1βγ promotes the interaction between pollen and stigma by mediating the biosynthesis of mitochondria and peroxidase in pollen, thus participating in the regulation of physiological processes such as pollination and seed germination [28]. It was found that rice seeds with an added AMPK activator 991 can specifically activate SnRK1; a low concentration of this can promote seed germination, while a high concentration can inhibit seed germination [29]. AMPK activator A-769662 regulates the activity of SnRK1 by upregulating amylase gene expression, downregulating the ABA-mediated signaling pathway, and stimulating ethylene synthesis, thus promoting seed germination [26]. Compared with the wild type (WT), the seed germination rate of Arabidopsis thaliana overexpressing Phyllostachys edulis PeSnRK1α was significantly improved under NaCl treatment [30].
SnRK2 is a unique subfamily of plants [30] that participates in ABA-dependent and ABA-independent abiotic stresses, regulating the growth and development processes of plants, such as osmotic stress, stomatal regulation, seed germination, and seedling growth [31,32,33]. SnRK2D/SnRK2.2, SnRK2I/SnRK2.3, and SnRK2E/SnRK2.6/OST1 are involved in the ABA signaling pathway, which is necessary for seed development, dormancy, and the germination of Arabidopsis thaliana [34,35]. TaABF, which is the phosphorylated substrate of wheat PKABA1, regulates the ripening and dormancy of wheat grains [36]. In Arabidopsis thaliana, it was found that SnRK2.2 and SnRK2.3 mutant plants were insensitive to ABA during seed germination, which indicated that these two protein kinases positively regulated ABA signal transduction during seed germination [37]. Similarly, the overexpression of ALSnRK2.6 can increase the oil content in plant seeds, shorten the flowering period, and increase the seed yield due to the increase in the seed number [38]. There is a negative regulatory factor—BTB—in Arabidopsis thaliana that responds to ABA signal intensity, which reduces the activity of AtSnRK2.3 and regulates ABA sensitivity during seed germination. BTB-A2.1, BTB-A2.2, and BTB-A2.3 antagonize SnRK2.3 and mediate the ABA response during seed germination [39]. Both abscisic acid (ABA) and jasmonic acid (JA) inhibit seed germination. The soybean SnRK2 gene (GmSnRK2s) was comprehensively identified and characterized. It was found that GmSnRK2.2 and GmSnRK2.16 were preferentially expressed in meristem and participated in the regulation of stem morphology [40]. GmSnRK2.7, GmSnRK2.14, GmSnRK2.15, and GmSnRK2.18 showed specific expression patterns during seed development [41]. Feng et al. [42] found that SnRK2 protein kinases (SnRK2.2, SnRK2.3, and SnRK2.6) can phosphorylate the RAV1 transcription factor. At the same time, RAV1 directly negatively regulates the expression of ABI3, ABI4, and ABI5, thus participating in the regulation of seed germination and seedling growth. The ABA-activated SnRK2D/SnRK2.2, SnRK2E/SnRK2.6/OST1, and SnRK2I/SnRK2.3 with functional redundancy in Arabidopsis thaliana were identified as key regulatory factors for seed development and germination [43]. Previous studies showed that the overexpression of TaSnRK2.4 led to the delay of Arabidopsis germination, and a similar phenotype was also observed when TaSnRK2.10 was overexpressed in rice [44]. AtSnRK2.6 has been proven to be related to the ABA signal, as well as salt, dehydration, and osmotic stress. It is also involved in regulating sucrose metabolism, carbohydrate metabolism, controlled seed development, seedling growth, and stomatal closure [45,46,47,48]. In rice, 10 members of the SnRK2 family were identified as osmotic stress-/ABA-activated protein kinases (SAPK1-10), which participated in many growth reactions [49]. For example, SAPK6 shows ABA insensitivity during tobacco seed germination and root elongation [50], while SAPK2 is resistant to ABA in rice [51]. SAPK1 and SAPK2 participate in seed germination and seedling growth under NaCl [52]. However, in the presence of ABA, SAPK10 stimulates the biosynthesis of JA and inhibits seed germination [41]. In addition, the SAPK10-bZIP72–AOC pathway inhibited rice seed germination [40], while the overexpression of SAPK8, SAPK9, and SAPK10 inhibited the germination of rice seeds [33].
SnRK3 is a plant-specific subfamily [31] that is involved in regulating biotic and abiotic stress responses and the Ca2+ signaling pathway [53]. SnRK3.16 and SnRK3.17 combine with CBL9 to participate in the ABA-independent stress response [54], playing a negative regulatory role for ABA during plant seed germination [55]. SnRK3.26 interacts with ABA signal regulators ABI1, ABI2, and ABI5, positively regulating plant seed germination [56]. The interaction between SnRK3.22 and ABI5 is involved in the ABA response, regulating seed germination, root elongation, and other growth and development processes [57]. In Arabidopsis thaliana, some other SnRK3 protein kinases, such as PKS3, PKS18, and CIPK3, play a role in ABA induction during seedling growth, stomatal closure, and seed germination [58,59,60]. The insertion mutant of oscipk31 is highly sensitive to ABA and stress (e.g., salt, mannitol, and glucose). When it is exposed to these abiotic stresses during the seed germination and seedling stage, the expression level of several stress response genes changes [61]. By analyzing the sensitivity of atcipk23 seeds to ABA, it was found that AtCIPK23 played a role in seed dormancy and the germination of Arabidopsis thaliana [62]. A phenotypic analysis of AtCIPK23 also showed that the deletion of atcipk23 did not significantly affect the hypocotyl elongation and seed germination of Arabidopsis thaliana [63,64]. It is found that NtCIPK23 may be used as an activator to promote nutrient transformation, chloroplast development, or photosynthesis establishment, thus actively promoting seed germination, cotyledon extension, and greening [65].
2.2. Mitogen-Activated Protein Kinase (MAPK)
As a specific serine/threonine protein kinase, MAPK is one of the largest transferases in eukaryotes [66]. The MAPK cascade consists of three kinds of enzymes, namely, MAPK, mitogen-activated protein kinases kinases (MAPKKs), and mitogen-activated protein kinases kinases kinases (MAPKKKs), and the ABA signal transduction pathway is also related to this cascade reaction [67]. The diversity of MAPK substrates and their different spatiotemporal expression patterns allow MAPK cascade modules to extensively participate in many processes of regulating plant growth and development [68,69,70]. A total of 20 MAPKs were found in Arabidopsis thaliana, 15 were found in rice, and 19 were found in maize [71,72,73]. In addition, MAPK has also been identified in banana, apple, tomato, and cucumber [74,75,76,77]. The MAPK cascade reaction not only regulates the development of stomata and flowers in plants but also regulates biotic and abiotic stress reactions [78]. It plays an important role in plant immunity [79,80], various biotic and abiotic stresses [81,82,83], and hormone cell division signal responses [84,85], as well as cell differentiation, growth, and development [86,87].
AtMKK1 and AtMPK6 are key molecules involved in ABA and the sugar regulation of seed germination in Arabidopsis thaliana [88]. MKK1 and MPK6 are downstream regulators of glucose signals during seed germination, and glucose promotes ABA synthesis through MKK1 and MPK6, thus inhibiting seed germination [88]. Similarly, rice OsMPK6 can maintain dormancy and inhibit seed germination by enhancing ABA synthesis and signal intensity [89,90]. MPK8 phosphorylates TCP14 by interacting with the transcription factor TCP14, which reacts with gibberellic acid (GA), enhancing its transcription activity and regulating the transition from dormancy to seed germination [90,91]. MKK3 is located in the MAPKK pathway and plays an important role in controlling the dormancy of grain seeds [92]. Xing et al. [88] showed that glucose may inhibit seed germination by regulating the increase in ABA content; AtMKK1 and AtMPK6 in MAPK in Arabidopsis thaliana are both key signals involved in ABA and sugar-regulating seed germination. AtMYB44 can also regulate the seed germination of Arabidopsis thaliana due to the action of the MPK3 and MPK6 kinases on the Ser53 and Ser145 of MYB44, respectively [93]. MKK3 is located in the MAPKK pathway and plays an important role in controlling the dormancy of grain seeds [91]. Wheat TaMKK3-A is located on chromosome 4A, and it is a candidate gene for seed dormancy site Phs1 [94]. The expression level of the wheat TaMKK3-A gene in MEL29 seeds was higher than that in MEL31, and the higher expression of TaMKK3-A promoted dormancy release [95]. However, Hordeum vulgare has two main quantitative trait loci of seed dormancy—SD1 and SD2 [96]—in which the Qsd2-AK locus determines the difference in seed dormancy among different varieties. Interestingly, MKK3 can interact with Qsd2-AK, thus regulating seed dormancy. In addition, N260 is critical for the activity of MKK3 kinase, and the substitution of N260T in this allele will reduce the activity of MKK3 kinase, leading to deepening dormancy, thus delaying seed germination [91]. In addition, both the Raf10 and Raf11 kinases also regulate seed dormancy in the MAPKKK pathway [97]. Compared with the WT, seeds of raf10 and raf11 mutants exhibit reduced dormancy and decreased sensitivity to ABA, while their overexpression leads to delayed seed germination and enhanced ABA sensitivity. Further study found that the expressions of positive regulatory genes of ABA signals ABI3 and ABI5 were upregulated in the seeds overexpressed with Raf10 and Raf11 [98]. Moreover, Raf10 and Raf11 can undergo autophosphorylation, and their kinase activities are inhibited by the MAPKKK inhibitor BAY 43-9006 [97], thus affecting their regulation of seed dormancy. It was found that protein kinase MAPK11 in tomato positively regulated ABA biosynthesis, thus negatively regulating seed germination [27]. Xiao et al. [99] found that the overexpression of OsMAPK12-1 can inhibit seed germination and seedling growth, indicating that OsMAPK12-1 negatively regulates rice growth and development. Lu et al. [100] found that the stimulation of exogenous ABA can activate the activity of AtMAPK3 in Arabidopsis thaliana, and AtMAPK3 may phosphorylate and activate ABI5 to inhibit seed germination and seedling growth. Although abscisic acid (ABA) typically triggers catalase (CAT) gene expression, the Arabidopsis Atmkk1 mutant exhibits impaired ABA-induced transcriptional activation of both the CAT1 and CAT3 isoforms. This molecular deficiency consequently attenuates ABA responsiveness during critical physiological processes, including seed germination and stomatal regulation, in the mutant plants [101]. Protein interaction between MKK1 and MPK6 in Arabidopsis thaliana regulates seed germination and plant resistance by participating in ABA-induced H2O2 accumulation [102]. Previous studies have reported that the germination process of barley seeds is related to the decrease in MAPKl kinase activity [103]. MPK6 can be activated by ABA, but this activation was suppressed in the mkk1 mutant. The results showed that the cascade pathway of MKK1-MPK6 participated in the accumulation of hydrogen peroxide induced by ABA, thus regulating seed germination and plant stress resistance [23,104].
2.3. Calcium-Dependent Protein Kinases (CDPKs)
Calcium ions (Ca2+) are essential signaling components in plant cells, particularly for coordinating stress adaptation mechanisms during exposure to adverse environmental conditions. When plants are subjected to external stress, the concentration of free Ca2+ in the cytoplasm will temporarily increase, and Ca2+ will combine with calcium-binding protein to decode Ca2+ signals and cause changes in the biochemical and physiological processes of cells to cope with various adversity stresses [105,106]. Transient fluctuations in cytosolic Ca2+ levels initiate calcium-mediated signaling events that are subsequently sensed and transduced by specialized calcium-binding proteins. This molecular decoding mechanism activates downstream signaling pathways, regulating critical cellular processes including enzymatic phosphorylation cascades and the transcriptional regulation of gene expression [107,108]. In plants, calcium-binding proteins can be divided into five categories—calcium-dependent protein kinases (CDPKs) and related protein kinases (CRKs), calmodulin proteins (CaMs), calmodulin-like proteins (CMLs), calcineurin B-like protein (CBLs), and CBL interacting protein kinase (CIPK) [109,110].
