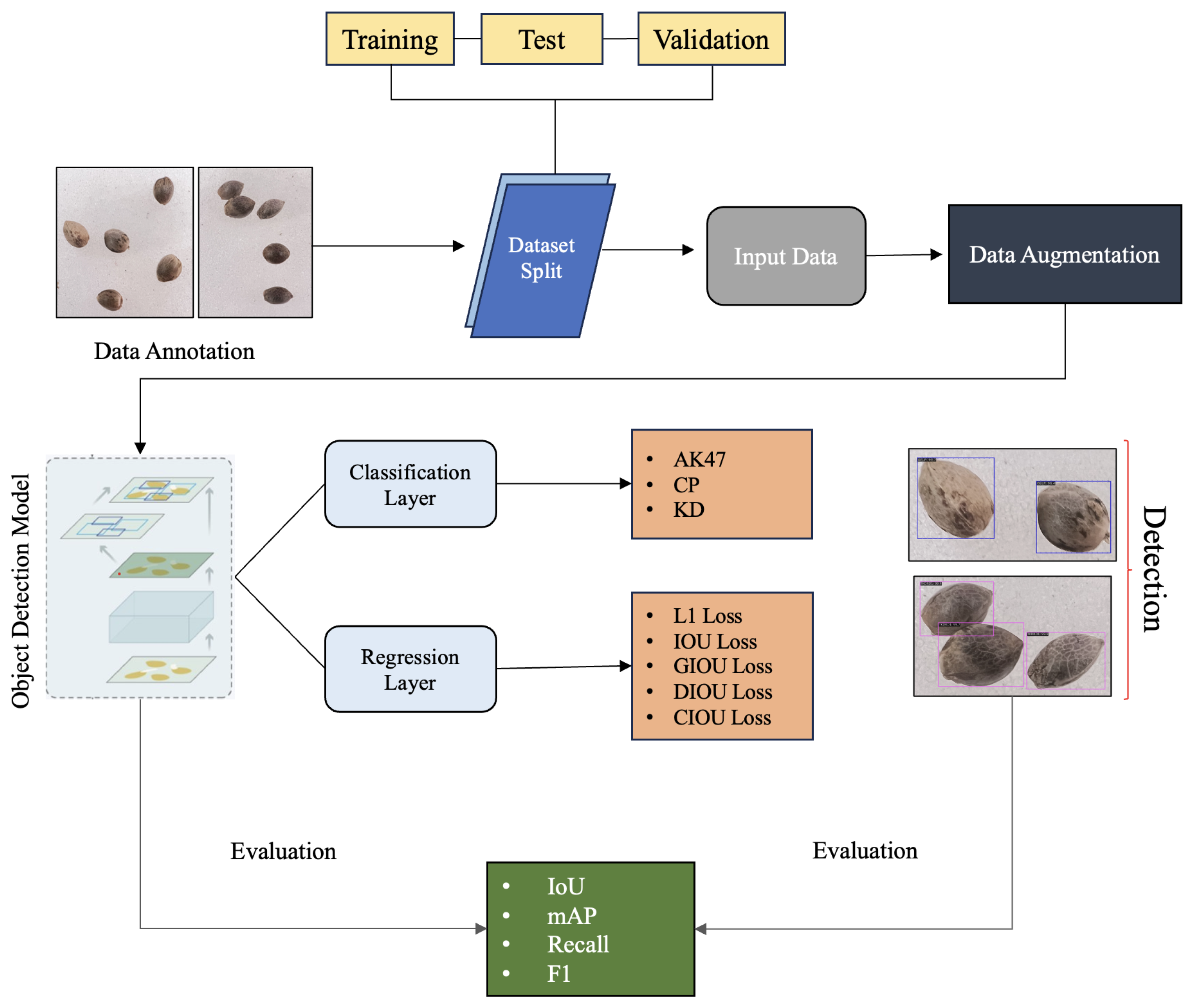
1. Introduction
Cannabis, a widely cultivated and consumed plant, has garnered significant attention in recent years due to its medicinal and recreational properties. With the increasing legalization and commercialization of cannabis products, there is a growing need for efficient and accurate methods to detect and classify cannabis seeds [
1]. Traditional manual sorting and classification processes are labor-intensive, time-consuming, and prone to human error. In response to these challenges, deep learning, a subset of artificial intelligence, has emerged as a promising solution. Integrating advanced technologies, such as deep learning and computer vision, in this sector is paramount. In the context of seed detection and classification, particularly for cannabis seeds, these technologies offer significant benefits. Automating manual processes can enhance productivity and quality control, ultimately improving efficiency in tasks such as seed sorting and quality testing. The detection of cannabis seeds can significantly reduce the labor and time required for seed identification, benefiting farmers and researchers alike. It allows for the precise classification of seeds, helping farmers identify the specific variety of cannabis they are dealing with. Cannabis seed detection holds particular significance compared to other seed detection processes due to the unique characteristics and legal considerations surrounding cannabis cultivation. Unlike many other crops, cannabis varieties vary significantly in their chemical composition, particularly in the concentration of psychoactive compounds such as THC. As a result, the accurate identification of cannabis seeds is crucial for regulatory compliance and ensuring that seeds are cultivated within legal limits.
