Recombinant Protein Expression and Its Biotechnological Applications in Chlorella spp.
Abstract
1. Introduction
2. Genetic Elements for Protein Expression in Chlorella spp.
2.1. Transformation Methods
2.2. Selectable Markers
2.3. Promoters and Terminators
2.4. Enhancers, Introns, and Signal Peptides
3. Applications for Recombinant Protein Expression
3.1. Model Proteins
| Protein (Gene) | Selection | Transfection Method | Expression Elements | Notes | Ref. |
|---|---|---|---|---|---|
| GUS | Hygromycin B | EP | PCaMV35S-gusA-TNOS | C. vulgaris | [76] |
| GUS | Kanamycin | MBP | PCaMV35S-gusA-TNOS | C. vulgaris | [77] |
| GUS | Geneticin | AMT | PCaMV35S-gus-TCaMV35S | C. vulgaris Contains IV2 intron from ST-LS1 | [48] |
| GUS | n.a. | EP | PNR-gus:NR-TNOS | C. ellipsoidea NR-GUS fusion protein Transient expression | [78] |
| GUS | Hygromycin B | AMT | PCaMV35S-gus-TNOS | C. sorokiniana | [79] |
| GUS | Phleomycin | PP + PEG | PUbi-gus-TNOS | C. ellipsoidea Stable for 10 months | [60] |
| GUS | n.a. | MPB | PCaMV35S-gus-TNOS | C. ellipsoidea Transient expression | [34] |
| GUS | n.a. | PP + EP | PCaMV35S-gus-TNOS | C. saccharophila Transient expression | [80] |
| GUS | Geneticin | MPB | PCaMV35S-gus-TNOS | C. kessleri | [81] |
| GUS | n.a. | EP | PCaMV35S-gus-TNOS | Chlorella sp. MACC/C95 Transient expression | [82] |
| GFP-GUS fusion | Hygromycin B | AMT | PCaMV35S-gfp:gusA-TNOS | C. vulgaris | [25] |
| Enhanced GFP (egfp) | Geneticin | PP + PEG | PCaMV35S-egfp-TNOS PNOS-gusA-TNOS | C. vulgaris | [31] |
| Cyan fluorescent protein (cfp), GFP (mgfp5) | Hygromycin B or Geneticin | PP + EP | PCaMV35S-mgfp5-TNOS PHSP70-cfp-TNOS | C. vulgaris | [49] |
| EGFP | Hygromycin B | HIV-TAT peptide + Triton X-100 | PCaMV35S-egfp-TNOS | C. vulgaris Used cell-penetrating peptide for transformation | [43] |
| GFP | Hygromycin B | EP | PCaMV35S-mgfp5-TNOS PCaMV35S-zCas9-NLS-TNOS | C. vulgaris sgRNA expressed using U6 promoter directed toward FAD Enhance lipid production | [41] |
| EGFP | Hygromycin B | AMT | PCaMV35S-mgfp5-TNOS | C. vulgaris Video method | [55] |
| EGFP | Geneticin | EP | PUbi1-egfp-TNOS | C. pyrenoidosa | [74] |
| mCherry EGFP AMP MSI99 | Phleomycin | EP | P3843-ble:mCherry-T8657 P8657-egfp-T8655 P3843-ble-2A-MSI99-2A-mCherry | Chlorella sp. MEM25 Used transcriptomics to design native promoter and terminator | [65] |
| Luciferase | n.a. | PP + PEG | PCaMV35S-luc-TNOS | C. ellipsoidea Transient expression | [75] |
| Luciferase | KClO3 | EP | PSIP-luc-TRbcS2 | C. vulgaris Identified new salt inducible promoter (SIP) | [46] |
3.2. Recombinant Protein Production
| Protein (Gene) | Expression Elements | Notes | Ref. |
|---|---|---|---|
| Human growth hormone (hGh) | PCaMV35S-hGH PRbcS2-hGH PCaMV35S+RbcS2-hGH | C. vulgaris Yield: 200–600 µg/L Extracellular, transient expression Also tested a Chlorella virus promoter and RbcS2 intron 1. | [67] |
| GUS, Neutrophil peptide-1 (NP1) | PCaMV35S-gus-TNOS PUbi-gus-TNOS PCaMV35S+Ubi-gus-TNOS PUbi-NP1-TNOS | C. ellipsoidea Enhancer (TMV Ω 5′UTR) doubled GUS activity | [68] |
| Flounder growth hormone (fGH) | PCaMV35S-fGH | C. ellipsoidea Yield: 400 µg/L Oral growth supplement for flounder | [32] |
| Trypsin-modulating oostatic factor (tmfA) | PCaMV35S-tmfA-TRbcS | C. dessicata Yield: 17–20 µg/3 × 108 cells Stable > 3 months | [84] |
| Infectious bursal disease virus protein 2 (IBDV vp2) | PCaMV35S-vp2-TOCS | C. pyrenoidosa Edible chicken vaccine | [26] |
| Human granulocyte colony-stimulating factor (hG-CSF) | PCvNDI1-hG-CSF-TRAmy3D | C. vulgaris Transit peptides from highly secreted proteins Nitrogen deficiency inducible promoter (CvNDI1) | [56] |
| SARS-CoV-2 receptor-binding domain (RBD), Basic fibroblast growth factor (bFGF) | PCaMV35S-RBD-TExt3 PCaMV35S-bFGF-TExt3 | C. vulgaris Yield: 1.14 µg/g RBD Yield: 1.61 ng/g bFGF Transient expression using geminiviral system Dual CaMV35S promoter Tobacco extension 3′UTR NbPsalK2T1-63 5′UTR, | [24] |
| bFGF | Prrn-bFGF-RBS-aph6-TpsbA | C. vulgaris Chloroplast integration using HR with 16S-trn1 and trnA-23S region C. reinhartii ribosomal RNA (rrn) promoter | [29] |
| Viral hemorrhagic septicemia virus glycoprotein (VHSV G) | PCaMV35S-VHSVG-TRbcS2 | C. vulgaris KClO3—Selection for NR KO Stable > 1 year Used triple HR | [47] |