Calmodulin CDPKs contain four conserved domains—the N-terminal variable domain, the kinase catalytic domain, the self-inhibitory linkage domain, and the C-terminal regulatory domain [111]—and contains the sensing activity of the EF-hand calcium binding motif and the response activity of the protein kinase domain [112]. In total, 34 CDPKs were identified in the Arabidopsis genome [113], 31 CDPKs in rice [114], 26 CDPKs in wheat [115], 29 CDPKs in foxtail millet, and 35 CDPKs in maize [116,117]. CDPKs can directly transduce Ca2+ signals into phosphorylation cascades, which endows CDPKs with the dual function of being a Ca2+ receptor and responder. Arabidopsis CDPKs participate in salt stress responses by regulating ABA-responsive element binding factors (ABFs), transcription factor-mediated ABA response gene expression, ABA-mediated anion channels (SLAC1 and SLAH3), and stomatal closure [118,119]. CDPKs are considered to be related to ABA signals, which are involved in regulating seed germination and plant development (Figure 2) [120]. ABA stimulated two homologous kinases, CDPK4 and CDPK11, in Arabidopsis thaliana, and the loss-of-function mutations of CPK4 and CPK11 resulted in pleiotropic ABA-insensitive phenotypes in seed germination. These two CDPKs were identified as important regulatory factors in the ABA signaling pathway, which provided clear genetic evidence for CDPK/Ca2þ to participate in ABA signal transduction in seed germination, seedling growth, stomatal movement, and plant response to salt stress [121]. CPK12 is a negative regulator of ABA signals during seed germination and post-germination growth [122]. The CDPK gene can also regulate the process of seed germination. During the early development of castor seeds (Ricinus communis L.), the expression of RcCDPK2 first increased and then decreased [123]. AtCDPK12 negatively regulates ABA signals during seed germination, and the interaction between AtCDPK9 and AtCDPK33 negatively regulates the response to ABA and drought stress [124,125,126,127]. AtCPK4, AtCPK11, and AtCPK12 are the regulatory factors during the seed germination of Arabidopsis thaliana [128,129]. Compared with the WT, CPK12-RNAi seeds are sensitive to ABA during germination. CPK12 phosphorylates ABI2 by interacting with ABI2, a negative regulatory protein of the ABA signaling pathway, and phosphorylates ABA-responsive transcription factors ABF1 and ABF4, downregulating their expression [129], thus positively regulating seed germination. In the study of rice, it was found that OsCDPK1 was involved in regulating the development of seeds, including starch content and seed size [130]. It was found that CDPK participated in the process of embryo development, seed formation, and the germination of sandalwood, showing obvious spatiotemporal accumulation and activity in the process of regulation [131]. The overexpression of grape CDPK1 in Arabidopsis thaliana promoted seed germination and growth [132]. PnCDPK1 mainly accumulates in petals and sepals, and it participates in the seed germination and morphogenesis of morning glory [133,134]. It was found that rice CDPK (OsSPK) was specifically expressed in the endosperm of immature rice seeds, which regulated the biosynthesis of storage substances in rice seeds. After silencing the gene, it was found that the content of storage substances such as starch in rice seeds would decrease. The seeds in the silenced rice plants with OsSPK became very soft, and further analysis found that the silenced rice plants could not use sucrose, suggesting that SPK was involved in the anabolic pathway of sucrose and the synthetic pathway of seed storage [135]. CDPK was found to be involved in tuber development, and the expression patterns of the StCDPK1 and StCDPK3 genes in potato were different in different stages of tuber development. StCDPK1 was only expressed at the top of the tuber, while the StCDPK3 gene was only expressed when the tuber was elongated [136]. Recently, it has been shown that the overexpression of maize ZmCPK4 in transgenic Arabidopsis thaliana enhances ABA sensitivity in seed germination [137]. In previous studies, it has been shown that PnCDPK1 is the main isoform of CDPK in morning glory seedlings, and it exists in proliferation and growth tissues. The transcription level of PnCDPK1 is very high in dry seeds and gradually decreases during seed germination and seedling growth, being negatively regulated by light [133,138]. However, different PnCDPK kinases (PnCDPK52) were active during the germination of morning glory seeds [139].
3. The Dynamic Regulation of Phosphatase on Seed Germination
Reversible protein phosphorylation mediated by protein kinase and protein phosphatase is a very old regulatory mechanism that was formed in the process of evolution; it has become the main way to regulate protein function and most biological processes, playing a key role in the overall life cycle [140,141,142]. Studies have found that phosphorylated proteins of eukaryotes are abundant in mitochondria, chloroplasts, nuclei, the cytoplasm, and the extracellular matrix [143,144,145].
3.1. PP2C
PP2C is a monomer protein phosphatase, which has no similarity with other protein phosphatases in the same family. The C-terminal catalytic domain is highly conserved, and it is an amino acid homologous region of PP2C. However, there is an extension area at the N-end, which is not conservative. It is predicted that this region may be the key region for regulating PP2C activity [146,147,148]. The PP2C family is widely distributed in various plants. According to whole-genome sequencing, about 10 PP2C genes were identified in green algae, and nearly 50 were identified in mosses and lycopodes [149]. There are more PP2C genes in higher plants; for example, there are 80 genes encoding PP2C in Arabidopsis thaliana, 90 in rice [150], 92 in tomato [151], 97 in maize [152], and 131 in rape [153]. Among the subfamilies of PP2C, subfamily A is the most widely studied. In Arabidopsis thaliana, ABI1 (ABA sensitive 1) and ABI2 were first found to play a negative regulatory role in ABA-related signal pathways, participating in the regulation of stomatal closure, seed germination, and plant growth [154,155,156]. Four members of family B of PP2C were identified in Arabidopsis thaliana. Among them, AP2C3 (Arabidopsis thaliana protein phosphatase 2C) and MAPKs are co-located in the nucleus and interact with MPK3, MPK4, and MPK6. Other closely related protein phosphatases, AP2C2 and AP2C4, are also MAPK phosphatases acting on MPK6, thus helping to maintain stomatal cells and regulate seed germination [157,158]. PP2C5, a member of the B subfamily in Arabidopsis thaliana, can inhibit the activities of MPK3 and MPK6, which can be used as MAPK phosphatases to positively regulate seed germination, stomatal closure, and ABA-induced gene expression [159].
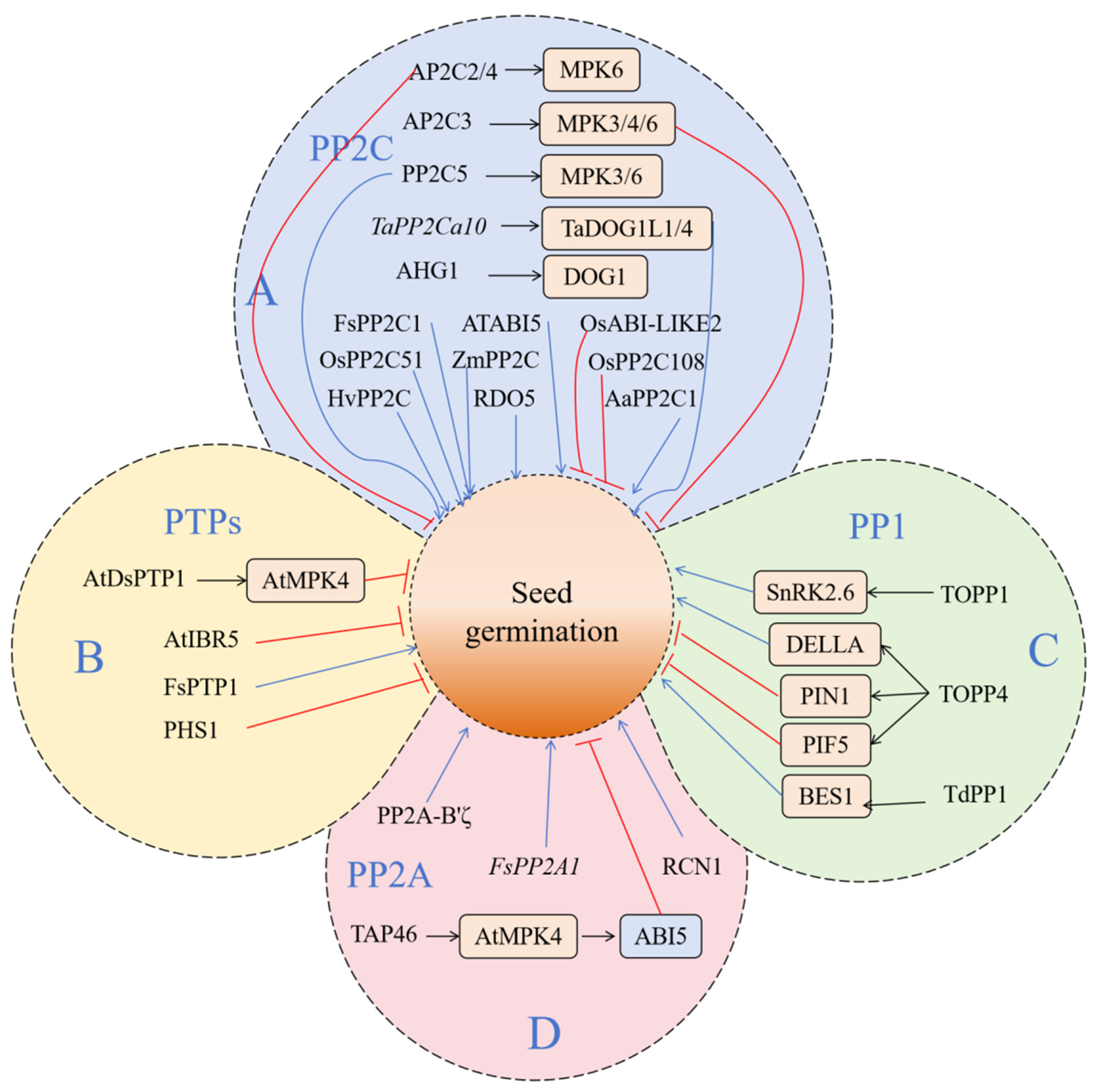
PP2C regulates the dephosphorylation of substrates and plays an important role in the ABA response to abiotic stress. It is widely involved in many physiological and biochemical processes, such as plant seed germination and leaf senescence, the stomatal opening and closing of guard cells, mechanical trauma response, and so on (Figure 3) [160]. The PP2C protein, which affects seed germination and dormancy, is mainly regulated by the ABA signaling pathway. A large number of PP2C proteins participate in seed germination as negative regulators of the ABA signaling pathway, including ABI1, ABI2, HAB1, HAB2, and HONSU [161,162]. AHG1 in Arabidopsis thaliana is a PP2C phosphatase. As a negative regulator in the ABA pathway, AHG1 can interact with the gene DOG1, encoding dormancy specificity and playing an important role in seed germination [163,164,165]. The Arabidopsis thaliana gene ATABI5, which belongs to the PP2C gene family, has also been proven to play a key role in the regulation of plant seed germination and early seedling growth [166]. In wheat, TaPP2Ca10, a member of subfamily A, can interact with TaDOG1L1 and TaDOG1L4. The heterologous expression of TaPP2Ca10 in Arabidopsis thaliana can reduce the sensitivity of seed germination to ABA and promote seed germination [167]. Lorenzo et al. [168] isolated a PP2C gene from beech, which is highly homologous to ABI1 and ABI2; it was named FsPP2C1. After ABA treatment, the expression of the FsPP2C gene in beech seeds increased significantly, and the seed germination rate was positively correlated with the expression of the FsPP2C1 gene. After overexpressing FsPP2C1 in Arabidopsis thaliana, it was found that its seeds were not sensitive to ABA. The dormancy degree of seeds decreased, and the germination of seeds was not inhibited by ABA. The tolerance of seeds to salt stress and osmotic pressure was also significantly improved in a certain period of time. The results showed that FsPP2C1 played a negative regulatory role in the ABA signal transduction pathway and in seed dormancy and germination. Recently, the analysis of the knock-out lines and overexpression lines of the OsPP2C51 gene showed that OsPP2C51 negatively regulated the ABA response and positively regulated rice seed germination [169]. Studies have shown that the overexpression of OsPP2C51 in sorghum leads to an increase in α-amylase expression and enzyme activity, which accelerates the catabolism of GA. The transcriptional inhibition of OsPP2C51 on OsbZIP109 (the homologous gene of rice ABI5) increases the level of bioactive GA, leading to a loss of dormancy and the enhanced germination of seeds [170]. The expression of HvPP2C in barley aleurone is regulated by GA and ABA. It shows the importance of PP2C in regulating the crosstalk between ABA and GA, thus regulating the balance between seed germination and dormancy [171]. Xiang et al. [172] found that RDO5 (Reduced Dormancy5), a member of the PP2C family, is a specific gene for seed dormancy that can regulate seed dormancy without affecting other plant characters. Arabidopsis thaliana PP2CA negatively regulates the ABA pathway, and the mutant is sensitive to exogenous ABA in rooting and seed germination, strongly regulating seed germination [173]. In Artemisia annua, AaPP2C1 can regulate secondary metabolites. In transgenic Arabidopsis, the rooting and seed germination of expression lines are insensitive to ABA, which negatively regulates ABA signals [174]. The maize ZmPP2C and ZmPP2C-A10 genes were expressed in Arabidopsis thaliana, which negatively regulated ABA signals. Under exogenous ABA treatment, the overexpressed lines were insensitive to rooting and seed germination, showing sensitivity to salt and drought [175,176]. Née et al. [177] found that a key PP2C protein phosphatase, AHG1 (ABA hypersensitive germination 1), which is involved in the ABA regulation of seed germination, can interact with DOG1, thus affecting the PP2C activity of AHG1 and regulating seed dormancy. AHG1 and AHG3 in Arabidopsis thaliana class A PP2C play an important role in seed germination. Both the ahg1 and ahg3 mutants show that the dormancy period of seeds is prolonged and the germination rate is slowed down during the seed germination stage [173,178]. OsABI-LIKE2 and OsPP2C108, members of rice class A PP2C, can also regulate ABA-mediated seed germination [179,180].