The growing popularity of cannabis for both medicinal and recreational purposes has led to increased demand for accurate seed detection methods to maintain product quality and consistency. Many studies have tried to define
Cannabis sativa L. based on its appearance and chemical properties. The accepted taxonomy identifies two subspecies—sativa and indica. Each subspecies has two main varieties—cultivated and wild. The most important varieties for medicine are
C. sativa ssp. sativa var. sativa (known as
C. sativa) and
C. sativa ssp. indica var. indica (known as
C. indica). There is also a third, less common variety called
C. sativa ssp. sativa var. spontanea, known as C. ruderalis.
C. indica is usually grown for recreational purposes, while
C. sativa is increasingly recognized for its potential medical applications [
2]. These distinctions are crucial in understanding the diverse applications of cannabis. Industrial hemp and marijuana are distinct subtypes of the
Cannabis sativa species, primarily differentiated by their use, chemistry, and cultivation methods. Industrial hemp is mainly based on the quantity of THC, the psychoactive component present in the plant. While 1% THC is usually enough to produce intoxication, several regions legally differentiate between marijuana and hemp based on the 0.3% THC threshold [
3]. Hemp is grown for industrial purposes, such as fibers, textiles, and seeds, and contains a very low level of the psychoactive compound
-tetrahydrocannabinol (THC) [
4]. In contrast, marijuana is cultivated for its THC-rich flowers and extracts, primarily for recreational or medicinal use, and it is selectively bred for high THC concentrations. The key difference lies in their intoxicating potential, with hemp having negligible THC content [
5].
Artificial intelligence methods have been increasingly applied to various aspects of cannabis agriculture in recent years. Seed classification has been a topic of extensive research, with studies employing various approaches including traditional manual methods [
6,
7,
8,
9], image processing techniques [
10,
11,
12,
13,
14,
15], and modeling techniques [
16,
17,
18,
19,
20,
21,
22,
23]. On the other hand, Sieracka et al. [
24] utilized artificial neural networks to predict industrial hemp seed yield based on cultivation data, showcasing the potential of AI in optimizing hemp production. In a different application, Bicakli et al. [
25] demonstrated the effectiveness of random forest models in distinguishing illegal cannabis crops from other vegetation using satellite imagery, which could aid in monitoring and regulation efforts. Ferentinos et al. [
26] introduced a deep learning system that leverages transfer learning to identify diseases, pests, and deficiencies in cannabis plant images, highlighting the potential of AI in early detection and intervention. More recently, Boonsri et al. [
23] applied deep learning-based object detection models to differentiate between male and female cannabis seeds from augmented seed image datasets, demonstrating the ability of AI to assist in gender-based seed sorting. Despite these advancements, applying deep learning techniques for cannabis seed variety detection and classification remains an area with untapped potential, warranting further research and exploration. However, traditional manual methods are often time-consuming and labor-intensive, relying on visual inspection and biochemical analysis. Image processing techniques have improved accuracy but struggle with handling variations in seed appearance and imaging conditions. Machine learning and deep learning approaches, categorized as modeling techniques, have achieved high accuracy but often lack robustness and primary data, particularly when classifying extremely similar seeds. Furthermore, many studies rely on a limited set of features for classification and fail to address the need for standardized evaluation metrics and benchmarks. Another key challenge that researchers faced was the variability in seed quality and genetics, which can impact the reproducibility and reliability of research findings. Ensuring seed purity and genetic stability is crucial but can be difficult due to the lack of standardized seed certification and analysis methods. However, to overcome these challenges, this research builds upon and significantly extends our previous work on cannabis seed variant detection using Faster R-CNN. While our previous study [
