Abstract
This study investigated the resistotyping of Salmonella spp. and Staphylococcus aureus from milk and milk products sold in Sabon Gari and Zaria local government areas in Kaduna State, Nigeria. A total of 122 isolates (Salmonella spp. (65) and Staphylococcus aureus (57)) were isolated from 400 milk and milk products. The isolates were subjected to antimicrobial susceptibility testing using the disc diffusion and E-test methods. The results obtained from the study indicated that 39 (31.967%) isolates were sensitive to all tested antibiotics, while 47 (38.525%) were resistant to a single antibiotic. Furthermore, 36 (29.508%) were resistant to two antibiotics, and none showed resistance to at least three antibiotics. None showed resistance to all four antibiotics. Resistance rates were most frequently observed in tetracycline with 80 isolates (65.574%), followed by ampicillin with 39 isolates (31.967%), and gentamicin and ciprofloxacin, both with 00 (00.000%) isolates. After making comparisons with the CLSI and EUCAST breakpoints, the resistance rate with CLSI was observed in tetracycline with 104 isolates (85.245%), followed by ampicillin with 39 isolates (66.393%), ciprofloxacin with 14 isolates (11.475%), and gentamicin with 06 isolates (04.918%). The resistance rate with EUCAST was tetracycline with 122 isolates (100.000%), followed by ampicillin with 110 isolates (90.164%), ciprofloxacin with 88 isolates (72.131%), and gentamicin with 17 isolates (13.934%). Based on these findings, it can be shown that Salmonella spp. and Staphylococcus aureus found in milk and milk products within Sabon Gari and Zaria local government areas have a high resistance to the antibiotics tested. It is imperative that urgent actions are taken to address the growing menace of AMR and prevent the spread of antibiotic-resistant pathogens.
1. Introduction
Milk is the fluid secreted by female mammals for nourishing offspring. It comprises a mixture of complex chemical substances which include fat, protein, lactose, and some mineral matters in the colloidal state in the form of a true solution [1].
Resistotyping involves grouping bacterial isolates based on resistance patterns to a set of randomly chosen antibiotics particular to specific strains via phenotypic methods. Antimicrobial resistance (AMR) is on the rise and has posed a major public health concern, severely limiting therapeutic options in clinical settings [2].
2. Materials and Methods
2.1. Study Area
The study area included two (2) local government areas (Soban Gari and Zaria) in Kaduna state (see Figure 1).
Figure 1.
Map of Zaria and Sabon Gari.
2.2. Isolation of Organisms
Four hundred (400) samples [100 each] of milk and milk products (Kindirimo, Nono, and yogurt) sold in Zaria and Sabon Gari were used.
2.2.1. Pre-Enrichment
Twenty-five (25) grams of the samples were weighed and poured into 225 mL of buff-ered peptone water and incubated at 37 °C for 24 h.
2.2.2. Selective Plating for Staphylococcus aureus
After incubation, a loopful of inoculum (from above) was inoculated on mannitol salt agar (MSA) plate, incubated at 37 ± 1 °C for 30 h and observed for growth. A yellow halo indicated Staphylococcus (S.) aureus.
2.2.3. Selective Plating for Salmonella spp.
To isolate Salmonella spp., one (1) ml of pre-enrichment culture was inoculated in Rappaport–Vassiliadis broth (RV) and incubated at 42 °C for 7 days. After the incubation, it was further cultured on selective agar plates of Salmonella–Shigella agar (SSA) at 37 °C for 48 h.