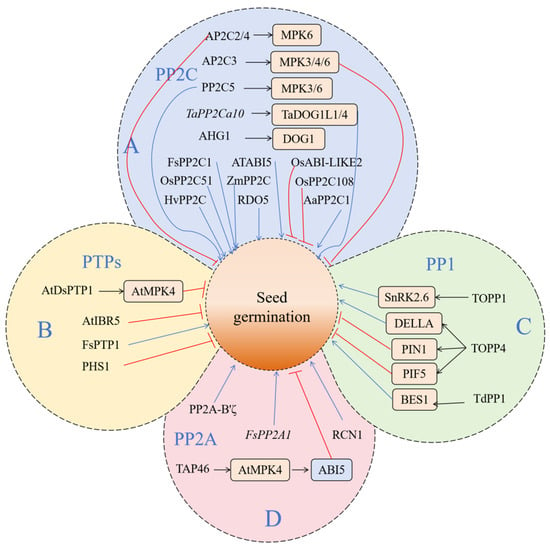
Figure 3.
The schematic diagram of key phosphatase signal pathways regulating seed germination. This figure shows how specific phosphatases can control the transformation process of seeds from dormancy to germination by regulating the activities of downstream kinases and transcription factors. (A) Protein phosphatase 2C (PP2C); (B) protein tyrosine phosphatase (PTP); (C) protein phosphatase 1 (PP1); (D) protein phosphatase 2A (PP2A). The blue and red lines indicate positive and negative effects, respectively.
3.2. PTP
PTPs are a family of phosphatases with diverse structures that contain highly conserved catalytic domains. In plants, the main target protein of PTPs is MAPKs. PTPs can be divided into two categories—tyrosine-specific PTPases (TsPTPs) and dual-specificity PTPases (DsPTPs) [181,182,183,184,185]. Current research shows that PTPs are involved in stomatal activity, growth and development, and the adversity stress of plants (Figure 3) [186]. As a negative regulatory factor, AtDsPTP1 is involved in the process of seed germination under osmotic stress. Its protein can promote the dephosphorylation of the phosphorylated substrate protein, which leads to the inactivation of AtMPK4 [187,188]. ABA participates in many physiological pathways of plants, such as seed dormancy and germination, fruit ripening, and adversity stress. AtDsPTP1 can regulate the accumulation of ABA and the expression of ABI family genes in the ABA signaling pathway. Plants mutated with this gene reported a reduced sensitivity to exogenous ABA [188]. AtIBR5 is a member of the double tyrosine protein phosphatase family found in Arabidopsis thaliana, which is involved in the regulation of ABA and auxin. AtIBR5 can positively regulate the expression levels of auxin and ABA [189,190]. In the presence of PAO, Arabidopsis seeds are hypersensitive to ABA-induced post-germination stagnation, which indicates that PTP is a negative regulator of ABA signals [191]. PTP can be reversibly inactivated under oxidative conditions and can participate in regulating the reaction of cells to redox changes [192]. Quettier et al. [193] used ABA to treat T-DNA inserted into the mutant seeds of the PTP gene in Arabidopsis thaliana; it was found that the seed germination rate of mutant PHS1-3 of the phs1 gene decreased greatly. In this mutant, the expression level of ABA-related genes is different from that of wild-type plants. After the ABA treatment of mutant and wild-type plants, the expression of PHS1 also increased significantly. This evidence shows that PTP family members are also involved in the regulation of the ABA pathway. In addition, it was found that the overexpression of FsPTP1 was carried out in Arabidopsis thaliana ecotype Cvi (Cape Verde Islands; seeds obtained from Nottingham Arabidopsis Stock Centre, UK), which is considered to be the best model for studying seed dormancy. This fact strengthens the important role of FsPTP1 as a negative regulator of ABA signal transduction and seed dormancy. The most specific tyrosine phosphatase involved in the ABA signal transduction pathway and the promotion of seed dormancy to germination was reported [194].
3.3. Potential Role of Other Phosphatase Families
In addition to PP2C and PTPs, protein phosphatase 1 (PP1) and protein phosphatase 2A (PP2A) may also participate in the regulation of seed germination. PP1 and PP2A constitute 90% of serine/threonine phosphatase [195]. PP1 is a member of the highly conserved serine/threonine protein phosphatase (PPP) family in eukaryotes, which exists widely in eukaryotes and catalyzes the dephosphorylation of many proteins. In animals and yeasts, it has been found that PP1 is involved in many important physiological processes, including cell division, carcinogenesis, immune response, and metabolism [195,196,197,198].
In plants, the PP1 family can form a huge complex with many important bioactive proteins to regulate the activities of cells (Figure 3) [199]. For example, TOPP1 regulates the auxin signaling pathway by combining with AUX/IAA, an important transcription factor in the auxin signaling pathway [199]. TOPP1 negatively regulates the ABA signaling pathway by inhibiting SnRK2.6 [200], indicating that the PP1 family plays a very important role in plant resistance to abiotic stresses such as salinity and drought. In Arabidopsis thaliana, the PP1 family protein TOPP4 can directly bind to the DELLA protein, a negative regulatory factor in the GA signal transduction pathway. Dephosphorylation can accelerate its degradation and promote GA signal transduction and plant growth and development [201]. Through the analysis of the abnormal morphology of epidermal cells of TOPP4-1, it was found that TOPP4 can dephosphorize PIN1. Adjusting the polarity orientation of PIN1 affects the polarity distribution of auxin and the polarity establishment of epidermal cells [202]. TOPP4-1 seedlings also showed an abnormal response to red light, including hypocotyl shortening and cotyledon angle increasing. Further study found that TOPP4 could interact with PIF5, which degraded PIF5 by dephosphorizing PIF5, and it affected the photomorphogenesis of plants [203]. In wheat, the expression of TdPP1 was significantly upregulated after salt treatment, suggesting that PP1 may be involved in the regulation of salt stress in wheat [204]. TdPP1 can also enhance the BR signal path by activating BES1 [205]. Applying the PP1 inhibitor Microcystin-LR to Vicia faba, it was found that TOPP may be involved in the regulation of ROS outbreak and mitosis [206,207].
PP2A is the main serine/threonine protein phosphatase in eukaryotic cells [208]. PP2A is highly conserved and widely expressed in the evolution process, mostly existing in the form of a trimer [209]. The heterotrimer of PP2A is composed of a scaffold subunit (A subunit), a regulatory subunit (B subunit), and a catalytic subunit (C subunit) [210]. In Arabidopsis thaliana, the PP2A family has 25 members, including 3 A subunits, 17 B subunits, and 5 C subunits [211]. PP2A not only regulates the development of plant cells but also plays an important role in a plant’s response to abiotic stress [212]. PP2A is also involved in the regulation of auxin transport in Arabidopsis thaliana [213] and rice seed germination (Figure 3) [214]. It was found that the B subunit of PP2A (regulatory subunit) PP2A-B’ζ positively regulated the process of seed germination and plant development. Its mutant pp2a-b’ζ was sensitive to ethylene. Further study found that PP2A-B’ζ negatively regulated the ethylene signaling pathway by interacting with CTR1 [215]. It was found that FsPP2A1 was a seed-specific expression gene. Gibberellin treatment can significantly increase the expression of FsPP2A1 and break seed dormancy. At the same time, OKA (okadaic acid), an inhibitor of phosphatase activity, can significantly reduce the expression of FsPP2A1 and inhibit seed germination. These results indicate that the PP2A complex may regulate the process of breaking its seed dormancy through the GA pathway [216]. TAP46 is a PP2A-related protein, which is highly expressed in seeds and induced by ABA. In the process of seed germination, the overexpression of TAP46 will decrease the activity of PP2A and increase ABA sensitivity, while the downregulation of TAP46 will increase the activity of PP2A and decrease ABA sensitivity [217]. The subtypes of the Arabidopsis thaliana PP2AA subunit are PDF1, PDF2, and RCN1, which combine the catalytic subunit and regulatory subunit B. RCN1 regulates auxin transport and geotropism. The sensitivity of the rcn1 mutant to NPA was enhanced. After NPA was applied, the root bending degree increased and the response to gravity decreased. However, it is not sensitive to ABA in seed germination and defense cell response. The mutant seedlings with a loss of function showed elongation defects of roots and hypocotyls, and the activity of PP2A decreased. Biochemistry analysis proved that RCN1 was the activator of PP2A activity [218].
4. Synergistic Regulatory Network of Kinase–Phosphatase
Among the known ABA core signaling pathways, members of the PYR/PYL/RCARs family have been widely studied as ABA receptors [219,220]. When plants are under environmental stress or during seed maturity, the ABA levels in plants are upregulated. ABA combined with its receptor PYLs to form a complex, recruited phosphatase PP2Cs, and released the inhibition of PP2Cs on kinase SnRK2s through phosphorylation. The activated SnRK2s activate the expression of downstream ABA-responsive genes by phosphorylating transcription factors such as AREB/ABFS (ABA-responsive element binding proteins/ABA-binding factors) and ABI5, which encourage plants to elicit ABA responses [221,222,223,224]. Under normal growth conditions, PP2Cs directly interact with SnRKs, negatively regulating their activity through dephosphorylation and blocking the ABA signaling pathway [225,226,227]. PYR/PYL/RCAR, PP2C, or SnRK2 mutants showed a strong ABA-insensitive phenotype in seed germination and root growth inhibition, which indicated that the PYR/PP2C/SnRK2 phosphorylation cascade played a role in dormancy regulation [35,43]. The inhibition of ABA on PP2Cs will lead to the release of SnRK1 kinase activity. When facing external stress, plants can quickly activate the expression of downstream genes related to transcription or metabolic reprogramming through SnRK1, thus maintaining the homeostasis of plants [23]. Wasilewska et al. [228] found that SnRK2s played a role downstream of PP2Cs. Arabidopsis ABI1 can interact with SRK2E/OST1/SnRK2.6 and inhibit the activity of SRK2E/OST1/SnRK2.6, and the interaction between them plays a key role in the stomatal closure of plants [48]. Kim et al. [229] found that SAPK2 in the rice SnRK2sII subfamily interacted with OsPP2C30, which induced and activated the expression of ABA-dependent genes. When ABA existed, this induced and activated degree was stronger.
In addition to the changes in the phosphorylation state of the ABA signal transduction pathway, other phosphorylation events also affect seed germination. FyPP1 and FyPP3PP6 phosphatases antagonize the SnRK2 kinase, which dephosphorizes and destabilizes ABI5 [230]. Raf10 and Raf11 MAP3K are positive regulators of dormancy and the expression of ABI3 and ABI5 [97]. In particular, Raf10 phosphorylates subclass III SnRK2 and subsequently phosphorylates ABI5, ABF2, and ABI3 TF to enhance their activities [98]. Phosphorylation also affects the signal transduction of GA. The stability of DELLA is also related to phosphorylation, because TOPP4 PP1 phosphatase directly binds to DELLA proteins RGA and GAI and dephosphorizes them, which promotes GA-dependent instability [201]. The activity of MYB44 TF in relation to seed germination also depends on its phosphorylation by MPK3 and MPK6 kinases [93]. Contrary to RAV1TF, it is inactivated after phosphorylation by the SnRK2 kinase [42]. Protein phosphatases of PP2Cs are also involved in the regulation of the MAPK cascade pathway, among which ABI1 can interact with MPK6 [215]. PP2C5 can interact with MPK3/4/6, and the activity of MPK3/6 in the mutant of PP2C5 is enhanced by ABA activation. AP2C1 and PP2C5 have high homology, and their single mutants all show the characteristics of large stomatal opening and are insensitive to ABA [159]. Brock et al. [159] used PP2C5, a member of the Arabidopsis protein phosphatase family (PP2C), to study its effect on seed germination. It was found that changing the level of PP2C5 affected the activation of MAPK. The results show that PP2C acts as an MAPK phosphatase that positively regulates seed germination, stomatal closure, and ABA-induced gene expression. PP2C5 interacts with MAPK3, MAPK4, and MAPK6 and dephosphorizes them, regulating stomatal closure, seed germination, and stress response [157,231,232,233].