| VHSV G | PSIP-VHSVG-TRbcS2 | C. vulgaris KClO3—Selection for NR KO Yield: 41.1 mg/10 g wet biomass Used to vaccinate fish | [45] |
| White spot syndrome virus (WSSV) VP28 | PCaMV35S-RBD-TRbcS | C. vulgaris KClO3—Selection for NR KO Used to vaccinate shrimp Used triple HR | [28] |
3.3. Metabolic Engineering
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AMPs | Antimicrobial peptides |
| AMT | Agrobacterium mediate transformation |
| APT | Adenine phosphoribosyltransferase |
| bFGF | Basic fibroblast growth factor |
| bZIP | Basic leucine zipper |
| CaMV35S | Cauliflower mosaic virus 35S |
| Cas9 | CRIPSR-associated protein 9 |
| CHO | Chinese hamster ovary |
| CP | Chloroplast |
| CRISPR | Clustered regularly interspaced short palindromic repeatsCTP—Chloroplast transit peptide |
| DGA | Diacylglycerol acyltransferase |
| EGFP | Enhanced green fluorescent protein |
| ELISA | enzyme-linked immunosorbent assay |
| EP | Electroporation |
| ER | Endoplasmic reticulum |
| Ext3 | Tobacco extension 3′UTR |
| FAD | ω-3 fatty acid desaturase |
| FBA | Fructose 1,6-bisphosphate adolase |
| fGH | Flounder growth hormone |
| GFP | Green fluorescent proteinGUS—β-glucuronidase |
| HEK | Human embryonic kidney |
| Hg-CSF | Human granulocyte colony-stimulating factor |
| hGH | Human growth factor |
| HIV | Human immunodeficiency virus |
| HSP70 | Heat shock protein 70 |
| HR | Homologous recombination |
| IBDV | Infectious bursal disease virus |
| KO | Knockout |
| MBP | Microparticle bombardment |
| mGFP5 | Modified green fluorescent protein 5 |
| MT | Mitochondria |
| NLS | Nuclear localization signal |
| NOS | Nopaline synthase |
| NP1 | Neutrophil peptide-1 |
| NR | Nitrate reductase |
| OCS | Octopine synthase |
| RbcS/L | Ribulose-1,5-bisphosphate carboxylase/oxygenase small/large subunit |
| RBD | Receptor binding domain |
| PDS | Phytoene desaturase |
| PP | Protoplasting |
| PEG | Polyethylene glycol transformation |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SDS-PAGE | Sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| Sf9/21 | Spodoptera frugiperda Sf9/21 cell |
| SIP | Salt inducible promoter |
| SV40 | Simian vacuolating virus 40 |
| Tag | T antigen |
| TAT | Transactivator of transcription |
| TF | Transcription factor |
| TMV | Tobacco mosaic virus |
| TMOF | Trypsin-modulating oostatic factor |
| Ubi | Maize ubiquitin |
| UTR | Untranslated region |
| VHSV | Viral hemorrhagic septicemia virus |
| WSSV | White spot syndrome virus |
| ZCD1 | Carotenoid cleavage dioxygenase |
References
- Rudge, S.R.; Ladisch, M.R. Industrial Challenges of Recombinant Proteins. In Current Applications of Pharmaceutical Biotechnology; Springer: Cham, Switzerland, 2019; pp. 1–22. [Google Scholar]
- Brondyk, W.H. Selecting an Appropriate Method for Expressing a Recombinant Protein. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2009; pp. 131–147. [Google Scholar]
- Torres-Tiji, Y.; Fields, F.J.; Yang, Y.; Heredia, V.; Horn, S.J.; Keremane, S.R.; Jin, M.M.; Mayfield, S.P. Optimized Production of a Bioactive Human Recombinant Protein from the Microalgae Chlamydomonas Reinhardtii Grown at High Density in a Fed-Batch Bioreactor. Algal Res. 2022, 66, 102786. [Google Scholar] [CrossRef]
- Slattery, S.S.; Giguere, D.J.; Stuckless, E.E.; Shrestha, A.; Briere, L.A.K.; Galbraith, A.; Reaume, S.; Boyko, X.; Say, H.H.; Browne, T.S.; et al. Phosphate-Regulated Expression of the SARS-CoV-2 Receptor-Binding Domain in the Diatom Phaeodactylum Tricornutum for Pandemic Diagnostics. Sci. Rep. 2022, 12, 7010. [Google Scholar] [CrossRef] [PubMed]
- Galarza, J.I.; Gimpel, J.A.; Rojas, V.; Arredondo-Vega, B.O.; Henríquez, V. Over-Accumulation of Astaxanthin in Haematococcus Pluvialis through Chloroplast Genetic Engineering. Algal Res. 2018, 31, 291–297. [Google Scholar] [CrossRef]
- Ibrahim, I.; Elbaily, Z. A Review: Importance of Chlorella and Different Applications. Alex. J. Vet. Sci. 2020, 65, 16. [Google Scholar] [CrossRef]
