Exploring the Interactions between Human microRNAs and the Ilheus Virus Genome
Abstract
:1. Introduction
2. Results
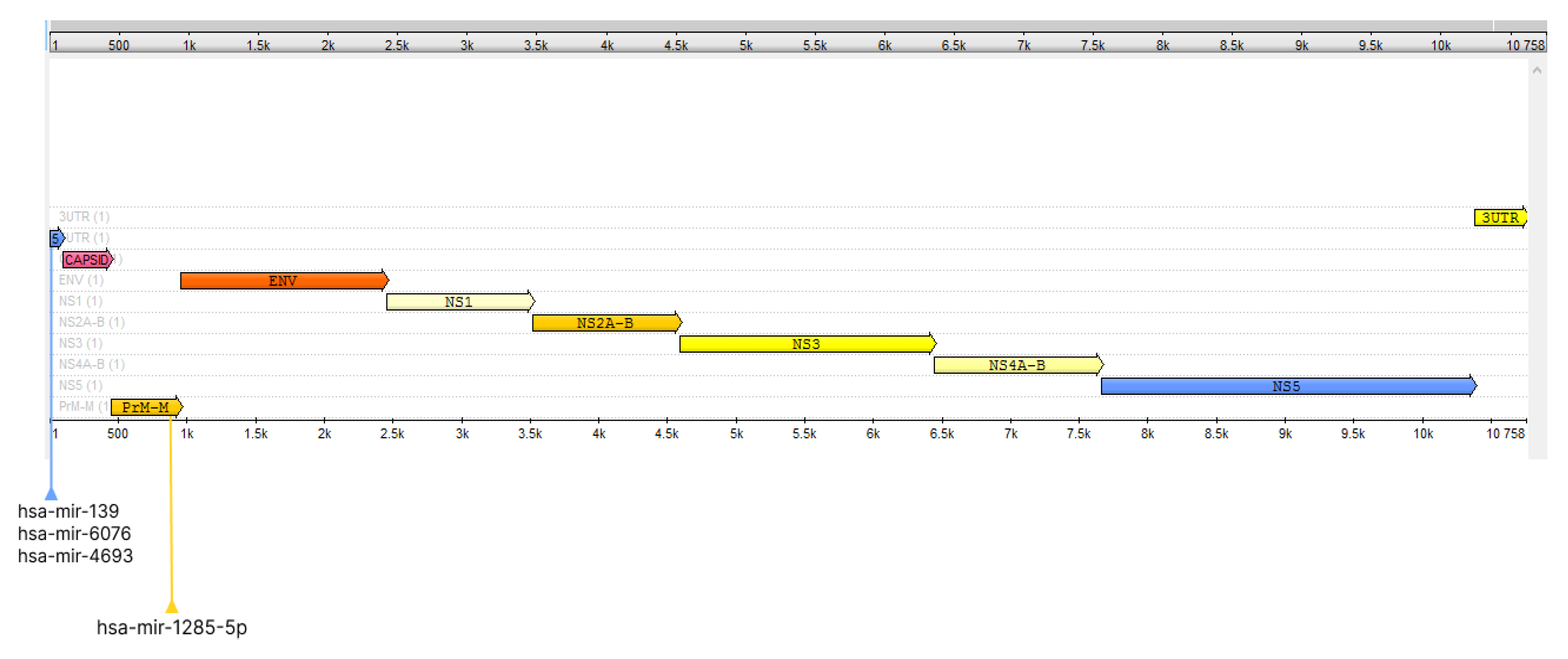
2.1. Human miRNAs Interact with Key Regions of the ILHV Genome
2.2. Human miRNAs Regulate Cellular Pathways Associated with Viral Replication, Cell Cycle Control, and Immune Signaling
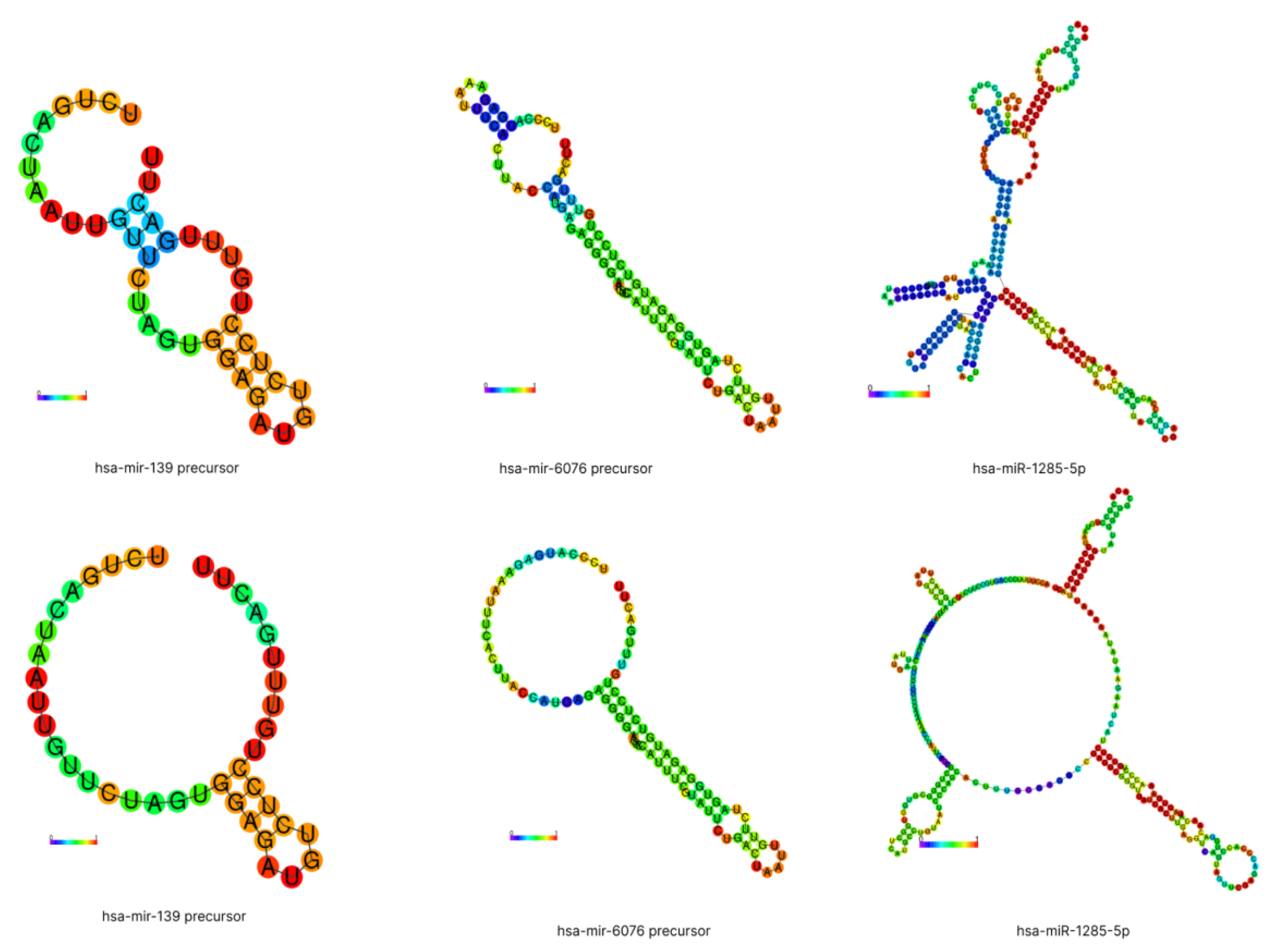
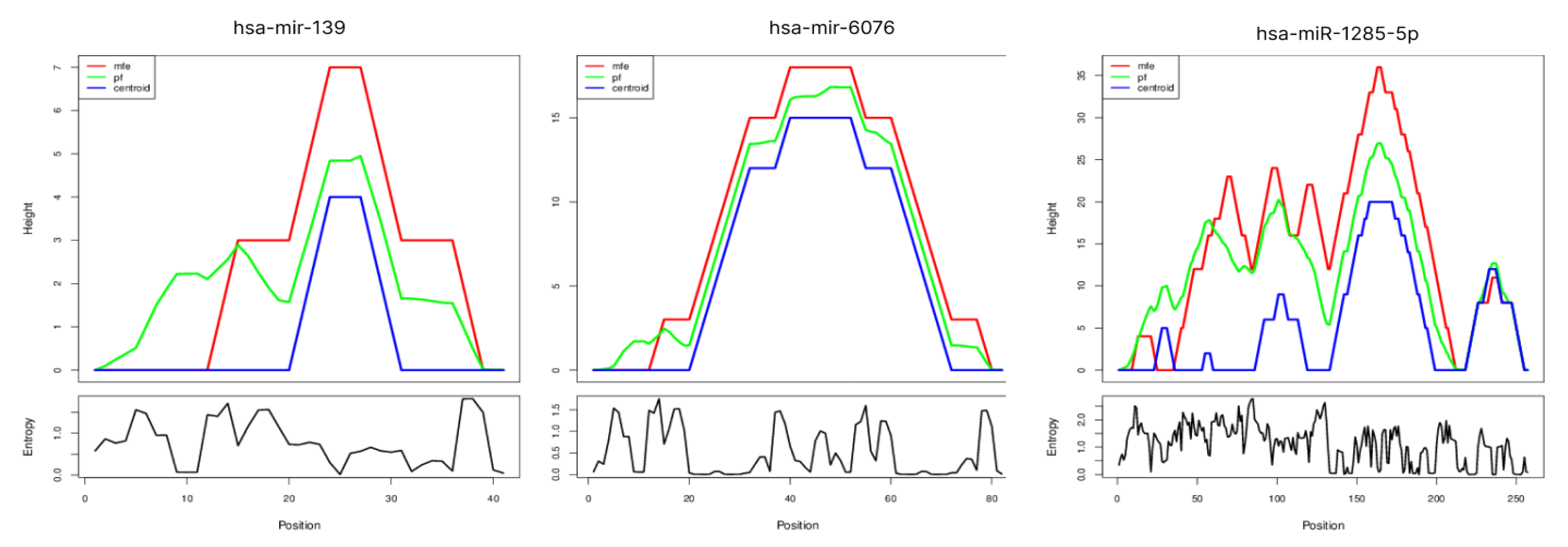
2.3. Secondary Structure of miRNAs
3. Discussion
4. Materials and Methods
4.1. Database and Alignment
4.2. miRNA Search
4.3. Determination of miRNA Targets and Functions
4.4. Secondary Structure
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| miRNA | Target Gene | Function |
|---|---|---|
| hsa-mir-139 | ATL2 | Allows identical protein-binding activity. Involved in Golgi organization and organization of the tubular network membrane of the endoplasmic reticulum. Located in the tubular network membrane of the endoplasmic reticulum. It is an integral component of the membrane. |
| CKB | Cytoplasmic enzyme involved in energy homeostasis | |
| SOCKS7 | Predicted to act upstream or within various processes, including brain development, adipocyte differentiation, and the insulin receptor signaling pathway. | |
| MAP2 | Involved in microtubule assembly, which is an essential step in neurogenesis. | |
| CDH20 | Calcium-dependent cell adhesion proteins; cadherins can thus contribute to the classification of heterogeneous cell types. | |
| FZD3 | Promotes neurogenesis by maintaining sympathetic neuroblasts in the cell cycle in a beta-catenin-dependent manner (by similarity). | |
| KLC2 | Microtubule-associated force-producing protein that plays a role in organelle transport. | |
| MGA | Functions as a dual-specificity transcription factor, regulating the expression of target genes of the MAX network and T-box family. Functions as a repressor or activator. | |
| TCF3 | Transcriptional regulator involved in early neuronal differentiation and mesenchymal-to-epithelial transition (by similarity). | |