27] focused solely on the Faster R-CNN architecture with a ResNet50 backbone and explored various loss functions, the current research broadens the scope considerably. We now aim to include a comprehensive comparison between Faster R-CNN and RetinaNet, incorporate additional backbone architectures (ResNet101 and ResNeXt101), and integrate the insights gained from our previous loss function analysis. This expanded investigation aims to provide a more thorough understanding of deep learning approaches for cannabis seed detection and classification. The dataset [
28] used in this study is a collection of cannabis seed variants from 17 different categories, which was used in our previous study [
27] as well. The main objective of this paper is to classify seeds of these 17 kinds of cannabis. Our study aims to fill previous research gaps by utilizing deep learning algorithms to precisely identify and outline the bounding box regions of various cannabis seed varieties. Key contributions of this research are given below:
Extension of our previous work on cannabis seed detection, incorporating additional object detection architectures (RetinaNet alongside Faster R-CNN) and expanding the analysis scope.
Unlike previous studies, which focused on limited seed varieties [
23], this research classifies seeds from 17 different cannabis varieties, providing in-depth metrics on detection accuracy and processing speed.
Integration of optimal loss functions identified in our earlier study, applied to an expanded set of model configurations.
This study employs state-of-the-art deep learning models, including ResNet 50, ResNet 101, ResNext 101, and RetinaNet, to enhance the accuracy and efficiency of cannabis seed detection and classification.
Validation and extension of our previous findings, offering refined insights into effective deep learning approaches for cannabis seed classification and detection.
The paper is structured as follows:
Section 2 delves into the related work, providing an overview of prior research in the field.
Section 3 outlines the dataset used, along with the data pre-processing steps and the training methodology employed.
Section 4 elaborates on the object detection models, including their architectures and the backbone networks utilized in our experiments. The experimental results are presented in
Section 5, where we discuss our findings and compare the performance of the various object detectors. In
Section 6, we presented the discussion, and, finally,
Section 7 presents the conclusions of this research.
2. Related Work
Several studies have already been done on seed classification. We can divide them into three categories: the traditional (manual) method, image processing, and modeling. The traditional method [
6,
7,
8,
9] of seed classification involves visual inspection, biochemical seed identification, machine vision, and DNA analysis for accurate categorization. Additionally, image processing methods are, again, divided into three categories: structural method [
10,
11,
12], threshold method [
13,
29,
30], and spectral method [
14,
15,
31]. The structural method analyzes patterns, shapes, and relationships between different image elements to extract meaningful information and enhance understanding. In contrast, the threshold method concerns setting a specific intensity level, or threshold, to segment an image into different regions based on pixel values. On the other hand, the spectral method involves the analysis of different wavelength bands within the electromagnetic spectrum. The modeling technique usually learns patterns and features from a labeled dataset. Machine learning and deep learning are the most common modeling techniques for classification. These models, often convolutional neural networks, extract hierarchical representations of an image. During training, the model adjusts its parameters to minimize the difference between predicted and actual labels. Once trained, the model can generalize its learned patterns to detect objects or features in new, unseen images. In seed detection and classification, several works have been conducted on machine learning [
16,
17,
18,
19,
32,
33], and deep learning [
20,
21,
22,
34,
35,
36,
37]. In
Figure 1, we summarize works conducted on seed detection and classification.