5. Research Challenges and Future Directions
5.1. Existing Research Bottlenecks
Although remarkable progress has been made in the research of kinases and phosphatases in seed germination regulation, there are still many challenges.
Most studies focus on the regulation of seed dormancy and germination by phosphorylation through mediating the ABA signal, but the regulatory mechanisms of GA synthesis, signal transduction, and other hormone pathways are still limited. Therefore, it is necessary to further explore the molecular mechanism of protein phosphorylation and kinase regulation modification interacting with other plant hormone pathways to regulate seed dormancy and germination in order to further enrich the understanding of seed dormancy and germination regulation. The network of signal pathway interaction has not been resolved—the antagonism between GA and ABA is the core regulation mechanism of seed germination, but the cross-regulation network between GA and ABA at the kinase/phosphatase level is still unclear. The intersection of kinase cascades is unknown. The target specificity of phosphatase is vague. The research on the dynamics of post-translation modification is insufficient. The tissue-specific signal transduction mechanism is missing, and the cross-regulation of epigenetic and GA signals is ignored. The role of the non-classical signaling pathway has not been explored. In addition to the DELLA-PIF core pathway, GA may indirectly regulate kinase activity through non-coding RNA or lipid signals (such as phosphatidic acid PA).
First of all, there are technical difficulties in the identification of the substrate proteins of kinases and phosphatases, and the functions of many modified proteins are not clear, which limits the in-depth understanding of regulatory networks. Second, the interaction mechanism between different kinase–phosphatases and that between them and other signal molecules is complex, and how to analyze the integrated regulation mechanism of multi-signal pathways is still a research difficulty. In addition, the current research focuses on model plants; research on important crops is relatively insufficient, which limits the application of research results in agricultural production. Protein phosphorylation plays an important role in the ABA signal transduction pathway. Most studies focus on the regulation of seed dormancy and germination by phosphorylation through mediating ABA signals. However, the regulatory mechanisms of GA synthesis, signal transduction, and other hormone pathways are still limited. Therefore, it is necessary to further explore the molecular mechanism of protein phosphorylation and kinase regulation modification interacting with other plant hormone pathways to regulate seed dormancy and germination, further enriching the understanding of seed dormancy and germination regulation.
Second, oxygen and water are indispensable elements in the process of seed germination. The relationship between protein phosphorylation, kinase regulation and modification, and oxygen/water use efficiency in the pathways of plants deserves further study. Therefore, systematically revealing the relationship between protein phosphorylation and kinase regulation modification and the oxygen/water pathway will greatly expand and deepen our understanding of the regulation mechanism of seed dormancy and germination. This field is also one of the most important research directions in seed biology.
Finally, at present, most studies focus on the regulation of plant development under abiotic stress by genes related to phosphorylation and kinase regulation. However, the precise molecular mechanism regulating seed dormancy and germination is still unknown to a large extent. At present, most of the knowledge about the structure, mechanism, and function of protein phosphatases and kinases comes from animals and fungi. Therefore, it is very important to study the molecular functions of plant protein kinases and phosphatases and to analyze their protein structures in order to systematically understand the molecular mechanism of protein phosphorylation-related genes regulating seed dormancy and germination.
5.2. Prospect of Frontier Technology Application
The rapid development of protein omics, phosphorylated protein omics, single-cell sequencing, and other technologies provides new means to analyze the kinase–phosphatase regulatory network. Using phosphorylated protein proteomics technology, the substrate proteins of kinase and phosphatase can be identified on a large scale, and the dynamic modification changes during seed germination can be revealed. At the same time, with the application of gene editing technologies such as CRISPR/Cas9, redundant kinase genes are simultaneously knocked out and key phosphatases are overexpressed, and the function of the kinase–phosphatase synergistic regulation network is deeply studied. In addition, artificial intelligence and machine learning technology can be used to analyze complex biological data. A synergistic model of kinase and phosphatase based on ordinary differential equations (ODEs) was established; it integrates 100+ known phosphorylation events (such as SnRK2-PP2C and CDK-PP1) to predict the target and regulatory network of kinase–phosphatase.
5.3. Agricultural Application Potential
- (1)
- The development of intelligent seed treatment technology: By designing small molecular regulators targeting kinase/phosphatase (such as kinase inhibitors or phosphatase activators), seed dormancy and germination can be accurately regulated.
- (2)
- Molecular marker-assisted breeding: By analyzing the allelic variation in key kinases (such as SnRK2) and phosphatases (such as PP2C), natural germplasm resources with high seed germination activity can be screened.
- (3)
- Construction of a synthetic biology regulation module: By coupling the light-/temperature-responsive promoter with the core kinase gene, environmentally intelligent seeds can be designed.
- (4)
- Accurate diagnosis system of seed vigor: The rapid detection chip based on phosphorylated protein groups can quantitatively evaluate the activity of the kinase network during seed storage.
It is of great application value for agricultural production to study the molecular mechanism of kinase–phosphatase networks regulating seed germination. By regulating the expression of related genes, the dormancy characteristics of crop seeds can be improved and the problem of seed pre-harvest germination can be solved. Additionally, the germination ability of seeds under adverse conditions can be enhanced to improve the stress resistance of crops. In addition, the development of new plant growth regulators based on kinase–phosphatase regulation mechanisms will also provide new technical means for agricultural production.
6. Conclusions
Through the phosphorylation and dephosphorylation of proteins, kinases and phosphatases play key roles in hormone signal transduction, environmental response, and the metabolic regulation of seed germination. They cooperate with each other to form a complex coordinated regulation network to ensure the seeds start germinating under suitable conditions. Although the current research has made some progress, it still faces many challenges in substrate identification, signal pathway integration, and agricultural application. In the future, combined with cutting-edge technology, an in-depth analysis of the kinase–phosphatase regulatory network will provide important theoretical support and technical guarantees for revealing the molecular mechanism of seed germination and promoting the development of agricultural production.
Author Contributions
B.W., H.L. and J.L.: methodology, investigation, data curation, formal analysis, visualization, writing—original draft, and writing—review and editing. R.L. and N.Y.: conceptualization, supervision, visualization, writing—review and editing, project administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science and Technology Innovation Program of Hunan Province (2024NK1010, 2023NK2002) and the National Natural Science Foundation of China (32171927).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the data are available on request.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Nonogaki, H.; Barrero, J.M.; Li, C. Editorial: Seed Dormancy, Germination, and Pre-harvest Sprouting. Front. Plant Sci. 2018, 9, 1783. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S. Triadimefon pretreatment protects newly assembled membrane system and causes up-regulation of stress proteins in salinity stressed Amaranthus lividus L. during early germination. J. Environ. Biol. 2008, 29, 805–810. [Google Scholar] [PubMed]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Bally, J.; Job, C.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef]
- Bewley, J.D.; Bradford, K.; Hilhorst, H.; Nonogaki, H. Seeds: Physiology of development, germination and dormancy. Seed Sci. Res. 2013, 23, 289. [Google Scholar]
- He, Y.; Zhao, J.; Feng, D.; Huang, Z.; Liang, J.; Zheng, Y.; Cheng, J.; Ying, J.; Wang, Z. RNA-Seq study reveals AP2-domain-containing signalling regulators involved in initial imbibition of seed germination in rice. Rice Sci. 2020, 27, 302–314. [Google Scholar]
- Bewley, J.D. Seed Germination and Dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef]
- Weitbrecht, K.; Müller, K.; Leubner-Metzger, G. First off the mark: Early seed germination. J. Exp. Bot. 2011, 62, 3289–3309. [Google Scholar] [CrossRef] [PubMed]
- Matilla, A.; Gallardo, M.; Puga-Hermida, M.I. Structural, physiological and molecular aspects of heterogeneity in seeds: A review. Seed Sci. Res. 2005, 15, 63–76. [Google Scholar] [CrossRef]
- Nonogaki, H.; Bassel, G.W.; Bewley, J.D. Germination: Still a Mystery. Plant Sci. 2010, 179, 574–581. [Google Scholar] [CrossRef]
- Ortiz-Espín, A.; Iglesias-Fernández, R.; Calderón, A.; Carbonero, P.; Sevilla, F.; Jiménez, A. Mitochondrial AtTrxo1 is transcriptionally regulated by AtbZIP9 and AtAZF2 and affects seed germination under saline conditions. J. Exp. Bot. 2017, 68, 1025–1038. [Google Scholar] [CrossRef]
- Daszkowska-Golec, A. Arabidopsis seed germination under abiotic stress as a concert of action of phytohormones. OMICS 2011, 5, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H. Seed dormancy and germination-emerging mechanisms and new hypotheses. Front. Plant Sci. 2014, 5, 233. [Google Scholar] [CrossRef]
- Gallardo, K.; Job, C.; Groot, S.P.; Puype, M.; Demol, H.; Vandekerckhove, J.; Job, D. Proteomics of Arabidopsis seed germination. A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 2002, 129, 823–837. [Google Scholar] [CrossRef]
- Rajjou, L.; Gallardo, K.; Debeaujon, I.; Vandekerckhove, J.; Job, C.; Job, D. The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 2004, 134, 1598–1613. [Google Scholar] [CrossRef]
- Yang, P.; Li, X.; Wang, X.; Chen, H.; Chen, F.; Shen, S. Proteomic analysis of rice (Oryza sativa) seeds during germination. Proteomics 2007, 7, 3358–3368. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Wang, B.C.; Jin, X.; Li, H.B.; Han, P.; Wei, K.H.; Zhang, X.M.; Zhu, Y.X. Proteomic analysis and extensive protein identification from dry, germinating Arabidopsis seeds and young seedlings. BMB Rep. 2005, 38, 650–660. [Google Scholar] [CrossRef]
- Margalha, L.; Valerio, C.; Baena-González, E. Plant SnRK1 Kinases: Structure, Regulation, and Function. Exp. Suppl. 2016, 107, 403–438. [Google Scholar] [PubMed]