| hsa-mir-4693 | KCNIP1 | Regulatory subunit of type A potassium channels. |
| ARRB2 | Associated with agonist-mediated desensitization of G protein-coupled receptors. | |
| hsa-mir-6076 | SMG9 | Encodes a regulatory subunit of the SMG1 complex, which plays a critical role in nonsense-mediated mRNA decay (NMD). |
| hsa-mir-1285-5p | SCP2 | Plays a crucial role in peroxisomal oxidation of branched-chain fatty acids. |
| SNAP25 | Involved in the molecular regulation of neurotransmitter release. | |
| MARCKS | Involved in cell motility, phagocytosis, membrane trafficking, and mitogenesis. | |
| EBI3 | This gene was identified for its induced expression in B lymphocytes in response to Epstein–Barr virus infection. Associates with IL27 to form interleukin IL-27, which acts in innate immunity. | |
| SH2D1A | Encodes a protein that plays an important role in bidirectional stimulation of T and B cells. |
Appendix B
References
- da Costa, V.G.; Saivish, M.V.; Lino, N.A.; Bittar, C.; de Freitas Calmon, M.; Nogueira, M.L.; Rahal, P. Clinical Landscape and Rate of Exposure to Ilheus Virus: Insights from Systematic Review and Meta-Analysis. Viruses 2022, 15, 92. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.J.; Andrade, C.D.; Kubiszeski, J.R.; Silva, D.J.; Barreto, E.S.; Massey, A.L.; Canale, G.R.; Bernardo, C.S.; Levi, T.; Peres, C.A.; et al. Detection of Ilheus virus in mosquitoes from southeast Amazon, Brazil. Trans. R. Soc. Trop. Med. Hyg. 2019, 113, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Plante, J.A.; Plante, K.S.; Popov, V.L.; Shinde, D.P.; Widen, S.G.; Buenemann, M.; Nogueira, M.L.; Vasilakis, N. Morphologic and genetic characterization of ilheus virus, a potential emergent flavivirus in the americas. Viruses 2023, 15, 195. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Jeng, S.; McWeeney, S.K.; Hirsch, A.J. A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, A.; Basu, A. Involvement of host microRNAs in flavivirus-induced neuropathology: An update. J. Biosci. 2022, 47, 1–18. [Google Scholar] [CrossRef]