While previous research on cannabis agriculture has employed deep learning methods that have demonstrated potential in the gender screening of cannabis seeds [
23], their effectiveness in discriminating between seed varieties has not been investigated. Additionally, this study has not categorized their research into multiple classes of seeds. In image processing, Ahmed et al. [
10] worked on applying X-ray CT scanning to pepper seed analysis, employing recycling, feature extraction, and classification to robustly categorize seeds into viable and nonviable groups. Pereira et al. [
11] addressed soybean seed quality challenges, introducing an image analysis framework that significantly enhanced vigor classification, achieving 81% accuracy. Meanwhile, Zhang et al. [
12] innovatively utilized deep learning and edge detection for internal crack detection in corn seeds, presenting the optimized S2ANet model with 95.6% average precision. However,
Table 1 shows a compact summary of the contributions of seed classification and detection by modeling techniques.
Table 1 provides an overview of various research contributions in the field of seed classification using machine learning and deep learning algorithms. Luo et al. [
34] introduced a nondestructive intelligent image recognition system employing deep CNN models like AlexNet and GoogLeNet, achieving a notable accuracy of 93.11% in detecting 140 weed seed species. Their study proposes a solution through nondestructive intelligent image recognition. An image acquisition system captures and segments images of single weed seeds, forming a dataset of 47,696 samples from 140 species. Their research emphasizes the importance of selecting a CNN model based on specific identification accuracy and time constraints. A notable limitation of this study is the robustness of the proposed detection method. Khan et al. [
16] worked on classifying dry beans using machine learning algorithms such as LR, NB, KNN, DT, RF, XGB, SVM, and MLP, achieving a high accuracy of 95.4%, with the limitation being the considerable time requirement for ignoring the bean suture axis. Dubey et al. [
36] applied an artificial neural network to classify wheat seed varieties with an accuracy of 88%, highlighting the space for improvement in accuracy. Cheng et al. [
21] used machine learning for rice seed defect detection, employing Principal Component Analysis and a back-propagation neural network with an accuracy range of 91–99%, suggesting potential improvements in the algorithm. Lawal [
39] proposed a deep learning model for fruit seed detection using YOLO, achieving 91.6% accuracy, with the need for accuracy enhancement specifically in the YOLO Muskmelon model. Heo et al. [
37] developed a high-throughput seed sorting system using YOLO, reaching an impressive accuracy of 99.81%, showcasing its potential for acquiring clean seed samples. Bi et al. [
38] focused on maize seed identification, employing a range of algorithms such as AlexNet, Vgg16, ResNet50, Visio-Transformer, and Swin-Transformer, achieving the best accuracy of 96.53%. However, their work identified a need for improvement in classifying extremely similar seeds. Madhavan et al. [
20] conducted post-harvest classification of wheat seeds using artificial neural networks and achieved an accuracy of 96.7%, but their study lacked primary data. Javanmardi et al. [
22] concentrated on corn seed classification, employing CNN and ANN, achieving a good accuracy of 98.1%. Ali et al. [
17] explored corn seed classification using various machine learning algorithms, including MLP, LB, RF, and BN, achieving a high accuracy of 98.93%, but faced limitations due to the scarcity of features for classification. Jamuna et al. [
19] focused on cotton seed quality classification at different growth stages, utilizing algorithms like NB, MLP, and J48, achieving an accuracy of 98.78%, with the limitation of lacking primary data in their study.
While seed analysis has been explored using various techniques, research specifically focused on cannabis seed detection and classification remains limited. Boonsri et al. [
23] made initial strides in this field by classifying a limited number of cannabis seed varieties using convolutional neural networks. However, their work, along with other studies in seed object detection, revealed several persistent gaps. These include limited publicly available datasets, inadequate methods for handling variations in seed appearance and imaging conditions, and a lack of standardized evaluation metrics and benchmarks. Additionally, existing approaches often rely on traditional computer vision methods that may struggle with complex seed shapes and textures. To address these research gaps, we initiated a comprehensive study on cannabis seed detection and classification. Our previous work [
27] laid the groundwork in this domain by exploring the use of Faster R-CNN for detecting and classifying 17 varieties of cannabis seeds. Using a locally sourced dataset from Thailand, we investigated various loss functions (L1, IoU, GIoU, DIoU, and CIoU) with a ResNet50 backbone, achieving a mAP score of 94.08% and an F1 score of 95.66%. This study emphasized the importance of accurate seed variant identification for precision breeding, regulatory compliance, and meeting diverse market demands. Building upon our previous research, the current study aims to further advance the field of cannabis seed detection and classification. We expand our investigation to include multiple object detection architectures and backbone networks, providing a more comprehensive analysis of state-of-the-art deep learning techniques in this domain.