- Smeekens, S.; Ma, J.; Hanson, J.; Rolland, F. Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 2010, 13, 273–278. [Google Scholar] [CrossRef]
- Hulsmans, S.; Rodriguez, M.; De Coninck, B.; Rolland, F. The SnRK1 Energy Sensor in Plant Biotic Interactions. Trends Plant Sci. 2016, 21, 648–661. [Google Scholar] [CrossRef]
- Jossier, M.; Bouly, J.P.; Meimoun, P.; Arjmand, A.; Lessard, P.; Hawley, S.; Grahame Hardie, D.; Thomas, M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 2009, 59, 316–328. [Google Scholar] [CrossRef]
- Cho, Y.H.; Hong, J.W.; Kim, E.C.; Yoo, S.D. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 2012, 158, 1955–1964. [Google Scholar] [CrossRef] [PubMed]
- Tsai, A.Y.; Gazzarrini, S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012, 69, 809–821. [Google Scholar] [CrossRef]
- Rodrigues, A.; Adamo, M.; Crozet, P.; Margalha, L.; Confraria, A.; Martinho, C.; Elias, A.; Rabissi, A.; Lumbreras, V.; González-Guzmán, M.; et al. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 2013, 25, 3871–3884. [Google Scholar] [CrossRef]
- Radchuk, R.; Emery, R.J.; Weier, D.; Vigeolas, H.; Geigenberger, P.; Lunn, J.E.; Feil, R.; Weschke, W.; Weber, H. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 2010, 61, 324–338. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.A.; Lin, C.C.; Lee, K.W.; Chen, J.L.; Huang, L.F.; Ho, S.L.; Liu, H.J.; Hsing, Y.I.; Yu, S.M. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 2007, 19, 2484–2499. [Google Scholar] [CrossRef]
- Hu, Y.; Bai, J.; Xia, Y.; Lin, Y.; Ma, L.; Xu, X.; Ding, Y.; Chen, L. Increasing SnRK1 activity with the AMPK activator A-769662 accelerates seed germination in rice. Plant Physiol. Biochem. 2022, 185, 155–166. [Google Scholar] [CrossRef]
- Song, J.; Shang, L.; Wang, X.; Xing, Y.; Xu, W.; Zhang, Y.; Wang, T.; Li, H.; Zhang, J.; Ye, Z. MAPK11 regulates seed germination and ABA signaling in tomato by phosphorylating SnRKs. J. Exp. Bot. 2021, 72, 1677–1690. [Google Scholar] [CrossRef]
- Gao, X.Q.; Liu, C.Z.; Li, D.D.; Zhao, T.T.; Li, F.; Jia, X.N.; Zhao, X.Y.; Zhang, X.S. The Arabidopsis KINβγ Subunit of the SnRK1 Complex Regulates Pollen Hydration on the Stigma by Mediating the Level of Reactive Oxygen Species in Pollen. PLoS Genet. 2016, 12, e1006228. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lin, Y.; Bai, J.; Xu, X.; Wang, Z.; Ding, C.; Ding, Y.; Chen, L. AMPK activator 991 specifically activates SnRK1 and thereby affects seed germination in rice. J. Exp. Bot. 2024, 75, 2917–2932. [Google Scholar] [CrossRef]
- Pan, L. Role of the Phyllostachys edulis SnRK1a gene in plant growth and stress tolerance. South Afr. J. Bot. 2020, 130, 414–421. [Google Scholar] [CrossRef]
- Emanuelle, S.; Hossain, M.I.; Moller, I.E.; Pedersen, H.L.; van de Meene, A.M.; Doblin, M.S.; Koay, A.; Oakhill, J.S.; Scott, J.W.; Willats, W.G.; et al. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015, 82, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e5. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, H.; Tian, S.; Chang, X.; Jing, R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. J. Exp. Bot. 2010, 61, 683–696. [Google Scholar] [CrossRef]
- Coello, P.; Hey, S.J.; Halford, N.G. The sucrose non-fermenting-1-related (SnRK) family of protein kinases: Potential for manipulation to improve stress tolerance and increase yield. J. Exp. Bot. 2011, 62, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef]
- Johnson, G.L.; Lapadat, R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 2002, 298, 1911–1912. [Google Scholar] [CrossRef]
- Kamiyama, Y.; Katagiri, S.; Umezawa, T. Growth promotion or osmotic stress response: How SNF1-Related Protein Kinase 2 (SnRK2) Kinases are activated and manage intracellular signaling in plants. Plants 2021, 10, 1443. [Google Scholar] [CrossRef]
- Zheng, Z.; Xu, X.; Crosley, R.A.; Greenwalt, S.A.; Sun, Y.; Blakeslee, B.; Wang, L.; Ni, W.; Sopko, M.S.; Yao, C.; et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 2010, 153, 99–113. [Google Scholar] [CrossRef]
- Cai, G.; Wang, Y.; Tu, G.; Chen, P.; Luan, S.; Lan, W. Type A2 BTB Members Decrease the ABA Response during Seed Germination by Affecting the Stability of SnRK2.3 in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 3153. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, Y.; Qiu, J.; Wang, H.; Wang, S.; Tang, L.; Tong, X.; Zhang, J. Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytol. 2020, 228, 1336–1353. [Google Scholar] [CrossRef]
- Zhao, W.; Cheng, Y.H.; Zhang, C.; Shen, X.J.; You, Q.B.; Guo, W.; Li, X.; Song, X.J.; Zhou, X.A.; Jiao, Y.Q. Genome-Wide Identification and Characterization of the GmSnRK2 Family in Soybean. Int. J. Mol. Sci. 2017, 18, 1834. [Google Scholar] [CrossRef]
- Feng, C.Z.; Chen, Y.; Wang, C.; Kong, Y.H.; Wu, W.H.; Chen, Y.F. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014, 80, 654–668. [Google Scholar] [CrossRef]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Li, Y.; Rehman, S.U.; Miao, L.; Zhang, Y.; Chen, X.; Yu, C.; Wang, J.; Li, C.; Jing, R. The Sucrose Non-Fermenting 1-Related Protein Kinase 2 (SnRK2) Genes Are Multifaceted Players in Plant Growth, Development and Response to Environmental Stimuli. Plant Cell Physiol. 2020, 61, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Wu, F.; Sheng, P.; Zhang, Z.; Zhang, X.; Guo, X.; Wang, J.; Cheng, Z.; Wang, J.; Wang, H.; et al. The SnRK2-APC/C(TE) regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef] [PubMed]
- Belin, C.; de Franco, P.O.; Bourbousse, C.; Chaignepain, S.; Schmitter, J.M.; Vavasseur, A.; Giraudat, J.; Barbier-Brygoo, H.; Thomine, S. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006, 141, 1316–1327. [Google Scholar] [CrossRef]
- Xie, T.; Ren, R.; Zhang, Y.Y.; Pang, Y.; Yan, C.; Gong, X.; He, Y.; Li, W.; Miao, D.; Hao, Q.; et al. Molecular mechanism for inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by protein phosphatase ABI1. J. Biol. Chem. 2012, 287, 794–802. [Google Scholar] [CrossRef]
- Yoshida, R.; Umezawa, T.; Mizoguchi, T.; Takahashi, S.; Takahashi, F.; Shinozaki, K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006, 281, 5310–5318. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef]
- Chae, M.J.; Lee, J.S.; Nam, M.H.; Cho, K.; Hong, J.Y.; Yi, S.A.; Suh, S.C.; Yoon, I.S. A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol. Biol. 2007, 63, 151–169. [Google Scholar] [CrossRef]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 Confers Abscisic Acid Sensitivity and Tolerance to Drought Stress in Rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [PubMed]
- Lou, D.; Wang, H.; Yu, D. The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice. BMC Plant Biol. 2018, 18, 203. [Google Scholar] [CrossRef]
- Kim, K.N.; Lee, J.S.; Han, H.; Choi, S.A.; Go, S.J.; Yoon, I.S. Isolation and characterization of a novel rice Ca2+-regulated protein kinase gene involved in responses to diverse signals including cold, light, cytokinins, sugars and salts. Plant Mol. Biol. 2003, 52, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, C.; Weinl, S.; Batistic, O.; Pandey, G.K.; Cheong, Y.H.; Schültke, S.; Albrecht, V.; Ehlert, B.; Schulz, B.; Harter, K.; et al. Alternative complex formation of the Ca-regulated protein kinase CIPK1 controls abscisic acid-dependent and independent stress responses in Arabidopsis. Plant J. 2006, 48, 857–872. [Google Scholar] [CrossRef]
- Pandey, G.K.; Grant, J.J.; Cheong, Y.H.; Kim, B.G.; Li, L.G.; Luan, S. Calcineurin-B-like protein CBL9 interacts with target kinase CIPK3 in the regulation of ABA response in seed germination. Mol. Plant. 2008, 1, 238–248. [Google Scholar] [CrossRef]
- Lyzenga, W.J.; Liu, H.; Schofield, A.; Muise-Hennessey, A.; Stone, S.L. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J. Exp. Bot. 2013, 64, 2779–2791. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hao, H.; Zhang, Y.; Bai, Y.; Zhu, W.; Qin, Y.; Yuan, F.; Zhao, F.; Wang, M.; Hu, J.; et al. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-Type Protein Kinase, Is Important for Abscisic Acid Responses in Arabidopsis through Phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 2015, 168, 659–676. [Google Scholar] [CrossRef]
- Guo, Y.; Xiong, L.; Song, C.P.; Gong, D.; Halfter, U.; Zhu, J.K. A calcium sensor and its interacting protein kinase are global regulators of abscisic acid signaling in Arabidopsis. Dev. Cell 2002, 3, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.; Zhang, C.; Chen, X.; Gong, Z.; Zhu, J.K. Constitutive activation and transgenic evaluation of the function of an arabidopsis PKS protein kinase. J. Biol. Chem. 2002, 277, 42088–42096. [Google Scholar] [CrossRef]
- Kim, K.N.; Cheong, Y.H.; Grant, J.J.; Pandey, G.K.; Luan, S. CIPK3, a calcium sensor-associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 2003, 15, 411–423. [Google Scholar] [CrossRef]
- Piao, H.L.; Xuan, Y.H.; Park, S.H.; Je, B.I.; Park, S.J.; Park, S.H.; Kim, C.M.; Huang, J.; Wang, G.K.; Kim, M.J.; et al. OsCIPK31, a CBL-interacting protein kinase is involved in germination and seedling growth under abiotic stress conditions in rice plants. Mol. Cells 2010, 30, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Footitt, S.; Ölçer-Footitt, H.; Hambidge, A.J.; Finch-Savage, W.E. A laboratory simulation of Arabidopsis seed dormancy cycling provides new insight into its regulation by clock genes and the dormancy-related genes DOG1, MFT, CIPK23 and PHYA. Plant Cell Environ. 2017, 40, 1474–1486. [Google Scholar] [CrossRef]
- Xu, J.; Li, H.D.; Chen, L.Q.; Wang, Y.; Liu, L.L.; He, L.; Wu, W.H. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 2006, 125, 1347–1360. [Google Scholar] [CrossRef]
- Straub, T.; Ludewig, U.; Neuhäuser, B. The Kinase CIPK23 Inhibits Ammonium Transport in Arabidopsis thaliana. Plant Cell 2017, 29, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; An, L.; Mao, J.; Aluko, O.O.; Ullah, Z.; Xu, F.; Liu, G.; Liu, H.; Wang, Q. The CBL-Interacting Protein Kinase NtCIPK23 Positively Regulates Seed Germination and Early Seedling Development in Tobacco (Nicotiana tabacum L.). Plants 2021, 10, 323. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, Y.; Li, P.; Jian, J.; Zhao, C.; Wen, G. Mitogen-Activated Protein Kinase and Substrate Identification in Plant Growth and Development. Int. J. Mol. Sci. 2022, 23, 2744. [Google Scholar] [CrossRef]
- Nakagami, H.; Pitzschke, A.; Hirt, H. Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci. 2005, 10, 339–346. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Komis, G.; Šamajová, O.; Ovečka, M.; Šamaj, J. Cell and Developmental Biology of Plant Mitogen-Activated Protein Kinases. Annu. Rev. Plant Biol. 2018, 69, 237–265. [Google Scholar] [CrossRef]
- Zheng, Y.; Deng, X.; Qu, A.; Zhang, M.; Tao, Y.; Yang, L.; Liu, Y.; Xu, J.; Zhang, S. Regulation of pollen lipid body biogenesis by MAP kinases and downstream WRKY transcription factors in Arabidopsis. PLoS Genet. 2018, 14, e1007880. [Google Scholar] [CrossRef]
- MAPK Group. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, D.; Wang, L.; Li, D. Genome-Wide Analysis of Mitogen Activated Protein Kinase Gene Family in Maize. Plant Mol. Biol. Rep. 2013, 31, 1446–1460. [Google Scholar] [CrossRef]
- Majeed, Y.; Zhu, X.; Zhang, N.; Ul-Ain, N.; Raza, A.; Haider, F.U.; Si, H. Harnessing the role of mitogen-activated protein kinases against abiotic stresses in plants. Front. Plant Sci. 2023, 14, 932923. [Google Scholar] [CrossRef] [PubMed]