- Lizzul, A.; Lekuona-Amundarain, A.; Purton, S.; Campos, L. Characterization of Chlorella sorokiniana, UTEX 1230. Biology 2018, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Sarkany, N.; Cui, Y. Biomass and Lipid Productivities of Chlorella vulgaris under Autotrophic, Heterotrophic and Mixotrophic Growth Conditions. Biotechnol. Lett. 2009, 31, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Peter, A.P.; Tan, X.; Lim, J.Y.; Chew, K.W.; Koyande, A.K.; Show, P.L. Environmental Analysis of Chlorella vulgaris Cultivation in Large Scale Closed System under Waste Nutrient Source. Chem. Eng. J. 2022, 433, 134254. [Google Scholar] [CrossRef]
- Parashar, A.; Shah, N.; Rane, M.; Shastri, Y. Biogas-Assisted Growth of Chlorella vulgaris in an Open Raceway Pond: Proof of Concept and Economic Assessment. Chem. Eng. Technol. 2023, 46, 1455–1463. [Google Scholar] [CrossRef]
- Hamilton, M.; Powers, S.; Napier, J.; Sayanova, O. Heterotrophic Production of Omega-3 Long-Chain Polyunsaturated Fatty Acids by Trophically Converted Marine Diatom Phaeodactylum tricornutum. Mar. Drugs 2016, 14, 53. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, W.; Jiang, S.; Wang, Y.; Chen, L.; Yang, G.; Liu, T. Heterotrophic Modification of Phaeodactylum Tricornutum Bohlin. Algal Res. 2023, 72, 103137. [Google Scholar] [CrossRef]
- Banerjee, A.; Ward, V. Production of Recombinant and Therapeutic Proteins in Microalgae. Curr. Opin. Biotechnol. 2022, 78, 102784. [Google Scholar] [CrossRef]
- Dehghani, J.; Movafeghi, A.; Mathieu-Rivet, E.; Mati-Baouche, N.; Calbo, S.; Lerouge, P.; Bardor, M. Microalgae as an Efficient Vehicle for the Production and Targeted Delivery of Therapeutic Glycoproteins against SARS-CoV-2 Variants. Mar. Drugs 2022, 20, 657. [Google Scholar] [CrossRef]
- Hayashi, K.; Morooka, N.; Yamamoto, Y.; Fujita, K.; Isono, K.; Choi, S.; Ohtsubo, E.; Baba, T.; Wanner, B.L.; Mori, H.; et al. Highly Accurate Genome Sequences of Escherichia coli K-12 Strains MG1655 and W3110. Mol. Syst. Biol. 2006, 2, 2006-0007. [Google Scholar] [CrossRef]
- Engel, S.R.; Wong, E.D.; Nash, R.S.; Aleksander, S.; Alexander, M.; Douglass, E.; Karra, K.; Miyasato, S.R.; Simison, M.; Skrzypek, M.S.; et al. New Data and Collaborations at the Saccharomyces Genome Database: Updated Reference Genome, Alleles, and the Alliance of Genome Resources. Genetics 2022, 220, iyab224. [Google Scholar] [CrossRef]
- Xu, X.; Nagarajan, H.; Lewis, N.E.; Pan, S.; Cai, Z.; Liu, X.; Chen, W.; Xie, M.; Wang, W.; Hammond, S.; et al. The Genomic Sequence of the Chinese Hamster Ovary (CHO)-K1 Cell Line. Nat. Biotechnol. 2011, 29, 735–741. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The Tobacco Genome Sequence and Its Comparison with Those of Tomato and Potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Joo, S.; Nishimura, Y.; Cronmiller, E.; Hong, R.H.; Kariyawasam, T.; Wang, M.H.; Shao, N.C.; El Akkad, S.-E.-D.; Suzuki, T.; Higashiyama, T.; et al. Gene Regulatory Networks for the Haploid-to-Diploid Transition of Chlamydomonas reinhardtii. Plant Physiol. 2017, 175, 314–332. [Google Scholar] [CrossRef]
- Craig, R.J.; Gallaher, S.D.; Shu, S.; Salomé, P.A.; Jenkins, J.W.; Blaby-Haas, C.E.; Purvine, S.O.; O’Donnell, S.; Barry, K.; Grimwood, J.; et al. The Chlamydomonas Genome Project, Version 6: Reference Assemblies for Mating-Type plus and Minus Strains Reveal Extensive Structural Mutation in the Laboratory. Plant Cell 2023, 35, 644–672. [Google Scholar] [CrossRef]
- Giguere, D.J.; Bahcheli, A.T.; Slattery, S.S.; Patel, R.R.; Browne, T.S.; Flatley, M.; Karas, B.J.; Edgell, D.R.; Gloor, G.B. Telomere-to-Telomere Genome Assembly of Phaeodactylum tricornutum. PeerJ 2022, 10, e13607. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum Genome Reveals the Evolutionary History of Diatom Genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Cecchin, M.; Marcolungo, L.; Rossato, M.; Girolomoni, L.; Cosentino, E.; Cuine, S.; Li-Beisson, Y.; Delledonne, M.; Ballottari, M. Chlorella vulgaris Genome Assembly and Annotation Reveals the Molecular Basis for Metabolic Acclimation to High Light Conditions. Plant J. 2019, 100, 1289–1305. [Google Scholar] [CrossRef]
- Malla, A.; Rosales-Mendoza, S.; Phoolcharoen, W.; Vimolmangkang, S. Efficient Transient Expression of Recombinant Proteins Using DNA Viral Vectors in Freshwater Microalgal Species. Front. Plant Sci. 2021, 12, 650820. [Google Scholar] [CrossRef]