- Miskiewicz, J.; Szachniuk, M. Discovering Structural Motifs in miRNA Precursors from the Viridiplantae Kingdom. Molecules 2018, 23, 1367. [Google Scholar] [CrossRef]
- Li, Z.; Xu, R.; Li, N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. 2018, 15, 68. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes. Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef]
- Casseb, S.M.; Simith, D.B.; Melo, K.F.; Mendonca, M.H.; Santos, A.C.; Carvalho, V.L.; Cruz, A.C.; Vasconcelos, P.F. Drosha, DGCR8, and Dicer mRNAs are down-regulated in human cells infected with dengue virus 4, and play a role in viral pathogenesis. Genet. Mol. Res. 2016, 15, gmr.15027891. [Google Scholar] [CrossRef]
- Seong, R.-K.; Lee, J.K.; Shin, O.S. Zika Virus-Induction of the Suppressor of Cytokine Signaling 1/3 Contributes to the Modulation of Viral Replication. Pathogens 2020, 9, 163. [Google Scholar] [CrossRef]
- Li, C.; Hu, J.; Hao, J.; Zhao, B.; Wu, B.; Sun, L.; Peng, S.; Gao, G.F.; Meng, S. Competitive virus and host RNAs: The interplay of a hidden virus and host interaction. Protein Cell. 2014, 5, 348–356. [Google Scholar] [CrossRef]
- Johnson, B.W.; Cruz, C.; Felices, V.; Espinoza, W.R.; Manock, S.R.; Guevara, C.; Olson, J.G.; Kochel, T.J. Ilheus virus isolate from a human, Ecuador. Emerg. Infect. Dis. 2007, 13, 956–958. [Google Scholar] [CrossRef]
- Uozaki, H.; Chong, J.M.; Fujimoto, E.; Itoh, M.; Saito, M.; Sakuma, K.; Sudo, M.; Ushiku, T.; Niki, T.; Nagai, H.; et al. Soft and hard keratin expression in Epstein-Barr-virus-associated gastric carcinoma. Anticancer Res. 2005, 25, 3183–3190. [Google Scholar]
- Saivish, M.V.; Pacca, C.C.; da Costa, V.G.; Menezes, G.d.L.; da Silva, R.A.; Nebo, L.; da Silva, G.C.D.; Milhim, B.H.G.d.A.; Teixeira, I.d.S.; Henrique, T.; et al. Caffeic Acid Has Antiviral Activity against Ilhéus Virus In Vitro. Viruses 2023, 15, 494. [Google Scholar] [CrossRef]