- Asif, M.H.; Lakhwani, D.; Pathak, S.; Bhambhani, S.; Bag, S.K.; Trivedi, P.K. Genome-wide identification and expression analysis of the mitogen-activated protein kinase gene family from banana suggest involvement of specific members in different stages of fruit ripening. Funct. Integr. Genomics. 2014, 14, 161–175. [Google Scholar] [CrossRef]
- Kong, F.; Wang, J.; Cheng, L.; Liu, S.; Wu, J.; Peng, Z.; Lu, G. Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene 2012, 499, 108–120. [Google Scholar] [CrossRef]
- Wang, J.; Pan, C.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 2015, 16, 386. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, R.; Luo, X.; Jiang, Z.; Shu, H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 2013, 531, 377–387. [Google Scholar] [CrossRef]
- Taj, G.; Agarwal, P.; Grant, M.; Kumar, A. MAPK machinery in plants: Recognition and response to different stresses through multiple signal transduction pathways. Plant Signal. Behav. 2010, 5, 1370–1378. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Genot, B.; Lang, J.; Berriri, S.; Garmier, M.; Gilard, F.; Pateyron, S.; Haustraete, K.; Van Der Straeten, D.; Hirt, H.; Colcombet, J. Constitutively Active Arabidopsis MAP Kinase 3 Triggers Defense Responses Involving Salicylic Acid and SUMM2 Resistance Protein. Plant Physiol. 2017, 174, 1238–1249. [Google Scholar] [CrossRef]
- Jiang, M.; Wen, F.; Cao, J.; Li, P.; She, J.; Chu, Z. Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. BMC Genom. 2015, 16, 228. [Google Scholar] [CrossRef]
- Çakır, B.; Kılıçkaya, O. Mitogen-activated protein kinase cascades in Vitis vinifera. Front. Plant Sci. 2015, 6, 556. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Sinha, A.K. Overexpression of constitutively active mitogen activated protein kinase kinase 6 enhances tolerance to salt stress in rice. Rice 2013, 6, 25. [Google Scholar] [CrossRef]
- Wang, G.; Lovato, A.; Liang, Y.H.; Wang, M.; Chen, F.; Tornielli, G.B.; Polverari, A.; Pezzotti, M.; Cheng, Z.M. Validation by isolation and expression analyses of the mitogen-activated protein kinase gene family in the grapevine (Vitis vinifera L.). Aust. J. Grape Wine Res. 2014, 20, 255–262. [Google Scholar] [CrossRef]
- Yue, H.; Li, Z.; Xing, D. Roles of Arabidopsis bax inhibitor-1 in delaying methyl jasmonate-induced leaf senescence. Plant Signal. Behav. 2012, 7, 1488–1489. [Google Scholar] [CrossRef]
- Zhao, F.Y.; Hu, F.; Zhang, S.Y.; Wang, K.; Zhang, C.R.; Liu, T. MAPKs regulate root growth by influencing auxin signaling and cell cycle-related gene expression in cadmium-stressed rice. Environ. Sci. Pollut. Res. 2013, 20, 5449–5460. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Fernie, A.R. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 2010, 5, 1210–1227. [Google Scholar] [CrossRef]
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 and AtMPK6 are involved in abscisic acid and sugar signaling in Arabidopsis seed germination. Plant Mol. Biol. 2009, 70, 725–736. [Google Scholar] [CrossRef]
- Xu, R.; Duan, P.; Yu, H.; Zhou, Z.; Zhang, B.; Wang, R.; Li, J.; Zhang, G.; Zhuang, S.; Lyu, J.; et al. Control of Grain Size and Weight by the OsMKKK10-OsMKK4-OsMAPK6 Signaling Pathway in Rice. Mol. Plant. 2018, 11, 860–873. [Google Scholar] [CrossRef]
- Zhang, W.; Cochet, F.; Ponnaiah, M.; Lebreton, S.; Matheron, L.; Pionneau, C.; Boudsocq, M.; Resentini, F.; Huguet, S.; Blázquez, M.Á.; et al. The MPK8-TCP14 pathway promotes seed germination in Arabidopsis. Plant J. 2019, 100, 677–692. [Google Scholar] [CrossRef]
- Tatematsu, K.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 2008, 53, 42–52. [Google Scholar] [CrossRef]
- Nakamura, S.; Pourkheirandish, M.; Morishige, H.; Kubo, Y.; Nakamura, M.; Ichimura, K.; Seo, S.; Kanamori, H.; Wu, J.; Ando, T.; et al. Mitogen-Activated Protein Kinase Kinase 3 Regulates Seed Dormancy in Barley. Curr. Biol. 2016, 26, 775–781. [Google Scholar] [CrossRef]
- Nguyen, X.C.; Hoang, M.H.; Kim, H.S.; Lee, K.; Liu, X.M.; Kim, S.H.; Bahk, S.; Park, H.C.; Chung, W.S. Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochem. Biophys. Res. Commun. 2012, 423, 703–708. [Google Scholar] [CrossRef]
- Martinez, S.A.; Shorinola, O.; Conselman, S.; See, D.; Skinner, D.Z.; Uauy, C.; Steber, C.M. Exome sequencing of bulked segregants identified a novel TaMKK3-A allele linked to the wheat ERA8 ABA-hypersensitive germination phenotype. TAG. Theoretical and applied genetics. Theor. Appl. Genet. 2020, 133, 719–736. [Google Scholar] [CrossRef]
- Torada, A.; Koike, M.; Ogawa, T.; Takenouchi, Y.; Tadamura, K.; Wu, J.; Matsumoto, T.; Kawaura, K.; Ogihara, Y. A Causal Gene for Seed Dormancy on Wheat Chromosome 4A Encodes a MAP Kinase Kinase. Curr. Biol. 2016, 26, 782–787. [Google Scholar] [CrossRef]
- Gong, X.; Li, C.; Zhou, M.; Bonnardeaux, Y.; Yan, G. Seed dormancy in barley is dictated by genetics, environments and their interactions. Euphytica 2014, 197, 355–368. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, M.H.; Kim, J.I.; Kim, S.Y. Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol. 2015, 56, 84–97. [Google Scholar] [CrossRef]
- Nguyen, Q.T.C.; Lee, S.J.; Choi, S.W.; Na, Y.J.; Song, M.R.; Hoang, Q.T.N.; Sim, S.Y.; Kim, M.S.; Kim, J.I.; Soh, M.S.; et al. Arabidopsis Raf-Like Kinase Raf10 Is a Regulatory Component of Core ABA Signaling. Mol. Cells 2019, 42, 646–660. [Google Scholar]
- Xiao, X.; Tang, Z.; Li, X.; Hong, Y.; Li, B.; Xiao, W.; Gao, Z.; Lin, D.; Li, C.; Luo, L.; et al. Overexpressing OsMAPK12-1 inhibits plant growth and enhances resistance to bacterial disease in rice. Funct. Plant Biol. 2017, 44, 694–704. [Google Scholar] [CrossRef]
- Lu, C.; Han, M.H.; Guevara-Garcia, A.; Fedoroff, N.V. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. USA 2002, 99, 15812–15817. [Google Scholar] [CrossRef]
- Zhang, A.; Jiang, M.; Zhang, J.; Tan, M.; Hu, X. Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 2006, 141, 475–487. [Google Scholar] [CrossRef]
- Xing, Y.; Jia, W.; Zhang, J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008, 54, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Testerink, C.; Vennik, M.; Kijne, J.W.; Wang, M.; Heimovaara-Dijkstra, S. Inactivation of a MAPK-like protein kinase and activation of a MBP kinase in germinating barley embryos. FEBS Lett. 2000, 484, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Jia, W.; Zhang, J. AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J. Exp. Bot. 2007, 58, 2969–2981. [Google Scholar] [CrossRef]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Wang, C.; Gao, Q.; Li, L.; Luan, S. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 2020, 6, 750–759. [Google Scholar] [CrossRef]
- McCormack, E.; Tsai, Y.C.; Braam, J. Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 2005, 10, 383–389. [Google Scholar] [CrossRef]
- Aldon, D.; Mbengue, M.; Mazars, C.; Galaud, J.P. Calcium Signalling in Plant Biotic Interactions. Int. J. Mol. Sci. 2018, 19, 665. [Google Scholar] [CrossRef]
- Batistič, O.; Kudla, J. Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 2012, 1820, 1283–1293. [Google Scholar] [CrossRef]
- Schulz, P.; Herde, M.; Romeis, T. Calcium-dependent protein kinases: Hubs in plant stress signaling and development. Plant Physiol. 2013, 163, 523–530. [Google Scholar] [CrossRef]
- Yip Delormel, T.; Boudsocq, M. Properties and functions of calcium-dependent protein kinases and their relatives in Arabidopsis thaliana. New Phytol. 2019, 224, 585–604. [Google Scholar] [CrossRef] [PubMed]
- Liese, A.; Romeis, T. Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta 2013, 1833, 1582–1589. [Google Scholar] [CrossRef]
- Baba, A.I.; Rigó, G.; Ayaydin, F.; Rehman, A.U.; Andrási, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T.; et al. Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef]
- Mall, T.K.; Dweikat, I.; Sato, S.J.; Neresian, N.; Xu, K.; Ge, Z.; Wang, D.; Elthon, T.; Clemente, T. Expression of the rice CDPK-7 in sorghum: Molecular and phenotypic analyses. Plant Mol. Biol. 2011, 75, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Li, A.L.; Zhu, Y.F.; Tan, X.M.; Wang, X.; Wei, B.; Guo, H.Z.; Zhang, Z.L.; Chen, X.B.; Zhao, G.Y.; Kong, X.Y.; et al. Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol. Biol. 2008, 66, 429–443. [Google Scholar] [CrossRef]
- Yu, T.F.; Zhao, W.Y.; Fu, J.D.; Liu, Y.W.; Chen, M.; Zhou, Y.B.; Ma, Y.Z.; Xu, Z.S.; Xi, Y.J. Genome-Wide Analysis of CDPK Family in Foxtail Millet and Determination of SiCDPK24 Functions in Drought Stress. Front. Plant Sci. 2018, 9, 651. [Google Scholar] [CrossRef] [PubMed]
- Weckwerth, P.; Ehlert, B.; Romeis, T. ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signalling. Plant Cell Environ. 2015, 38, 544–558. [Google Scholar] [CrossRef]
- Edel, K.H.; Kudla, J. Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 2016, 33, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, D.; Yang, B.; Liu, W.Z.; Mu, B.; Song, H.; Chen, B.; Li, Y.; Ren, D.; Deng, H.; et al. Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J. Exp. Bot. 2020, 71, 188–203. [Google Scholar] [CrossRef]
- Yu, X.C.; Li, M.J.; Gao, G.F.; Feng, H.Z.; Geng, X.Q.; Peng, C.C.; Zhu, S.Y.; Wang, X.J.; Shen, Y.Y.; Zhang, D.P. Abscisic acid stimulates a calcium-dependent protein kinase in grape berry. Plant Physiol. 2006, 140, 558–579. [Google Scholar] [CrossRef]
- Zhu, S.Y.; Yu, X.C.; Wang, X.J.; Zhao, R.; Li, Y.; Fan, R.C.; Shang, Y.; Du, S.Y.; Wang, X.F.; Wu, F.Q.; et al. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 2007, 19, 3019–3036. [Google Scholar] [CrossRef]
- Zhao, R.; Sun, H.L.; Mei, C.; Wang, X.J.; Yan, L.; Liu, R.; Zhang, X.F.; Wang, X.F.; Zhang, D.P. The Arabidopsis Ca(2+) -dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 2011, 192, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Fedosejevs, E.T.; Gerdis, S.A.; Ying, S.; Pyc, M.; Anderson, E.M.; Snedden, W.A.; Mullen, R.T.; She, Y.M.; Plaxton, W.C. The calcium-dependent protein kinase RcCDPK2 phosphorylates sucrose synthase at Ser11 in developing castor oil seeds. Biochem. J. 2016, 473, 3667–3682. [Google Scholar] [CrossRef] [PubMed]
- Dammann, C.; Ichida, A.; Hong, B.; Romanowsky, S.M.; Hrabak, E.M.; Harmon, A.C.; Pickard, B.G.; Harper, J.F. Subcellular targeting of nine calcium-dependent protein kinase isoforms from Arabidopsis. Plant Physiol. 2003, 132, 1840–1848. [Google Scholar] [CrossRef]
- Oh, M.H.; Wu, X.; Kim, H.S.; Harper, J.F.; Zielinski, R.E.; Clouse, S.D.; Huber, S.C. CDPKs are dual-specificity protein kinases and tyrosine autophosphorylation attenuates kinase activity. FEBS Lett. 2012, 586, 4070–4075. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Milla, M.A.; Uno, Y.; Chang, I.F.; Townsend, J.; Maher, E.A.; Quilici, D.; Cushman, J.C. A novel yeast two-hybrid approach to identify CDPK substrates: Characterization of the interaction between AtCPK11 and AtDi19, a nuclear zinc finger protein. FEBS Lett. 2006, 580, 904–911. [Google Scholar] [CrossRef]
- Chen, D.H.; Liu, H.P.; Li, C.L. Calcium-dependent protein kinase CPK9 negatively functions in stomatal abscisic acid signaling by regulating ion channel activity in Arabidopsis. Plant Mol. Biol. 2019, 99, 113–122. [Google Scholar] [CrossRef]
- Choi, H.I.; Park, H.J.; Park, J.H.; Kim, S.; Im, M.Y.; Seo, H.H.; Kim, Y.W.; Hwang, I.; Kim, S.Y. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 2005, 139, 1750–1761. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, X.F.; Zhang, D.P. CPK12: A Ca2+-dependent protein kinase balancer in abscisic acid signaling. Plant Signal. Behav. 2011, 6, 1687–1690. [Google Scholar] [CrossRef]