- Cha, T.S.; Yee, W.; Aziz, A. Assessment of Factors Affecting Agrobacterium-Mediated Genetic Transformation of the Unicellular Green Alga, Chlorella vulgaris. World J. Microbiol. Biotechnol. 2012, 28, 1771–1779. [Google Scholar] [CrossRef]
- Reddy, P.H.; Johnson, A.M.A.; Kumar, J.K.; Naveen, T.; Devi, M.C. Heterologous Expression of Infectious Bursal Disease Virus VP2 Gene in Chlorella Pyrenoidosa as a Model System for Molecular Farming. Plant Cell Tissue Organ. Cult. 2017, 131, 119–126. [Google Scholar] [CrossRef]
- Zhang, J.; Hao, Q.; Bai, L.; Xu, J.; Yin, W.; Song, L.; Xu, L.; Guo, X.; Fan, C.; Chen, Y.; et al. Overexpression of the Soybean Transcription Factor GmDof4 Significantly Enhances the Lipid Content of Chlorella ellipsoidea. Biotechnol. Biofuels 2014, 7, 128. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, S.H.; Kim, J.O.; Lee, T.K.; Jang, I.K.; Choi, T.J. Efficacy of White Spot Syndrome Virus Protein VP28-Expressing Chlorella vulgaris as an Oral Vaccine for Shrimp. Viruses 2023, 15, 2010. [Google Scholar] [CrossRef]
- Bolaños-Martínez, O.C.; Malla, A.; Rosales-Mendoza, S.; Vimolmangkang, S. Construction and Validation of a Chloroplast Expression Vector for the Production of Recombinant Proteins in Chlorella vulgaris. Front. Mar. Sci. 2022, 9, 884897. [Google Scholar] [CrossRef]
- Yang, B.; Liu, J.; Ma, X.; Guo, B.; Liu, B.; Wu, T.; Jiang, Y.; Chen, F. Genetic Engineering of the Calvin Cycle toward Enhanced Photosynthetic CO2 Fixation in Microalgae. Biotechnol. Biofuels 2017, 10, 229. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, J.; Liu, B.; Sun, P.; Ma, X.; Jiang, Y.; Wei, D.; Chen, F. Development of a Stable Genetic System for Chlorella vulgaris-A Promising Green Alga for CO2 Biomitigation. Algal Res. 2015, 12, 134–141. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, Y.T.; Cho, J.J.; Bae, J.H.; Hur, S.B.; Hwang, I.; Choi, T.J. Stable Integration and Functional Expression of Flounder Growth Hormone Gene in Transformed Microalga, Chlorella ellipsoidea. Mar. Biotechnol. 2002, 4, 63–73. [Google Scholar] [CrossRef]
- Wang, K.; Gao, Z.; Wang, Y.; Meng, C.; Li, J.; Qin, S.; Cui, Y. The Chloroplast Genetic Engineering of a Unicellular Green Alga Chlorella vulgaris with Two Foreign Peptides Co-Expression. Algal Res. 2021, 54, 102214. [Google Scholar] [CrossRef]
- Ying, C.I.; Wen-bin, L.; Qin-hua, B.; Yong-ru, S. Study On Transient Expression Of Gus Gene In Chlorella ellipsoidea (Chlorophyta) by Using Biolistic Particle Delivery System. Chin. J. Oceanol. Limnol. 1998, 16, 47–49. [Google Scholar] [CrossRef]
- Nawkarkar, P.; Kapase, V.U.; Chaudhary, S.; Kajla, S.; Kumar, S. Heterogeneous Diacylglycerol Acyltransferase Expression Enhances Lipids and PUFA in Chlorella Species. GCB Bioenergy 2023, 15, 1240–1254. [Google Scholar] [CrossRef]
- Liu, J.; Sun, Z.; Gerken, H.; Huang, J.; Jiang, Y.; Chen, F. Genetic Engineering of the Green Alga Chlorella Zofingiensis: A Modified Norflurazon-Resistant Phytoene Desaturase Gene as a Dominant Selectable Marker. Appl. Microbiol. Biotechnol. 2014, 98, 5069–5079. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Lauersen, K.J. Gene Delivery Technologies with Applications in Microalgal Genetic Engineering. Biology 2021, 10, 265. [Google Scholar] [CrossRef]
- Gelvin, S.B. Agrobacterium-Mediated Plant Transformation: The Biology behind the “Gene-Jockeying” Tool. Microbiol. Mol. Biol. Rev. 2003, 67, 16–37. [Google Scholar] [CrossRef]
- Zhang, R.; Patena, W.; Armbruster, U.; Gang, S.S.; Blum, S.R.; Jonikas, M.C. High-Throughput Genotyping of Green Algal Mutants Reveals Random Distribution of Mutagenic Insertion Sites and Endonucleolytic Cleavage of Transforming DNA. Plant Cell 2014, 26, 1398–1409. [Google Scholar] [CrossRef]
- Kim, J.; Chang, K.S.; Lee, S.; Jin, E. Establishment of a Genome Editing Tool Using CRISPR-Cas9 in Chlorella vulgaris UTEX395. Int. J. Mol. Sci. 2021, 22, 480. [Google Scholar] [CrossRef]
- Lin, W.R.; Ng, I.S. Development of CRISPR/Cas9 System in Chlorella vulgaris FSP-E to Enhance Lipid Accumulation. Enzyme Microb. Technol. 2020, 133, 109458. [Google Scholar] [CrossRef]