- Kincaid, R.P.; Sullivan, C.S. Virus-Encoded microRNAs: An Overview and a Look to the Future. PLoS Pathog. 2012, 8, e1003018. [Google Scholar] [CrossRef]
- Byk, L.A.; Gamarnik, A.V. Properties and Functions of the Dengue Virus Capsid Protein. Annu. Rev. Virol. 2016, 3, 263–281. [Google Scholar] [CrossRef]
- Monel, B.; Rajah, M.M.; Hafirassou, M.L.; Sid Ahmed, S.; Burlaud-Gaillard, J.; Zhu, P.P.; Nevers, Q.; Buchrieser, J.; Porrot, F.; Meunier, C.; et al. Atlastin Endoplasmic Reticulum-Shaping Proteins Facilitate Zika Virus Replication. J. Virol. 2019, 93, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Hara, H.; Aizaki, H.; Matsuda, M.; Shinkai-Ouchi, F.; Inoue, Y.; Murakami, K.; Shoji, I.; Kawakami, H.; Matsuura, Y.; Lai, M.M.C.; et al. Involvement of Creatine Kinase B in Hepatitis C Virus Genome Replication through Interaction with the Viral NS4A Protein. J. Virol. 2009, 83, 5137–5147. [Google Scholar] [CrossRef] [PubMed]
- Avdoshina, V.; Mahoney, M.; Gilmore, S.F.; Wenzel, E.D.; Anderson, A.; Letendre, S.L.; Imamichi, T.; Fischer, N.O.; Mocchetti, I. HIV influences microtubule associated protein-2: Potential marker of HIV-associated neurocognitive disorders. AIDS 2020, 34, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Karousis, E.D.; Mühlemann, O. Nonsense-Mediated mRNA Decay Begins Where Translation Ends. Cold Spring Harb. Perspect. Biol. 2018, 11, a032862. [Google Scholar] [CrossRef]
- Mocquet, V.; Neusiedler, J.; Rende, F.; Cluet, D.; Robin, J.P.; Terme, J.M.; Duc Dodon, M.; Wittmann, J.; Morris, C.; Le Hir, H.; et al. The human T-lymphotropic virus type 1 tax protein inhibits nonsense-mediated mRNA decay by interacting with INT6/EIF3E and UPF1. J. Virol. 2012, 86, 7530–7543. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shi, W.; Song, R.; Long, W.; Guo, H.; Yuan, S.; Zhang, T. Divergent patterns of genic copy number variation in KCNIP1 gene reveal risk locus of type 2 diabetes in Chinese population. Endocr. J. 2018, 65, 537–545. [Google Scholar] [CrossRef]