- Jiang, J.Z.; Kuo, C.H.; Chen, B.H.; Chen, M.K.; Lin, C.S.; Ho, S.L. Effects of OsCDPK1 on the Structure and Physicochemical Properties of Starch in Developing Rice Seeds. Int. J. Mol. Sci. 2018, 19, 3247. [Google Scholar] [CrossRef]
- Frattini, M.; Morello, L.; Breviario, D. Rice calcium-dependent protein kinase isoforms OsCDPK2 and OsCDPK11 show different responses to light and different expression patterns during seed development. Plant Mol. Biol. 1999, 41, 753–764. [Google Scholar] [CrossRef]
- Yu, X.C.; Zhu, S.Y.; Gao, G.F.; Wang, X.J.; Zhao, R.; Zou, K.Q.; Wang, X.F.; Zhang, X.Y.; Wu, F.Q.; Peng, C.C.; et al. Expression of a grape calcium-dependent protein kinase ACPK1 in Arabidopsis thaliana promotes plant growth and confers abscisic acid-hypersensitivity in germination, postgermination growth, and stomatal movement. Plant Mol. Biol. 2007, 64, 531–538. [Google Scholar] [CrossRef]
- Jaworski, K.; Pawełek, A.; Kopcewicz, J.; Szmidt-Jaworska, A. The calcium-dependent protein kinase (PnCDPK1) is involved in Pharbitis nil flowering. J. Plant Physiol. 2012, 169, 1578–1585. [Google Scholar] [CrossRef]
- Pawełek, A.; Szmidt-Jaworska, A.; Świeżawska, B.; Jaworski, K. Genomic structure and promoter characterization of the CDPK kinase gene expressed during seed formation in Pharbitis nil. J. Plant Physiol. 2015, 189, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Kunieda, N.; Omura, Y.; Ibe, H.; Kawasaki, T.; Takano, M.; Sato, M.; Furuhashi, H.; Mujin, T.; Takaiwa, F.; et al. Rice SPK, a calmodulin-like domain protein kinase, is required for storage product accumulation during seed development: Phosphorylation of sucrose synthase is a possible factor. Plant Cell 2002, 14, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Santin, F.; Bhogale, S.; Fantino, E.; Grandellis, C.; Banerjee, A.K.; Ulloa, R.M. Solanum tuberosum StCDPK1 is regulated by miR390 at the posttranscriptional level and phosphorylates the auxin efflux carrier StPIN4 in vitro, a potential downstream target in potato development. Physiol Plant. 2017, 159, 244–261. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, D.; Wang, L.; Pan, J.; Liu, Y.; Kong, X.; Zhou, Y.; Li, D. A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signaling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2013, 71, 112–120. [Google Scholar] [CrossRef]
- Jaworski, K.; Pawełek, A.; Szmidt-Jaworska, A.; Kopcewicz, J. Expression of calcium-dependent protein kinase gene (PnCDPK1) is affected by various light conditions in Pharbitis nil seedlings. J. Plant Growth Regul. 2010, 29, 316–327. [Google Scholar] [CrossRef]
- Jaworski, K.; Szmidt-Jaworska, A.; Kopcewicz, J. Two calcium dependent protein kinases are differently regulated by light and have different activity patterns during seedling growth in Pharbitis nil. Plant Growth Regul. 2011, 65, 369–379. [Google Scholar] [CrossRef]
- Moorhead, G.B.; De, W.V.; Templeton, G.; Kerk, D. Evolution of protein phosphatases in plants and animals. Biochem. J. 2009, 417, 401–409. [Google Scholar] [CrossRef]
- Olsen, J.V.; Vermeulen, M.; Santamaria, A.; Kumar, C.; Miller, M.L.; Jensen, L.J.; Gnad, F.; Cox, J.; Jensen, T.S.; Nigg, E.A. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 2010, 3, ra3. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T.; Pawson, T. The evolution of protein phosphorylation. Preface. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2012, 367, 2512. [Google Scholar]
- Ito, J.; Taylor, N.L.; Castleden, I.; Weckwerth, W.; Millar, A.H.; Heazlewood, J.L. A survey of the Arabidopsis thaliana mitochondrial phosphoproteome. Proteomics 2009, 9, 229–4240. [Google Scholar] [CrossRef]
- Reiland, S.; Messerli, G.; Baerenfaller, K.; Gerrits, B.; Endler, A.; Grossmann, J.; Gruissem, W.; Baginsky, S. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009, 150, 889–903. [Google Scholar] [CrossRef]
- Tagliabracci, V.S.; Engel, J.L.; Wen, J.; Wiley, S.E.; Worby, C.A.; Kinch, L.N.; Xiao, J.; Grishin, N.V.; Dixon, J.E. Secreted kinase phosphorylates extracellular proteins that regulate biomineralization. Science 2012, 336, 1150–1153. [Google Scholar] [CrossRef]
- Bhaskara, G.B.; Wen, T.N.; Nguyen, T.T.; Verslues, P.E. Protein Phosphatase 2Cs and Microtubule-Associated Stress Protein 1 Control Microtubule Stability, Plant Growth, and Drought Response. Plant Cell 2017, 29, 169–191. [Google Scholar] [CrossRef] [PubMed]
- Mackintosh, R.W.; Davies, S.P.; Clarke, P.R.; Weekes, J.; Gillespie, J.G.; Gibb, B.J.; Hardie, D.G. Evidence for a protein kinase cascade in higher plants. 3-Hydroxy-3-methylglutaryl-CoA reductase kinase. Eur. J. Biochem. 1992, 209, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.L.; Leube, M.P.; Grill, E. Molecular cloning in Arabidopsis thaliana of a new protein phosphatase 2C (PP2C) with homology to ABI1 and ABI2. Plant Mol. Biol. 1998, 38, 879–883. [Google Scholar] [CrossRef]
- Fuchs, S.; Grill, E.; Meskiene, I.; Schweighofer, A. Type 2C protein phosphatases in plants. FEBS J. 2013, 280, 681–693. [Google Scholar] [CrossRef]
- Xue, T.; Wang, D.; Zhang, S.; Ehlting, J.; Ni, F.; Jakab, S.; Zheng, C.; Zhong, Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom. 2008, 9, 550. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Ni, L.; Xia, X.; Chen, S.; Zhang, Y.; Lang, M.; Li, M.; Liu, B.; Pan, Y.; Li, J.; et al. Genome-Wide Analysis of the Protein Phosphatase 2C Genes in Tomato. Genes 2022, 13, 604. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Yuan, S.; Chen, J.; Chen, Y.; Li, Z.; Lin, W.; Zhang, Y.; Liu, J.; Lin, W. Molecular evolution and lineage-specific expansion of the PP2C family in Zea mays. Planta 2019, 250, 1521–1538. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.; Ke, H.; Hu, C.M.; Naseri, E.; Haider, M.S.; Ayaz, A.; Amjad Khan, W.; Wang, J.; Hou, X. Genome-Wide Identification, Evolution, and Transcriptional Profiling of PP2C Gene Family in Brassica rapa. BioMed Res. Int. 2019, 2019, 2965035. [Google Scholar] [CrossRef]
- Singh, A.; Pandey, A.; Srivastava, A.K.; Tran, L.S.; Pandey, G.K. Plant protein phosphatases 2C: From genomic diversity to functional multiplicity and importance in stress management. Crit. Rev. Biotechnol. 2016, 36, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.; Merlot, S.; Giraudat, J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 1997, 9, 759–771. [Google Scholar][Green Version]
- Roy, S.J.; Tucker, E.J.; Tester, M. Genetic analysis of abiotic stress tolerance in crops. Curr. Opin. Plant Biol. 2011, 14, 232–239. [Google Scholar] [CrossRef]
- Umbrasaite, J.; Schweighofer, A.; Kazanaviciute, V.; Magyar, Z.; Ayatollahi, Z.; Unterwurzacher, V.; Choopayak, C.; Boniecka, J.; Murray, J.A.; Bogre, L.; et al. MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PLoS ONE 2010, 5, e15357. [Google Scholar] [CrossRef]
- Wang, H.; Wang, W.; Li, W.; Chen, X.; Lu, C.; Li, M.; Chen, Y. Research progress of cold resistance and cultivation practice for Cassava moving northward. Chin. J. Trop. Crops 2016, 37, 1437–1443. [Google Scholar]
- Brock, A.K.; Willmann, R.; Kolb, D.; Grefen, L.; Lajunen, H.M.; Bethke, G.; Lee, J.; Nürnberger, T.; Gust, A.A. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 2010, 153, 1098–1111. [Google Scholar] [CrossRef]
- Martin, M.; Kettmann, R.; Dequiedt, F. Recent insights into protein phosphatase 2A structure and regulation: The reasons why PP2A is no longer considered as a lazy passive housekeeping enzyme. Biotechnol. Agron. Soc. Environ. 2010, 14, 243–252. [Google Scholar]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Lee, Y.; Park, J.; Lee, N.; Choi, G. HONSU, a protein phosphatase 2C, regulates seed dormancy by inhibiting ABA signaling in Arabidopsis. Plant Cell Physiol. 2013, 54, 555–572. [Google Scholar] [CrossRef]
- Stone, J.M.; Collinge, M.A.; Smith, R.D.; Horn, M.A.; Walker, J.C. Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 1994, 266, 793–795. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Tsuchiya, W.; Moresco, J.J.; Hayashi, Y.; Satoh, K.; Kaiwa, N.; Irisa, T.; Kinoshita, T.; Schroeder, J.I.; Yates, J.R., 3rd; et al. Control of seed dormancy and germination by DOG1-AHG1 PP2C phosphatase complex via binding to heme. Nat. Commun. 2018, 9, 2132. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Heo, S.; Lee, S. Inhibition of Type 2C Protein Phosphatases by ABA Receptors in Abscisic Acid-Mediated Plant Stress Responses. Methods Mol. Biol. 2022, 2462, 1–16. [Google Scholar]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef]
- Yu, X.; Han, J.; Li, L.; Zhang, Q.; Yang, G.; He, G. Wheat PP2C-a10 regulates seed germination and drought tolerance in transgenic Arabidopsis. Plant Cell Rep. 2020, 39, 635–651. [Google Scholar] [CrossRef]
- Lorenzo, O.; Rodríguez, D.; Nicolás, G.; Rodríguez, P.L.; Nicolás, C. A new protein phosphatase 2C (FsPP2C1) induced by abscisic acid is specifically expressed in dormant beechnut seeds. Plant Physiol. 2001, 125, 1949–1956. [Google Scholar] [CrossRef]
- Bhatnagar, N.; Min, M.K.; Choi, E.H.; Kim, N.; Moon, S.J.; Yoon, I.; Kwon, T.; Jung, K.H.; Kim, B.G. The protein phosphatase 2C clade A protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol. Biol. 2017, 93, 389–401. [Google Scholar] [CrossRef]
- Cantoro, R.; Crocco, C.D.; Benech-Arnold, R.L.; Rodríguez, M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 2013, 64, 5721–5735. [Google Scholar] [CrossRef]
- Chen, K.; An, Y.C. Transcriptional Responses to Gibberellin and Abscisic Acid in Barley Aleurone. J. Integr. Plant Biol. 2006, 48, 591–612. [Google Scholar] [CrossRef]
- Xiang, Y.; Nakabayashi, K.; Ding, J.; He, F.; Bentsink, L.; Soppe, W.J. Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell 2014, 26, 4362–4375. [Google Scholar] [CrossRef]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef]
- Zhang, F.; Fu, X.; Lv, Z.; Shen, Q.; Yan, T.; Jiang, W.; Wang, G.; Sun, X.; Tang, K. Type 2C phosphatase 1 of Artemisia annua L. is a negative regulator of ABA signaling. Biomed Res. Int. 2014, 2014, 521794. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Hu, X.; Song, J.; Zong, X.; Li, D.; Li, D. Over-expression of a Zea mays L. protein phosphatase 2C gene (ZmPP2C) in Arabidopsis thaliana decreases tolerance to salt and drought. J. Plant Physiol. 2009, 166, 531–542. [Google Scholar] [CrossRef]
- Xiang, Y.; Sun, X.; Gao, S.; Qin, F.; Dai, M. Deletion of an Endoplasmic Reticulum Stress Response Element in a ZmPP2C-A Gene Facilitates Drought Tolerance of Maize Seedlings. Mol. Plant. 2017, 10, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Née, G.; Kramer, K.; Nakabayashi, K.; Yuan, B.; Xiang, Y.; Miatton, E.; Finkemeier, I.; Soppe, W.J.J. DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 2017, 8, 72. [Google Scholar] [CrossRef]
- Nishimura, N.; Yoshida, T.; Kitahata, N.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J. 2007, 50, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Egawa, C.; Oohashi, M.; Meguro-Maoka, A.; Shimosaka, E.; Sato, Y. Ectopic expression of mutated type 2C protein phosphatase OsABI-LIKE2 decreases abscisic acid sensitivity in Arabidopsis and rice. Sci. Rep. 2018, 8, 12320. [Google Scholar] [CrossRef]
- Singh, A.; Jha, S.K.; Bagri, J.; Pandey, G.K. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE 2015, 10, e0125168. [Google Scholar] [CrossRef]