- Cao, S.; Xue, J.; Chen, X.; An, X.; Zhang, X. Magnetic Nanoparticles Mediate the Transformation of Antimicrobial Peptides HeM into Chlorella ellipsoidea. J. Appl. Phycol. 2020, 32, 3913–3921. [Google Scholar] [CrossRef]
- Gadamchetty, P.; Mullapudi, P.L.V.; Sanagala, R.; Markandan, M.; Polumetla, A.K. Genetic Transformation of Chlorella vulgaris Mediated by HIV-TAT Peptide. 3 Biotech 2019, 9, 139. [Google Scholar] [CrossRef]
- Ahmed, E.G.; Sung Kwon, L.; Sang, M.S.; Seung, H.Y.; Gyuhwa, C. Development of Stable Marker-Free Nuclear Transformation Strategy in the Green Microalga Chlorella vulgaris. Afr. J. Biotechnol. 2015, 14, 2715–2723. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, S.Y.; Kim, K.H.; Yoo, S.S.; Lee, T.K.; Choi, T.J. High-Level Expression of Recombinant VHSV Glycoprotein Using Transformed, C. Vulgaris and Verification of Vaccine Efficacy. Vaccines 2023, 11, 1205. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kim, S.H.; Abdellaoui, N.; Choi, T.J. Isolation and Characterization of a Salt Inducible Promoter from Chlorella vulgaris PKVL7422. J. Microbiol. Biotechnol. 2023, 33, 955–963. [Google Scholar] [CrossRef]
- Kim, M.J.; Kim, S.H.; Kim, Y.R.; Choi, T.J. Enhancement of Chlorella Transformation Efficacy by Insert Fragmentation. Algal Res. 2023, 72, 103146. [Google Scholar] [CrossRef]
- Yedahalli, S.; Rehmann, L.; Bassi, A. High Throughput Screening of β-Glucuronidase (GUS) Reporter in Transgenic Microalgae Transformed by Agrobacterium tumefaciens. Algal Res. 2018, 33, 328–336. [Google Scholar] [CrossRef]
- Kumar, M.; Jeon, J.; Choi, J.; Kim, S.R. Rapid and Efficient Genetic Transformation of the Green Microalga Chlorella vulgaris. J. Appl. Phycol. 2018, 30, 1735–1745. [Google Scholar] [CrossRef]
- Miki, B.; McHugh, S. Selectable Marker Genes in Transgenic Plants: Applications, Alternatives and Biosafety. J. Biotechnol. 2004, 107, 193–232. [Google Scholar] [CrossRef]
- Vicens, Q.; Westhof, E. Crystal Structure of Geneticin Bound to a Bacterial 16S Ribosomal RNA A Site Oligonucleotide. J. Mol. Biol. 2003, 326, 1175–1188. [Google Scholar] [CrossRef]
- Brönstrup, M.; Sasse, F. Natural Products Targeting the Elongation Phase of Eukaryotic Protein Biosynthesis. Nat. Prod. Rep. 2020, 37, 752–762. [Google Scholar] [CrossRef]
- Wirmer, J.; Westhof, E. Molecular Contacts Between Antibiotics and the 30S Ribosomal Particle. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2006; Volume 415, pp. 180–202. [Google Scholar]
- Liu, J.; Sun, Y.; Zhang, L.; Li, X.; He, Z.; Zhou, C.; Han, J. Screening of Antibiotics to Obtain Axenic Cell Cultures of a Marine Microalga Chrysotila Roscoffensis. Front. Bioeng. Biotechnol. 2023, 11, 1218031. [Google Scholar] [CrossRef] [PubMed]
- Roushan, M.R.; Chen, C.; Ahmadi, P.; Ward, V.C.A. Agrobacterium tumefaciens-Mediated Genetic Engineering of Green Microalgae, Chlorella vulgaris. J. Vis. Exp. 2023, e65382. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Choi, J.; Jeon, J.; Kumar, M.; Lee, J.; Jeong, W.J.; Kim, S.R. The Establishment of New Protein Expression System Using N Starvation Inducible Promoters in Chlorella. Sci. Rep. 2020, 10, 12713. [Google Scholar] [CrossRef] [PubMed]
- Borovinskaya, M.A.; Shoji, S.; Fredrick, K.; Cate, J.H.D. Structural Basis for Hygromycin B Inhibition of Protein Biosynthesis. RNA 2008, 14, 1590–1599. [Google Scholar] [CrossRef] [PubMed]
- Gangl, D.; Zedler, J.A.Z.; Rajakumar, P.D.; Martinez, E.M.R.; Riseley, A.; Włodarczyk, A.; Purton, S.; Sakuragi, Y.; Howe, C.J.; Jensen, P.E.; et al. Biotechnological Exploitation of Microalgae. J. Exp. Bot. 2015, 66, 6975–6990. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, S.; Sanda, S.; Uraguchi, Y.; Nakagawa, S.; Sawayama, S. Overexpression of the DOF-Type Transcription Factor Enhances Lipid Synthesis in Chlorella vulgaris. Appl. Biochem. Biotechnol. 2019, 189, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Zhang, Y.; Chen, X.; Zhang, P.; Ma, S. Development of a New Method for Genetic Transformation of the Green Alga Chlorella ellipsoidea. Mol. Biotechnol. 2013, 54, 211–219. [Google Scholar] [CrossRef]