2.3. Antimicrobial Susceptibility Test (AST)/Resistotyping
The AST for ampicillin (AMP), ciprofloxacin (CIP), gentamicin (GEN), and tetracycline (TET) were performed concurrently with the disc diffusion test (Bioanalyse, Ankara, Turkey) using AM30g, CIP05g, GM10g, and TE10g concentrations, respectively, and the E-test (HiMedia, Mumbai, India) using 0.016–256 g/mL concentrations [3]. Mueller–Hinton agar plates (HiMedia, India) were made fresh for each test. The plates were inoculated with standardized inoculum (0.5 McFarland standard) of isolates and incubated at 37 °C for 24 h. The minimum inhibitory concentration (MIC) and zone of inhibition (ZOI) values of AMP, CIP, GEN, and TET were interpreted as S (susceptible), I (intermediate), or R (resistant) based on the breakpoints indicated by the reference standards for the Clinical and Laboratory Standards Institute (CLSI) on http://em100.edaptivedocs.net/ (accessed on 21 October 2023) [4] and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) on http://www.eucast.org/ (accessed on 21 October 2023) [5].
3. Results
3.1. Organisms Isolated
A total of 122 isolates (Salmonella spp. (65) and S. aureus (57)) were isolated from the samples.
3.2. The ZOI Results
Figure 2a shows the ZOI for the antibiotic AMP against Salmonella spp. using the CLSI (R ≤ 13, I 14–16, S ≥ 17) and EUCAST (R < 14, S ≥ 14) breakpoints. CLSI showed that 53 (81.538%) of the isolates were R, while 02 (03.077%) were I and 10 (15.385%) were S, whereas EUCAST indicated 53 (81.538%) were R and 12 (18.462%) were S.
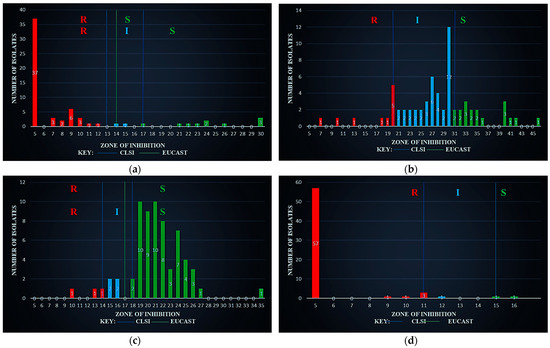
Figure 2.
Zone of inhibition against Salmonella spp. (a) using ampicillin (AM30µg), (b) using ciprofloxacin (CIP05µg), (c) using gentamicin (GM10µg), and (d) using tetracycline (TE10µg).
Figure 2b shows that the CLSI breakpoints for ZOI (CIP) on Salmonella spp. are R ≤ 20, I 21–30, and S ≥ 31; however, EUCAST had no breakpoints available. Based on CLSI, 10 (15.385%) were R, 37 (56.923%) I, and 18 (27.692%) S.
Figure 2c shows the ZOI (GEN) on Salmonella spp. with CLSI breakpoints R ≤ 14, I 15–17, and S ≥ 18; about 03 (04.615%) were R, 04 (06.154%) I and 58 (89.231%) S, whereas, on EUCAST (R < 17, S ≥ 17), 07 (10.769%) were R and 58 (89.231) S.
Figure 2d shows the ZOI (TET) with a CLSI breakpoint of R ≤ 11, I 12–14, S ≥ 15. There were no EUCAST breakpoints for the tetracycline. However, the CLSI gave 62 R (95.385%), 01 I (01.538%), and 02 S (03.077%).
Figure 3a shows the ZOI results for the antibiotic AMP against S. aureus with the CLSI breakpoints (R < 22, S ≥ 22). Moreover, EUCAST has no breakpoints. Based on CLSI, 30 (52.632%) of the isolates were R and 27 (47.368%) were S.
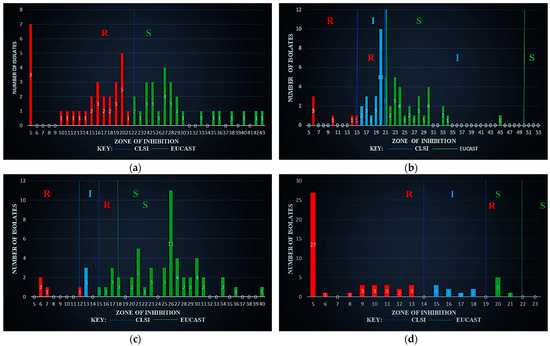
Figure 3.