- Wen, Q.; Li, Y.; Han, Z.; Liu, H.; Zhang, S.; Chen, Y.; He, J.; Du, X.; Fu, Y.; Zhang, L.; et al. β-Arrestin 2 Regulates Inflammatory Responses against Mycobacterium tuberculosis Infection through ERK1/2 Signaling. J. Immunol. 2021, 206, 2623–2637. [Google Scholar] [CrossRef]
- Herrero, L.; Monroy, N.; González, M.E. HIV-1 Vpu Protein Mediates the Transport of Potassium in Saccharomyces cerevisiae. Biochemistry 2012, 52, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, M.; Li, L.; Qian, G.; Wang, Y.; Chen, Z.; Liu, J.; Fang, C.; Huang, F.; Guo, D.; et al. β-arrestin 2 as an activator of cGAS-STING signaling and target of viral immune evasion. Nat. Commun. 2020, 11, 6000. [Google Scholar] [CrossRef] [PubMed]
- Aryal, M.; Lin, D.; Regan, K.; Du, S.; Shi, H.; Alvarado, J.J.; Ilina, T.V.; Andreotti, A.H.; Smithgall, T.E. The HIV-1 protein Nef activates the Tec family kinase Btk by stabilizing an intermolecular SH3-SH2 domain interaction. Sci. Signal. 2022, 15, eabn8359. [Google Scholar] [CrossRef]
- Fernández-Pato, A.; Virseda-Berdices, A.; Resino, S.; Ryan, P.; Martínez-González, O.; Pérez-García, F.; Martin-Vicente, M.; Valle-Millares, D.; Brochado-Kith, O.; Blancas, R.; et al. Plasma miRNA profile at COVID-19 onset predicts severity status and mortality. Emerg. Microbes Infect. 2022, 11, 676–688. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information (NCBI). National Library of Medicine (US), National Center for Biotechnology Information, Bethesda, MD, USA. 1988. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 6 April 2017).