- Kanchugal, P.S.; Selmer, M. Structural Recognition of Spectinomycin by Resistance Enzyme ANT(9) from Enterococcus Faecalis. Antimicrob. Agents Chemother. 2020, 64, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Siibak, T.; Peil, L.; Xiong, L.; Mankin, A.; Remme, J.; Tenson, T. Erythromycin- and Chloramphenicol-Induced Ribosomal Assembly Defects Are Secondary Effects of Protein Synthesis Inhibition. Antimicrob. Agents Chemother. 2009, 53, 563–571. [Google Scholar] [CrossRef]
- Niu, Y.F.; Zhang, M.H.; Xie, W.H.; Li, J.N.; Gao, Y.F.; Yang, W.D.; Liu, J.S.; Li, H.Y. A New Inducible Expression System in a Transformed Green Alga, Chlorella vulgaris. Genet. Mol. Res. 2011, 10, 3427–3434. [Google Scholar] [CrossRef]
- Fernández-Rodríguez, R.; Garro-Monge, G.; Guerrero-Barrantes, M.; Gómez-Espinoza, O. Alpha-Tubulin Promoter from Chlorella vulgaris Allows Genetic Transformation of Green Coccoid Microalga. Rev. Tecnol. En Marcha 2020, 33, 27–36. [Google Scholar] [CrossRef]
- Gu, X.; Deng, Y.; Wang, A.; Gan, Q.; Xin, Y.; Paithoonrangsarid, K.; Lu, Y. Engineering a Marine Microalga Chlorella sp. as the Cell Factory. Biotechnol. Biofuels Bioprod. 2023, 16, 133. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, M.L.L.; Luke, G.; Mehrotra, A.; Li, X.; Hughes, L.E.; Gani, D.; Ryan, M.D. Analysis of the Aphthovirus 2A/2B Polyprotein ‘Cleavage’ Mechanism Indicates Not a Proteolytic Reaction, but a Novel Translational Effect: A Putative Ribosomal ‘Skip’. J. Gen. Virol. 2001, 82, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, R.L.; Nakamura, M. Expression of Human Growth Hormone by the Eukaryotic Alga, Chlorella. Curr. Microbiol. 1999, 38, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Y.; Sun, Y.; Zhang, L.; Li, W. Highly Efficient Expression of Rabbit Neutrophil Peptide-1 Gene in Chlorella ellipsoidea Cells. Curr. Genet. 2001, 39, 365–370. [Google Scholar] [CrossRef]
- Norashikin, M.N.; Loh, S.H.; Aziz, A.; Cha, T.S. Metabolic Engineering of Fatty Acid Biosynthesis in Chlorella vulgaris Using an Endogenous Omega-3 Fatty Acid Desaturase Gene with Its Promoter. Algal Res. 2018, 31, 262–275. [Google Scholar] [CrossRef]
- Park, S.; Lee, Y.; Lee, J.H.; Jin, E.S. Expression of the High Light-Inducible Dunaliella LIP Promoter in Chlamydomonas Reinhardtii. Planta 2013, 238, 1147–1156. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.L.; Harikrishna, J.A.; Abu Bakar, F.; Yeo, C.C.; Cha, T.S. Heterologous Expression of the Streptococcus Pneumoniae YoeB and PezT Toxin Genes Is Lethal in Chlorella vulgaris. Algal Res. 2016, 19, 21–29. [Google Scholar] [CrossRef]
- Jefferson, R.A. Assaying Chimeric Genes in Plants: The GUS Gene Fusion System. Plant Mol. Biol. Report. 1987, 5, 387–405. [Google Scholar] [CrossRef]
- Margolin, W. Green Fluorescent Protein as a Reporter for Macromolecular Localization in Bacterial Cells. Methods 2000, 20, 62–72. [Google Scholar] [CrossRef]
- Run, C.; Fang, L.; Fan, J.; Fan, C.; Luo, Y.; Hu, Z.; Li, Y. Stable Nuclear Transformation of the Industrial Alga Chlorella Pyrenoidosa. Algal Res. 2016, 17, 196–201. [Google Scholar] [CrossRef]
- Jarvis, E.E.; Brown, L.M. Transient Expression of Firefly Lueiferase in of the Green Alga Chlorella ellipsoidea. Curr. Genet. 1991, 19, 317–321. [Google Scholar] [CrossRef]
- Chow, K.C.; Tung, W.L. Electrotransformation of Chlorella vulgaris. Plant Cell Rep. 1999, 18, 778–780. [Google Scholar] [CrossRef]
- Talebi, A.F.; Tohidfar, M.; Tabatabaei, M.; Bagheri, A.; Mohsenpor, M.; Mohtashami, S.K. Genetic Manipulation, a Feasible Tool to Enhance Unique Characteristic of Chlorella vulgaris as a Feedstock for Biodiesel Production. Mol. Biol. Rep. 2013, 40, 4421–4428. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Sun, Y.; Li, X.; Zhang, L.; Li, W.; Wang, Y. Rapid Isolation and Functional Analysis of Promoter Sequences of the Nitrate Reductase Gene from Chlorella ellipsoidea. J. Appl. Phycol. 2004, 16, 11–16. [Google Scholar] [CrossRef]
- Sharma, P.K.; Goud, V.V.; Yamamoto, Y.; Sahoo, L. Efficient Agrobacterium tumefaciens-Mediated Stable Genetic Transformation of Green Microalgae, Chlorella sorokiniana. 3 Biotech 2021, 11, 196. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, M.; Horáková, I.; Honda, H.; Xing, X.; Shiragami, N.; Unno, H. Introduction of Foreign DNA into Chlorella Saccharophila by Electroporation. Biotechnol. Tech. 1994, 8, 821–826. [Google Scholar] [CrossRef]