6. Discussion
This work showcases the contrasting performance characteristics of the two popular object detection models, RetinaNet and Faster R-CNN, in the context of detecting and classifying 17 different cannabis seed varieties. While both models demonstrated impressive capabilities, there were some differences in their performance across various evaluation metrics.
In terms of the mean average precision (mAP) metric evaluated over the IoU range of 0.5 to 0.95, the RetinaNet model with the ResNet101 backbone (mR101) emerged as the top performer with a mAP of 0.9458, slightly outperforming its Faster R-CNN counterpart, mFR50, which achieved a mAP of 0.9408. This suggests that the RetinaNet architecture, with its dedicated object classification and bounding box regression subnetworks, may have an edge in precisely localizing objects when high overlap with the ground truth is required. However, when the IoU threshold was relaxed to 0.5, the Faster R-CNN model with the ResNet50 backbone (mFR50) surpassed all other models, attaining a mAP of 0.9428. This performance advantage indicates that the Faster R-CNN architecture, with its region proposal network and region-of-interest pooling, might be better suited for detecting objects with less stringent overlap requirements. Across both models, the smaller ResNet50 backbone consistently demonstrated superior recall capabilities, outperforming the larger ResNet101 and ResNeXt101 backbones. This trend was observed in the average recall scores, where mR50 and mFR50 achieved 0.982 and 0.973, respectively, compared to their larger counterparts. The lightweight ResNet50 architecture’s ability to maintain high recall rates is particularly advantageous in applications where missing true positive detections is undesirable, such as in seed classification tasks. F1 scores, which balance precision and recall, followed a similar pattern, with the ResNet50-based models (mR50 and mFR50) outperforming their larger counterparts. This consistency across multiple metrics highlights the effectiveness of the ResNet50 backbone in achieving a well-rounded performance for the given task.
In terms of real-time inference speed in
Figure 9, the larger backbones generally exhibited slower performance compared to the ResNet50 models. The mFRX101 model achieved the fastest inference speed of 57.1 ms per image (17.5 FPS), closely followed by mFR50 at 59.5 ms per image (16.8 FPS). However, the lightweight mR50 model demonstrated the best balance between performance and speed, with an inference time of 62.1 ms per image (16.1 FPS) while maintaining competitive accuracy and recall scores. When examining the per-class detection performance, both models exhibited varying strengths and weaknesses across different classes. The mR50 model demonstrated superior performance in detecting challenging classes like ‘CP’ at both strict and lenient IoU thresholds. Conversely, the mFR101 model excelled in classes like ‘HKRKU’, ‘HSSNTT1’, and ‘KKV’ at the stricter IoU range. These observations highlight the importance of carefully evaluating model performance on a per-class basis, as different architectures and backbones may excel at detecting specific seed varieties or characteristics. Interestingly, the larger ResNeXt101 backbone did not consistently outperform the ResNet architectures in either the RetinaNet or Faster R-CNN models. While the mRX101 model achieved competitive results in some classes, its overall performance was slightly lower than the ResNet models across most metrics. This observation suggests that the increased complexity of the ResNeXt101 architecture may not necessarily translate into improved performance for this specific task, and careful model selection and tuning are crucial.