- Tonks, N.K.; Neel, B.G. Combinatorial control of the specificity of protein tyrosine phosphatases. Curr. Opin. Cell Biol. 2001, 13, 182–195. [Google Scholar] [CrossRef]
- Kerk, D.; Bulgrien, J.; Smith, D.W.; Barsam, B.; Veretnik, S.; Gribskov, M. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002, 129, 908–925. [Google Scholar] [CrossRef]
- Denu, J.M.; Stuckey, J.A.; Saper, M.A.; Dixon, J.E. Form and function in protein dephosphorylation. Cell 1996, 87, 361–364. [Google Scholar] [CrossRef]
- Kyriakis, J.M.; Avruch, J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001, 81, 807–869. [Google Scholar] [CrossRef]
- Owens, D.M.; Keyse, S.M. Differential regulation of MAP kinase singaling by dual-specificity protein phosphatases. Oncogene 2007, 26, 3203–3213. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.B.; Wulfsen, I.; Utku, E.; Sausbier, U.; Sausbier, M.; Wieland, T.; Ruth, P.; Korth, M. Dual role of protein kinase C on BK channel regulation. Proc. Natl. Acad. Sci. USA 2010, 107, 8005–8010. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Huang, Y.; Kieber, J.; Luan, S. Identification of a dual-specificity protein phosphatase that inactivates a MAP kinase from Arabidopsis. Plant J. 1998, 16, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liu, Y.; Ye, N.; Zhu, G.; Chen, M.; Jia, L.; Xia, Y.; Shi, L.; Jia, W.; Zhang, J. AtDsPTP1 acts as a negative regulator in osmotic stress signalling during Arabidopsis seed germination and seedling establishment. J. Exp. Bot. 2015, 66, 1339–1353. [Google Scholar] [CrossRef]
- Huang, H.J.; Lin, Y.M.; Huang, D.D.; Takahashi, T.; Sugiyama, M. Protein tyrosine phosphorylation during phytohormone-stimulated cell proliferation in Arabidopsis Hypocotyls. Plant Cell Physiol. 2003, 44, 770–775. [Google Scholar] [CrossRef]
- Monroe-Augustus, M.; Zolman, B.K.; Bartel, B. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 2003, 15, 2979–2991. [Google Scholar] [CrossRef]
- Reyes, D.; Rodríguez, D.; Nicolás, G.; Nicolás, C. Evidence of a role for tyrosine dephosphorylation in the control of postgermination arrest of development by abscisic acid in Arabidopsis thaliana L. Planta 2006, 223, 381–385. [Google Scholar] [CrossRef]
- Dixon, D.P.; Fordham-Skelton, A.P.; Edwards, R. Redox regulation of a soybean tyrosine-specific protein phosphatase. Biochemistry 2005, 44, 7696–7703. [Google Scholar] [CrossRef] [PubMed]
- Quettier, A.L.; Bertrand, C.; Habricot, Y.; Miginiac, E.; Agnes, C.; Jeannette, E.; Maldiney, R. The phs1-3 mutation in a putative dual-specificity protein tyrosine phosphatase gene provokes hypersensitive responses to abscisic acid in Arabidopsis thaliana. Plant J. 2006, 47, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Ramírez, A.; Rodríguez, D.; Reyes, D.; Jiménez, J.A.; Nicolás, G.; Nicolás, C. Functional analysis in Arabidopsis of FsPTP1, a tyrosine phosphatase from beechnuts, reveals its role as a negative regulator of ABA signaling and seed dormancy and suggests its involvement in ethylene signaling modulation. Planta 2011, 234, 589–597. [Google Scholar] [CrossRef]
- Ceulemans, H.; Bollen, M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004, 84, 1–39. [Google Scholar] [CrossRef]
- Grallert, A.; Boke, E.; Hagting, A.; Hodgson, B.; Connolly, Y.; Griffiths, J.R.; Smith, D.L.; Pines, J.; Hagan, I.M. A PP1-PP2A phosphatase relay controls mitotic progression. Nature 2015, 517, 94–98. [Google Scholar] [CrossRef]
- Nie, H.; Zheng, Y.; Li, R.; Guo, T.B.; He, D.; Fang, L.; Liu, X.; Xiao, L.; Chen, X.; Wan, B.; et al. Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-α in rheumatoid arthritis. Nat. Med. 2013, 19, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Li, J.; Xu, M.; Zhao, Q.; Hou, Y.; Yao, L.; Zhong, Y.; Chou, P.C.; Zhang, W.; Zhou, P.; et al. PKCε phosphorylates MIIP and promotes colorectal cancer metastasis through inhibition of RelA deacetylation. Nat. Commun. 2017, 8, 939. [Google Scholar] [CrossRef]
- Stubbs, M.D.; Tran, H.T.; Atwell, A.J.; Smith, C.S.; Olson, D.; Moorhead, G.B. Purification and properties of Arabidopsis thaliana type 1 protein phosphatase (PP1). Biochim. Biophys. Acta 2001, 1550, 52–63. [Google Scholar] [CrossRef]
- Hou, Y.J.; Zhu, Y.; Wang, P.; Zhao, Y.; Xie, S.; Batelli, G.; Wang, B.; Duan, C.G.; Wang, X.; Xing, L.; et al. Type One Protein Phosphatase 1 and Its Regulatory Protein Inhibitor 2 Negatively Regulate ABA Signaling. PLoS Genet. 2016, 12, e1005835. [Google Scholar] [CrossRef]
- Qin, Q.; Wang, W.; Guo, X.; Yue, J.; Huang, Y.; Xu, X.; Li, J.; Hou, S. Arabidopsis DELLA protein degradation is controlled by a type-one protein phosphatase, TOPP4. PLoS Genet. 2014, 10, e1004464. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Qin, Q.; Yan, J.; Niu, Y.; Huang, B.; Guan, L.; Li, Y.; Ren, D.; Li, J.; Hou, S. TYPE-ONE PROTEIN PHOSPHATASE4 regulates pavement cell interdigitation by modulating PIN-FORMED1 polarity and trafficking in Arabidopsis. Plant Physiol. 2015, 167, 1058–1075. [Google Scholar] [CrossRef]
- Yue, J.; Qin, Q.; Meng, S.; Jing, H.; Gou, X.; Li, J.; Hou, S. TOPP4 Regulates the Stability of PHYTOCHROME INTERACTING FACTOR5 during Photomorphogenesis in Arabidopsis. Plant Physiol. 2016, 170, 1381–1397. [Google Scholar] [CrossRef]
- Bradai, M.; Mahjoubi, H.; Chini, A.; Chabouté, M.E.; Hanin, M.; Ebel, C. Genome wide identification of wheat and Brachypodium type one protein phosphatases and functional characterization of durum wheat TdPP1a. PLoS ONE 2018, 13, e0191272. [Google Scholar] [CrossRef]
- Bradai, M.; Amorim-Silva, V.; Belgaroui, N.; Esteban Del Valle, A.; Chabouté, M.E.; Schmit, A.C.; Lozano-Duran, R.; Botella, M.A.; Hanin, M.; Ebel, C. Wheat Type One Protein Phosphatase Participates in the Brassinosteroid Control of Root Growth via Activation of BES1. Int. J. Mol. Sci. 2021, 22, 10424. [Google Scholar] [CrossRef]
- Garda, T.; Kónya, Z.; Tándor, I.; Beyer, D.; Vasas, G.; Erdődi, F.; Vereb, G.; Papp, G.; Riba, M.; M-Hamvas, M.; et al. Microcystin-LR induces mitotic spindle assembly disorders in Vicia faba by protein phosphatase inhibition and not reactive oxygen species induction. J. Plant. Physiol. 2016, 199, 1–11. [Google Scholar] [CrossRef]
- Garda, T.; Kónya, Z.; Freytag, C.; Erdődi, F.; Gonda, S.; Vasas, G.; Szücs, B.; M-Hamvas, M.; Kiss-Szikszai, A.; Vámosi, G.; et al. Allyl-Isothiocyanate and Microcystin-LR Reveal the Protein Phosphatase Mediated Regulation of Metaphase-Anaphase Transition in Vicia faba. Front. Plant Sci. 2018, 9, 1823. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.T.; Brewis, N.D.; Hughes, V.; Mann, D.J. Protein serine/threonine phosphatases; an expanding family. FEBS Lett. 1990, 268, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Kamibayashi, C.; Estes, R.; Lickteig, R.L.; Yang, S.I.; Craft, C.; Mumby, M.C. Comparison of heterotrimeric protein phosphatase 2A containing different B subunits. J. Biol. Chem. 1994, 269, 20139–20148. [Google Scholar] [CrossRef]
- Terol, J.; Bargues, M.; Carrasco, P.; Pérez-Alonso, M.; Paricio, N. Molecular characterization and evolution of the protein phosphatase 2A B’ regulatory subunit family in plants. Plant Physiol. 2002, 129, 808–822. [Google Scholar] [CrossRef]
- Farkas, I.; Dombrádi, V.; Miskei, M.; Szabados, L.; Koncz, C. Arabidopsis PPP familyof serine/threonine phosphatases. Trends Plant Sci. 2007, 12, 169–176. [Google Scholar] [CrossRef]
- Máthé, C.; M-Hamvas, M.; Freytag, C.; Garda, T. The Protein Phosphatase PP2A Plays Multiple Roles in Plant Development by Regulation of Vesicle Traffic-Facts and Questions. Int. J. Mol. Sci. 2021, 22, 975. [Google Scholar] [CrossRef] [PubMed]
- Garbers, C.; DeLong, A.; Deruére, J.; Bernasconi, P.; Söll, D. Amutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 1996, 15, 2115–2124. [Google Scholar] [CrossRef]
- Chang, M.; Wang, B.; Chen, X.; Wu, R. Molecular characterization of catalytic-subunit cDNA sequences encoding protein phosphatases 1 and 2A and study of their roles in the gibberellin-dependent Osamy-c expression in rice. Plant Mol. Biol. 1999, 39, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Shen, G.; Wijewardene, I.; Cai, Y.; Esmaeili, N.; Sun, L.; Zhang, H. The B’ζ subunit of protein phosphatase 2A negatively regulates ethylene signaling in Arabidopsis. Plant Physiol. Biochem. 2021, 169, 81–91. [Google Scholar] [CrossRef] [PubMed]
- González-García, M.P.; Rodríguez, D.; Nicolás, C.; Nicolás, G.; Lorenzo, O. A protein phosphatase 2A from Fagus sylvatica is regulated by GA3 and okadaic acid in seeds and related to the transition from dormancy to germination. Physiol. Plantarum. 2006, 128, 153–162. [Google Scholar] [CrossRef]
- Hu, R.; Zhu, Y.; Shen, G.; Zhang, H. TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiol. 2014, 164, 721–734. [Google Scholar] [CrossRef]
- Kwak, J.M.; Moon, J.H.; Murata, Y.; Kuchitsu, K.; Leonhardt, N.; DeLong, A.; Schroeder, J.I. Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 2002, 14, 2849–2861. [Google Scholar] [CrossRef]
- Nishimura, N.; Hitomi, K.; Arvai, A.S.; Rambo, R.P.; Hitomi, C.; Cutler, S.R.; Schroeder, J.I.; Getzoff, E.D. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 2009, 326, 1373–1379. [Google Scholar] [CrossRef]
- Santiago, J.; Dupeux, F.; Round, A.; Antoni, R.; Park, S.Y.; Jamin, M.; Cutler, S.R.; Rodriguez, P.L.; Márquez, J.A. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 2009, 462, 665–668. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, K.E.; Nishimura, N.; Hitomi, K.; Getzoff, E.D.; Schroeder, J.I. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010, 24, 1695–1708. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef]
- West, G.M.; Pascal, B.D.; Ng, L.M.; Soon, F.F.; Melcher, K.; Xu, H.E.; Chalmers, M.J.; Griffin, P.R. Protein conformation ensembles monitored by HDX reveal a structural rationale for abscisic acid signaling protein affinities and activities. Structure 2013, 21, 229–235. [Google Scholar] [CrossRef]
- Yu, F.; Wu, Y.; Xie, Q. Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 2015, 20, 569–575. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Wasilewska, A.; Vlad, F.; Sirichandra, C.; Redko, Y.; Jammes, F.; Valon, C.; Frei Dit Frey, N.; Leung, J. An update on abscisic acid signaling in plants and more. Mol. Plant. 2008, 1, 198–217. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef]
- Dai, M.; Xue, Q.; Mccray, T.; Margavage, K.; Chen, F.; Lee, J.H.; Nezames, C.D.; Guo, L.; Terzaghi, W.; Wan, J.; et al. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 2013, 25, 517–534. [Google Scholar] [CrossRef]
- Shenolikar, S. Protein serine/threonine phosphatases--new avenues for cell regulation. Annu. Rev. Cell Biol. 1994, 10, 55–86. [Google Scholar] [CrossRef] [PubMed]
- Sidonskaya, E.; Schweighofer, A.; Shubchynskyy, V.; Kammerhofer, N.; Hofmann, J.; Wieczorek, K.; Meskiene, I. Plant resistance against the parasitic nematode Heterodera schachtii is mediated by MPK3 and MPK6 kinases, which are controlled by the MAPK phosphatase AP2C1 in Arabidopsis. J. Exp. Bot. 2016, 67, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Shubchynskyy, V.; Boniecka, J.; Schweighofer, A.; Simulis, J.; Kvederaviciute, K.; Stumpe, M.; Mauch, F.; Balazadeh, S.; Mueller-Roeber, B.; Boutrot, F.; et al. Protein phosphatase AP2C1 negatively regulates basal resistance and defense responses to Pseudomonas syringae. J. Exp. Bot. 2017, 68, 1169–1183. [Google Scholar] [PubMed][Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).