- Rose, R.; Golosova, O.; Sukhomlinov, D.; Tiunov, A.; Prosperi, M. Flexible design of multiple metagenomics classification pipelines with UGENE. Bioinformatics 2018, 35, 1963–1965. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kudla, M.; Gutowska, K.; Synak, J.; Weber, M.; Bohnsack, K.S.; Lukasiak, P.; Villmann, T.; Blazewicz, J.; Szachniuk, M. Virxicon: A lexicon of viral sequences. Bioinformatics 2020, 36, 5507–5513. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; Van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Kostoulas, N.; Vergoulis, T.; Georgakilas, G.; Reczko, M.; Maragkakis, M.; Paraskevopoulou, M.D.; Prionidis, K.; Dalamagas, T.; Hatzigeorgiou, A.G. Hatzigeorgiou DIANA miRPath v.2.0: Investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012, 40, W498–W504. Available online: https://dianalab.e-ce.uth.gr/html/universe/index.php?r=mirpath (accessed on 8 September 2023).
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Yi, H.C.; You, Z.H.; Cheng, L.; Zhou, X.; Jiang, T.H.; Li, X.; Wang, Y.B. Learning distributed representations of RNA and protein sequences and its application for predicting lncRNA-protein interactions. Comput. Struct. Biotechnol. J. 2019, 18, 20–26. [Google Scholar] [CrossRef]
| Region | miRNA | Sequence |
|---|---|---|
| 5′ cap | hsa-mir-139 | UCUGACUAAUUGUUCUAGUGGAGAUGUCUCCUGUUUGACUU |
| hsa-mir-6076 | ||
| hsa-mir-4693 | ||
| prM | hsa-mir-1285-5p | ACCUUUUCCCAGUGCCUUCUUCUGCUUAUGUC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, J.B.; Casseb, S.M.M. Exploring the Interactions between Human microRNAs and the Ilheus Virus Genome. SynBio 2023, 1, 194-203. https://doi.org/10.3390/synbio1030014
Souza JB, Casseb SMM. Exploring the Interactions between Human microRNAs and the Ilheus Virus Genome. SynBio. 2023; 1(3):194-203. https://doi.org/10.3390/synbio1030014
Chicago/Turabian StyleSouza, Joyhare Barbosa, and Samir Mansour Moraes Casseb. 2023. "Exploring the Interactions between Human microRNAs and the Ilheus Virus Genome" SynBio 1, no. 3: 194-203. https://doi.org/10.3390/synbio1030014
APA StyleSouza, J. B., & Casseb, S. M. M. (2023). Exploring the Interactions between Human microRNAs and the Ilheus Virus Genome. SynBio, 1(3), 194-203. https://doi.org/10.3390/synbio1030014