Zone of inhibition against S. aureus (a) using ampicillin (AM30µg), (b) using ciprofloxacin (CIP05µg), (c) using gentamicin (GM10µg), and (d) using tetracycline (TE10µg).
Figure 3b shows the ZOI results for the antibiotic CIP against S. aureus with the CLSI breakpoints (R ≤ 15, I 16–20 S ≥ 21) and EUCAST breakpoints (R ≤ 21, I 22–49 S ≥ 50). Based on CLSI, 06 (10.526%) of the isolates were R, with 19 (33.333%) I and 32 (56.140%) S, whereas, on EUCAST, 25 (43.860%) were R, 32 (56.140%) I, and 00 (00.000%) S.
Figure 3c shows the ZOI results for the antibiotic GEN against S. aureus with the CLSI breakpoints (R ≤ 12, I 13–14 S ≥ 15) and EUCAST breakpoints (R < 18, S ≥ 18). Based on CLSI, 04 (07.018%) of the isolates were R, with 03 (05.263%) I and 50 (87.719%) S, whereas, on EUCAST, 14 (24.561%) were R and 43 (75.439%) S.
Figure 3d shows the ZOI results for the antibiotic TET against S. aureus with the CLSI breakpoints (R ≤ 14, I 15–18 S ≥ 19) and EUCAST breakpoints (R < 22, S ≥ 22). Based on CLSI, 43 (75.439%) of the isolates were R, with 08 (14.035%) I and 06 (10.526%) S, whereas, on EUCAST, 57 (100.000%) were R, with 00 (00.000%) S.
3.3. AST Summary
The data presented in Table 1 shows that Salmonella spp. and S. aureus displayed four distinct antimicrobial resistance patterns (ARPs) across Groups I, L, O, and P. The largest R population of 44 (36.066%) was in Group O with primary resistance to TET, followed by Group P (31.967%) in which all were S, Group I (29.508%) where AMP and TET were R and Group L with the least (02.459%) where only AMP was R. From these four (4) groups, representatives were selected for the E-test MIC, i.e., 2 per group to give a total of 16 organisms.

Table 1.
AST summary based on ZOI.
3.4. E-Test Summary
Table 2 highlights the E-test pattern for the MIC breakpoints for Salmonella spp. regarding CLSI (AMP S ≤ 08 I16 R ≥ 32; CIP S ≤ 0.25 I0.5 R ≥ 01; GEN S ≤ 02 I04 R ≥ 08; TET S ≤ 04 I08 R ≥ 16). A variation was specifically observed for Salmonella spp. Selected isolates from Groups I and L were S to AMP, despite earlier being R. The selected organisms that were earlier CIP S, became R and TET resistance was confirmed. While for S. aureus, CLSI (AMP S ≤ 04 I–R ≥ 08, CIP S ≤ 01 I02 R ≥ 04, GEN S ≤ 04 I08 R ≥ 16, TET S ≤ 04 I08 R ≥ 16). A variation was also specifically observed for S. aureus, and selected isolates from Groups I and L, which were S to AMP, became R, while Group O now showed S when it was previously R to only TET.

Table 2.
E-test summary.
4. Discussion
The study conducted by Tamba et al., 2016 [6], on Salmonella isolates showed resistance rates for AMP (85.7%), TET (35.7%), CIP (00.0%), and GEN (00.0%). Their findings indicated that AMP is the most resistant drug. Our study shows there is an increase in resistance rates among Salmonella isolates on all the other drugs tested. TET has 95.385% on CLSI breakpoint only, CIP has 15.385%, and GEN has 04.615%, respectively, on both CLSI and EUCAST. While AMP shows a drop from 85.7% to 81.538%, TET has an alarming jump from 35.7% to 95.385%.