Table 8 provides a comprehensive overview of the performance metrics for both RetinaNet and Faster R-CNN models across different backbone architectures, including our previous work’s results. This comparison clearly demonstrates the advancements made in our current study and underscores the value of our expanded methodology. Our previous work, which utilized Faster R-CNN with a ResNet50 backbone, achieved respectable results with a mAP@0.5:0.95 of 0.9408, an F1 score of 0.9566, and an inference speed of 16.8 FPS. However, our current study has yielded significant improvements across multiple metrics. The RetinaNet model with a ResNet101 backbone achieved the highest mAP@0.5:0.95 of 0.9458, representing a 0.5 percentage point improvement over our previous best. This enhancement in accuracy is crucial for precise cannabis seed detection and classification. Furthermore, our best model in this study (RetinaNet with ResNet101) achieved an impressive average recall of 0.985, compared to 0.973 in our previous work. This 1.2 percentage point improvement indicates a substantial reduction in false negatives, ensuring more comprehensive seed detection.
The F1 score, which balances precision and recall, saw a notable improvement from 0.9566 to 0.9650 with our best-performing model. This 0.84 percentage point increase demonstrates a well-rounded enhancement in overall detection performance. By expanding our study to include RetinaNet, we have discovered that this one-stage detector consistently outperforms Faster R-CNN in accuracy metrics for our specific task. This finding provides valuable insights for future research and applications in cannabis seed detection. Our evaluation of different backbones (ResNet50, ResNet101, and ResNeXt101) across both architectures has revealed that, while ResNet101 generally offers the best accuracy, ResNeXt101 can provide speed advantages, particularly with Faster R-CNN. While our previous Faster R-CNN model maintained a competitive speed of 16.8 FPS, our current study offers a range of options balancing speed and accuracy. For instance, Faster R-CNN with ResNeXt101 achieves the highest speed of 17.5 FPS, while the RetinaNet models offer superior accuracy with a slight trade-off in speed.
This research uniquely extends our previous findings by providing a comprehensive comparison between one-stage (RetinaNet) and two-stage (Faster R-CNN) detectors for cannabis seed detection, which was not explored in our earlier work. It offers insights into the performance of different backbone architectures, allowing for more informed model selection based on specific application requirements. We have demonstrated tangible improvements in key metrics (mAP, recall, and F1 score) over our previous best results, validating the effectiveness of our expanded methodology. Additionally, we have explored the speed–accuracy trade-offs in greater depth, which is crucial for practical applications in cannabis seed detection and classification. Therefore, this study not only builds upon our previous work but significantly expands the scope of analysis in cannabis seed detection. By achieving improved accuracy, recall, and F1 scores, while also providing a range of models with different speed–accuracy balances, we have advanced the field of automated cannabis seed classification. These findings offer valuable insights for both researchers and practitioners in agriculture technology, particularly in the rapidly evolving cannabis industry.
7. Conclusions
This research significantly extends our previous work on cannabis seed detection and classification, demonstrating the effective application of advanced deep learning models, specifically Faster R-CNN and RetinaNet, across various backbone architectures. Our expanded methodology has yielded notable improvements in detection accuracy and efficiency, addressing critical needs in the rapidly evolving cannabis industry. Our findings reveal that the RetinaNet model with the ResNet101 backbone achieved the highest mean average precision (mAP) of 0.9458 at the IoU range of 0.5 to 0.95, surpassing both our previous results (mAP of 0.9408) and the current Faster R-CNN implementations. RetinaNet models consistently demonstrated superior performance across key metrics, including recall and F1 score, indicating their effectiveness in minimizing missed detections and balancing precision with recall. This study provides valuable insights into the trade-offs between model architectures and backbones. While RetinaNet models excelled in accuracy, Faster R-CNN, particularly with the ResNeXt101 backbone, offered advantages in inference speed, achieving up to 17.5 FPS. This comprehensive evaluation enables more informed model selection based on specific application requirements. A key limitation remains the variability in seed quality and genetics, which can impact reproducibility. Future work could explore ensemble techniques and transformer models for further performance enhancements. Our improved method has vast potential applications, particularly in cannabis agriculture, and it could extend to other agricultural sectors. Our enhanced automated seed analysis can significantly improve productivity, consistency, and regulatory adherence in cannabis seed classification. This study not only addressed a critical research gap but also significantly advanced our previous findings in automating seed analysis. By leveraging and comparing advanced deep learning models, we contribute to improving efficiency and reliability in cannabis seed classification, providing a robust foundation for future research and practical applications in agriculture.