- El-Sheekh, M.M. Stable Transformation of the Intact Cells of the Chlorella Kessleri with High Velocity Microprojectiles. Biol. Plant 1999, 42, 209–216. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Su, Q.; Gao, X. Transient Expression of the GUS Gene in a Unicellular Marine Green Alga, Chlorella sp. MACC/C95, via Electroporation. Biotechnol. Bioprocess Eng. 2007, 12, 180–183. [Google Scholar] [CrossRef]
- Fei, X.; Xiao, S.; Huang, X.; Li, Z.; Li, X.; He, C.; Li, Y.; Zhang, X.; Deng, X. Control of Aedes Mosquito Populations Using Recombinant Microalgae Expressing Short Hairpin RNAs and Their Effect on Plankton. PLoS Negl. Trop. Dis. 2023, 17, e0011109. [Google Scholar] [CrossRef]
- Borovsky, D.; Sterner, A.; Powell, C.A. Cloning and Expressing Trypsin Modulating Oostatic Factor in Chlorella Desiccata to Control Mosquito Larvae. Arch. Insect Biochem. Physiol. 2016, 91, 17–36. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Peng, H.; Liu, J.; Chen, F.; Zhou, Y.; Ma, X.; Chen, H.; Wang, K. Chlorella sp. Transgenic with Scy-Hepc Enhancing the Survival of Sparus macrocephalus and Hybrid Grouper Challenged with Aeromonas hydrophila. Fish Shellfish Immunol. 2018, 73, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, H.J.; Su, C.H.; Chien, L.J. Accumulation of Lipid Production in Chlorella Minutissima by Triacylglycerol Biosynthesis-Related Genes Cloned from Saccharomyces Cerevisiae and Yarrowia Lipolytica. J. Microbiol. 2012, 50, 526–534. [Google Scholar] [CrossRef] [PubMed]
- Chien, L.J.; Hsu, T.P.; Huang, C.C.; Teng, K.; Hsieh, H.J. Novel Codon-Optimization Genes Encoded in Chlorella for Triacylglycerol Accumulation. Energy Procedia 2015, 75, 44–55. [Google Scholar] [CrossRef]
- Yan, J.; Kuang, Y.; Gui, X.; Han, X.; Yan, Y. Engineering a Malic Enzyme to Enhance Lipid Accumulation in Chlorella Protothecoides and Direct Production of Biodiesel from the Microalgal Biomass. Biomass Bioenergy 2019, 122, 298–304. [Google Scholar] [CrossRef]
- Ma, R.; Lin, X. Vitreoscilla Hemoglobin Gene (Vgb) Improves Lutein Production in Chlorella vulgaris. Chin. J. Oceanol. Limnol. 2014, 32, 390–396. [Google Scholar] [CrossRef]
- Lou, S.; Wang, L.; He, L.; Wang, Z.; Wang, G.; Lin, X. Production of Crocetin in Transgenic Chlorella vulgaris Expressing Genes CrtRB and ZCD1. J. Appl. Phycol. 2016, 28, 1657–1665. [Google Scholar] [CrossRef]
- Lin, W.R.; Lai, Y.C.; Sung, P.K.; Tan, S.I.; Chang, C.H.; Chen, C.Y.; Chang, J.S.; Ng, I.S. Enhancing Carbon Capture and Lipid Accumulation by Genetic Carbonic Anhydrase in Microalgae. J. Taiwan Inst. Chem. Eng. 2018, 93, 131–141. [Google Scholar] [CrossRef]
- Lee, H.; Shin, W.-S.; Kim, Y.U.; Jeon, S.; Kim, M.; Kang, N.K.; Chang, Y.K. Enhancement of Lipid Production under Heterotrophic Conditions by Overexpression of an Endogenous BZIP Transcription Factor in Chlorella sp. HS2. J. Microbiol. Biotechnol. 2020, 30, 1597–1606. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, D.; Zhang, J.; Chen, Y.; Liu, X.; Fan, C.; Wang, R.R.C.; Hou, Y.; Hu, Z. Overexpression of the Transcription Factor AtLEC1 Significantly Improved the Lipid Content of Chlorella ellipsoidea. Front. Bioeng. Biotechnol. 2021, 9, 626162. [Google Scholar] [CrossRef]
- Liu, Y.; Huo, X.; Yu, S.; Dong, W.; Xue, Y.; Liu, C. Quantitative Proteomic Analysis to Understand the Role of Arabidopsis Thaliana LEAFY COTYLEDON 2 in Promoting Lipid Accumulation in Chlorella sorokiniana by Upregulating Photosynthetic Proteins and G3PDH. J. Appl. Phycol. 2022, 34, 3035–3046. [Google Scholar] [CrossRef]
- Lin, H.D.; Liu, B.H.; Kuo, T.T.; Tsai, H.C.; Feng, T.Y.; Huang, C.C.; Chien, L.F. Knockdown of PsbO Leads to Induction of HydA and Production of Photobiological H2 in the Green Alga Chlorella sp. DT. Bioresour. Technol. 2013, 143, 154–162. [Google Scholar] [CrossRef] [PubMed]
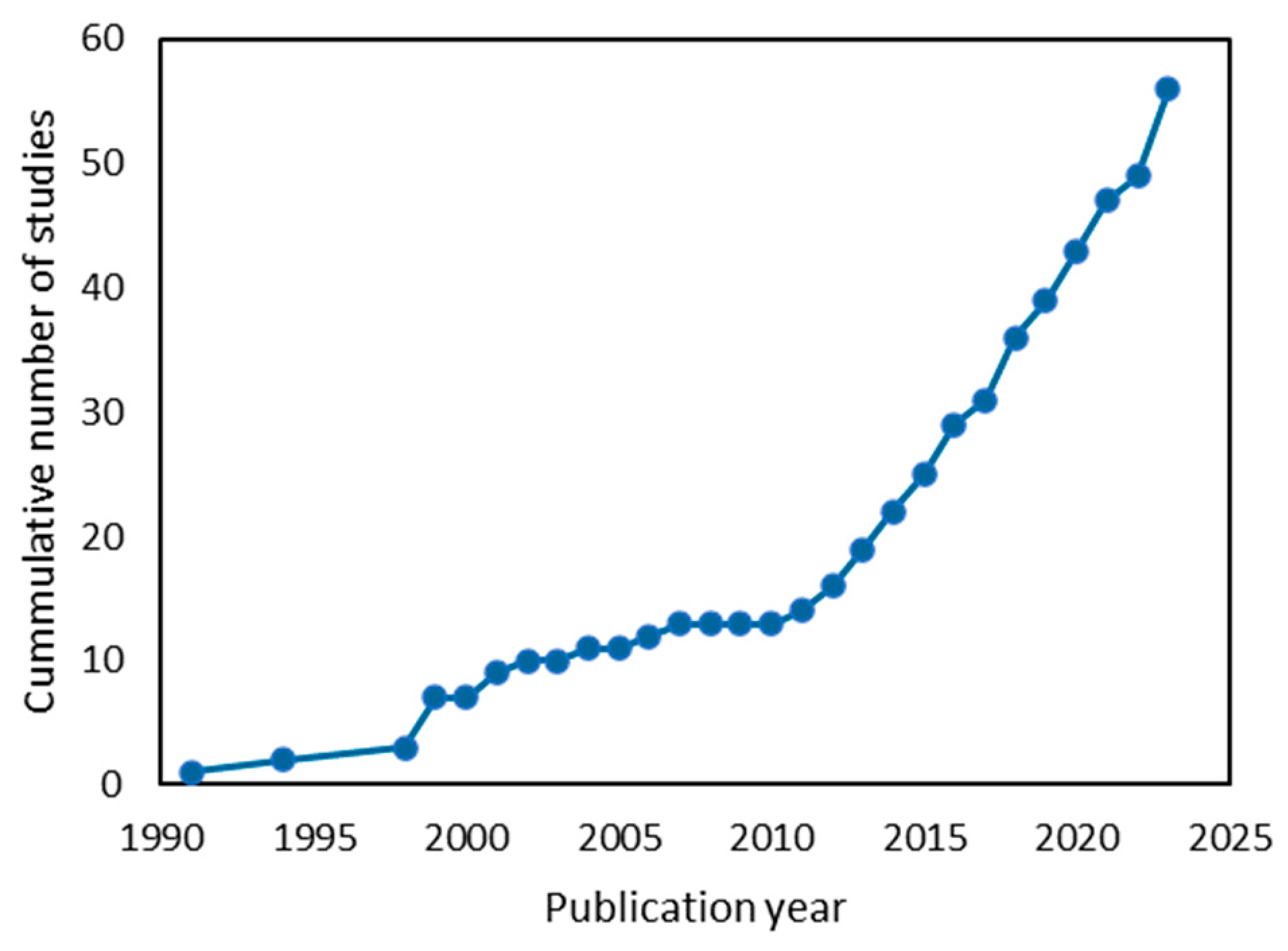
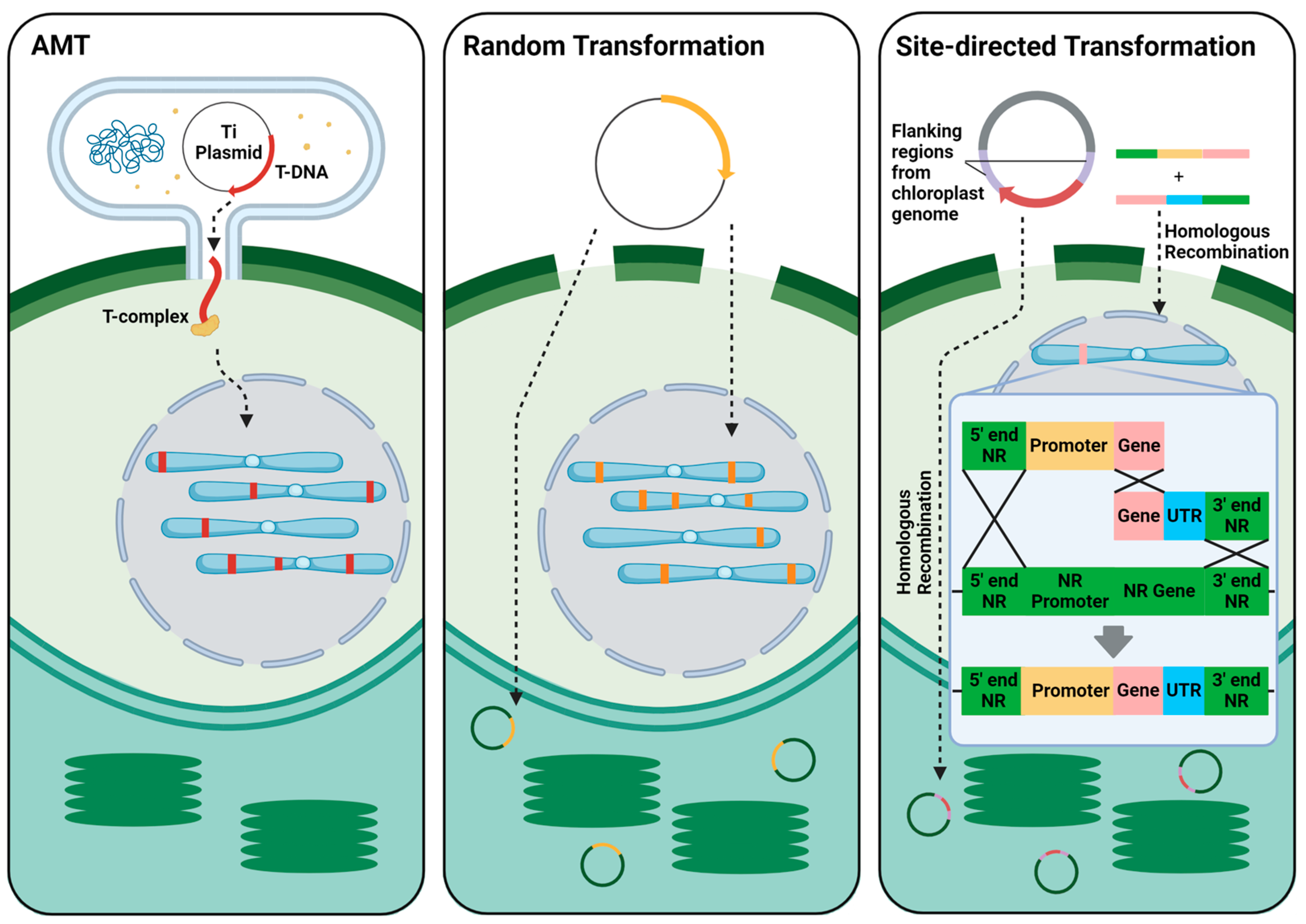

| Species | Genome Size (Mb) | Chromosomal Arrangement (Ploidy of Vegetative Cells) | Ref. |
|---|---|---|---|
| E. coli K12 | 4.6 | 1 chromosome (n = 1) | [15] |
| S. cerevisiae | 12.1 | 16 chromosomes + MT, (mostly n = 1) | [16] |
| Cricetulus griseus (CHO-K1) | 2450 | 21 chromosomes + MT (n = 2) | [17] |