In the case of S. aureus isolates, Umaru et al., 2013 [7], used the CIP, GEN, and TET along with others. The resistance rates were reported for TET (55.5%), CIP (38.9%), GEN (11.1%), and oxacillin (100.0%) which can be substituted for AMP [4], thus making AMP, the most resistant drug in that study. This study shows there is an increase in resistance rates among S. aureus isolates on all the drugs tested, with TET on the CLSI (75.439%) and on EUCAST (100.000%), AMP (52.632%) on CLSI only, CIP on the CLSI (10.526%) and on EUCAST (43.860%), and GEN on CLSI (10.526%) and on EUCAST (43.860%). The same pattern is observed here of an alarming jump in TET (55.5% to 75.439%) and a drop in AMP (100.0% to 52.632%).
As shown in Table 1, half of the selected isolates are resistant to AMP and TET, with all of them being susceptible to CIP and GEN. However, as shown in Table 2, AMP in Salmonella spp. was 25% R, S. aureus was 50% R, TET was 37.5% R for both, CIP was 50% R and 25% R, respectively, and GEN was 00% R and 12.5% R, respectively.
In summary, recent research has revealed a surge in antibiotic resistance, particularly in TET, which is the most resistant drug for both organisms. AMP, CIP, and GEN follow in that sequence. Prompt measures must be taken to tackle the escalating issue of antimicrobial resistance (AMR) and curb the proliferation of antibiotic-resistant pathogens.
Author Contributions
Conceptualization, M.M.F., S.A.A., M.B.T. and H.M.K.; methodology, M.M.F., S.A.A., M.B.T. and H.M.K.; validation, S.A.A., M.B.T. and H.M.K.; formal analysis, M.M.F., S.A.A., M.B.T. and H.M.K., J.S.O. and R.F.; investigation, M.M.F., S.A.A., M.B.T., H.M.K., J.S.O. and R.F.; resources, M.M.F. and R.F.; data curation, M.M.F., J.S.O. and R.F.; writing—original draft preparation, M.M.F., J.S.O. and R.F.; writing—review and editing, M.M.F., S.A.A., M.B.T., H.M.K., J.S.O. and R.F.; visualization, M.M.F., S.A.A., M.B.T. and H.M.K., J.S.O. and R.F.; supervision, S.A.A., M.B.T. and H.M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Original data are available from the authors at request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Mehta, B.M. Chemical Composition of Milk and Milk Products. In Handbook of Food Chemistry; Springer: Heidelberg, Germany, 2015; pp. 511–554. ISBN 978-3-642-36605-5. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E. A 10-year single-center experience on Stenotrophomonas maltophilia resistotyping in Szeged, Hungary. Eur. J. Microbiol. Immunol. 2020, 10, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Yang, X.; Yang, Y.; Guo, Y.; Yin, D.; Ding, L.; Wu, S.; Zhu, D.; Hu, F. Assessment of Ceftazidime-Avibactam 30/20-Μg Disk, Etest versus Broth Microdilution results when tested against Enterobacterales clinical isolates. Microbiol. Spectr. 2022, 10, e01092-21. [Google Scholar] [CrossRef] [PubMed]
- CLSI eClipse Ultimate Access—Powered by Edaptive Technologies. Available online: http://em100.edaptivedocs.net/dashboard.aspx (accessed on 21 October 2023).
- EUCAST: Clinical Breakpoints and Dosing of Antibiotics. Version 13.1. 2023. Available online: https://www.eucast.org/clinical_breakpoints. (accessed on 21 October 2023).
- Tamba, Z.; Bello, M.; Raji, M.A. Occurrence and antibiogram of Salmonella spp. in raw and fermented milk in Zaria and environs. Bangl. J. Vet. Med. 2016, 14, 103–107. [Google Scholar] [CrossRef]
- Umaru, G.A.; Kabir, J.; Umoh, V.J.; Bello, M.; Kwaga, J.K.P. Methicillin-Resistant Staphylococcus aureus (MRSA) in fresh and fermented milk in Zaria and Kaduna, Nigeria. Int. J. Drug Res. Technol. 2013, 3, 67–75. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).