| N. tabacum | 3600 | 24 chromosomes + MT + CP, (n = 2) | [18] |
| C. reinhardtii | 111 | 17 chromosomes + MT + CP, (mostly n = 1) | [19,20] |
| P. tricornutum | 27.5 | 25 chromosomes + MT + CP, (n = 2) | [21,22] |
| C. vulgaris | 40.2 | 14 scaffolds + MT + CP, (n = 1) | [23] |
| C. sorokiniana | 38.6 | 13 chromosomes + MT + CP, (n = 1) | GCA_025917655.1 |
| Antibiotic/Herbicide (Genes) | Conc. | C. vulgaris | C. ellipsoidae | C. sorokiniana |
|---|---|---|---|---|
| Geneticin/G418 (nptII) | 15–500 µg/mL | + | + | + |
| Hygromycin B (hpt) | 15–500 µg/mL | + | n.d. | + |
| Kanamycin (nptII) | 15–50 µg/mL | + | n.d. | n.d. |
| Chloramphenicol (cat) | 200 µg/mL | + | n.d. | n.d. |
| Phleomycin or Zeocin (ble) | 1–10 µg/mL | + | + | n.d. |
| Paromomycin (nptII) | 10 µg/mL | + | n.d. | n.d. |
| Protein (Gene) | Expression Elements | Notes | Ref. |
|---|---|---|---|
| YoeB toxin GFP fusion (yeoB:gfp) PezT toxin GFP fusion (pezT:gfp) | POlexA-yeoB:gfp-TT3A POlexA-pezT:gfp-TT3A | C. vulgaris XVE/OlexA estrogen inducible promoter system Stable for > 1 year | [71] |
| NZ2114 (ant1), piscidin-4 (ant-2) | PrbcL-ant1-RBS-ant2-TpsbA | C. vulgaris Chloroplast integration using HR with 16S-trn1 and trnA-23S region | [33] |
| Heliomicin (HeM) | PCaMV35S-HeM-TNOS | C. ellipsoidea MNP—Magnetic nanoparticle-mediated transformation | [42] |
| Hepcidin (hepc), scygonadin (scy), and fusion (scy:hepc) | PCaMV35S-hepc-TNOS PCaMV35S-scy-TNOS PCaMV35S-hepc:scy-TNOS | Chlorella sp. Production of AMPs for animal feed | [85] |
| Proteins (Genes) | Expression Elements | Notes | Ref. |
|---|---|---|---|
| Lipid accumulation-associated enzymes | PCaMV35S-gene-TNOS PRbcS2-gene-TNOS | C. minutissima Include homology regions Up to 5 genes expressed together | [86] |
| Vitreoscilla hemoglobin (vgb) | PCaMV35S-vgb-TCYC1 | C. vulgaris Increase cell respiration efficiency | [89] |
| TF GmDof4 | PUbi-GmDof4-TNOS | C. ellipsoidea Enhance lipid production | [27] |
| Lipid accumulation-associated enzymes | TCaMV35S | Chlorella sp. Promoter not specified | [87] |
| β-carotene hydroxylase 1 (crtRB) Carotenoid cleavage dioxygenase (ZCD1) | PCaMV35S-crtRB-TNOS & PUbi-ZCD1-TNOS | C. vulgaris Produce crocetin from saffron crocus | [90] |
| EGFP (egfp), Fructose 1,6-bisphosphate aldolase (fba) | PCaMV35S-egfp-TNOS PCaMV35S-fba-TNOS | C. vulgaris Chloroplast localization Increase phototrophic growth rate | [30] |
| Carbonic anhydrase (Mica) | PCaMV35S-Mica-TNOS | C. sorokiniana Improve CO2 capture | [91] |
| Omega-3 desaturase (FAD), GFP/GUS fusion | PFAD-ω-3 FAD-TFAD PCaMV35S-mgfp5:gusA-TFAD | C. vulgaris Enhance lipid production | [69] |
| Carbonic anhydrase | PCaMV35S-Mica-TNOS | C. vulgaris Improve CO2 capture | [91] |
| DNA binding with one finger (DOF)-type TF | PHSP+RbcS-ble-2A-DOF-TRbcS | C. vulgaris Enhance lipid production | [59] |
| Malic enzyme (me) | PCaMV35S-g-TNOS | C. protothecoides Enhance lipid production | [88] |
| Basic leucine zipper (bZIP)-TF (ZIP1) | PHSP70-ZIP1-TNOS | Chlorella sp. HS2 C-terminal FLAG tag Enhance lipid production | [92] |
| TF Lec1 | PUbi-Lec1-TNOS | C. ellipsoidea Enhance lipid production | [93] |
| TF Lec2 | PCaMV35S-Lec2 | C. sorokiniana Increase relative electron transfer rate Enhance lipid production | [94] |
| Diacylglycerol acyltransferase (DGA) | PHsp70-RbcS2-dga-TNOS | C. sorokiniana Enhance lipid production | [35] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.; Ward, V.C.A. Recombinant Protein Expression and Its Biotechnological Applications in Chlorella spp. SynBio 2024, 2, 223-239. https://doi.org/10.3390/synbio2020013
Chen C, Ward VCA. Recombinant Protein Expression and Its Biotechnological Applications in Chlorella spp. SynBio. 2024; 2(2):223-239. https://doi.org/10.3390/synbio2020013
Chicago/Turabian StyleChen, Chuchi, and Valerie C. A. Ward. 2024. "Recombinant Protein Expression and Its Biotechnological Applications in Chlorella spp." SynBio 2, no. 2: 223-239. https://doi.org/10.3390/synbio2020013
APA StyleChen, C., & Ward, V. C. A. (2024). Recombinant Protein Expression and Its Biotechnological Applications in Chlorella spp. SynBio, 2(2), 223-239. https://doi.org/10.3390/synbio2020013






